Abstract
Background
Cationic polymers have been accepted as effective nonviral vectors for gene delivery with low immunogenicity unlike viral vectors. However, the lack of organ or cell specificity sometimes hampers their application and the modification of polymeric vectors has also shown successful improvements in achieving cell-specific targeting delivery and in promoting intracellular gene transfer efficiency.
Methods
A folic acid-conjugated stearic acid-grafted chitosan (FA-CS-SA) micelle, synthesized by a 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide-coupling reaction, was designed for specific receptor-mediated gene delivery.
Results
Due to the cationic properties of chitosan, the micelles could compact the plasmid DNA (pDNA) to form micelle/pDNA complexes nanoparticles. The particle size and zeta potential of the FA-CS-SA/pDNA complexes with different N/P ratios were 100–200 nm and −20 to −10 mV, respectively. The DNase I protection assay indicated that the complexes can efficiently protect condensed DNA from enzymatic degradation by DNase I. A cytotoxicity study indicated that the micelles exhibited less toxicity in comparison with LipofectamineTM 2000. Using SKOV3 and A549 as model tumor cells, the cellular uptake of micelles was investigated.
Conclusion
It was found that cellular uptake of FA-CS-SA in SKOV3 cells with higher folate receptor expression was faster than that in A549 cells with a short incubation time. Luciferase assay and green fluorescent protein detection were used to confirm that FA-CS-SA could be an effective gene vector. Transfection efficiency of the FA-CS-SA/pDNA complexes in SKOV3 cells was enhanced up to 2.3-fold compared with that of the CS-SA/pDNA complexes. However, there was no significant difference between the transfection efficiencies of the two complexes in A549 cells. Importantly, the transfection efficiency of FA-CS-SA/pDNA decreased with free FA pretreatment in SKOV3 cells. It was concluded that the increase in transfection efficiency of the FA-CS-SA/pDNA complexes was attributed to folate receptor-mediated endocytosis.
Introduction
Gene therapy is considered to be a powerful approach to the treatment of congenital and acquired diseases, by producing bioactive agents or stopping abnormal functioning of cells, such as genetic disorders or uncontrolled proliferation of cells. However, the lack of effective vectors is a major barrier to progress in human gene delivery.Citation1 Virus vectors are very effective in terms of transfection efficiency, but their application in the human body is often frustrated by immunogenicity, potential infectivity, complicated production, and inflammation.Citation2 The limitations of viral vectors have led to the evaluation and development of alternative vectors based on nonviral systems, such as liposomesCitation3 and nanoparticles.Citation4 Cationic nanoparticles have emerged as one of the most attractive carriers because of their ability to form complexes with negatively charged small interfering RNA (siRNA) and their high transfection efficiency.Citation5 Chitosan has been considered to be a good gene carrier candidate, because it is known to be a biodegradable, biocompatible, low immunogenicity, and nontoxic material.Citation6,Citation7 Chitosan, as a cationic natural polysaccharide, can be complexed with negatively charged DNA by ionic interactions and protect DNA against DNase degradation.Citation8,Citation9 However, the low transfection efficiency and low specificity of chitosan must be overcome for its use in gene delivery. In addition, chitosan is poorly soluble because its amino groups are only partially protonated at physiological pH 7.4. Low molecular weight chitosan has been focused on for gene delivery systems because of its good solubility in physiological conditions, higher gene transfection, and more efficient enzymatic protection.Citation10–Citation12 Moreover, chitosan contains reactive amino and hydroxyl groups, thus many chitosan derivatives can be obtained by chemically altering its properties under mild reaction conditions.Citation13,Citation14 One approach to the improvement of transfection efficiency involves conjugating hydrophobic units with the polysaccharide backbone through amino groups or hydroxyl groups.Citation15
An ideal gene carrier system should efficiently accumulate in specific target tissues with minimal toxicity to nontarget tissues.Citation16 In order to increase the transfection efficiency of polymer/DNA complexes to tumor cells specifically, various targeting ligands, including antibodies, growth factors, peptides, and folate have been conjugated to several polymers.Citation17–Citation19 Folic acid (FA) has emerged as an optimal ligand for targeting the cell membrane and allows nanoparticle endocytosis via the folate receptor for higher transfection yields. At present, the folate receptor is a major focus in multiple experimental approaches to tumor-targeted therapy. The folate receptor is known to be overexpressed on many human cancer cell surfaces, such as the brain, kidney, lung, ovarian, and breast cancers, but is highly restricted in most normal tissues.Citation20 When folate is covalently linked to a molecule via its carboxyl moiety, its affinity for its cell surface receptor (Kd about 10–9 M) remains essentially unaltered. Some novel polymers have been synthesized, like folate-polylysineCitation21 and folate-polyethylenimine,Citation22, Citation23 all of which specifically increase gene transfection efficiency in target cells compared with nontargeted polymers.
In our previous research, novel self-aggregated micelles were formed by an amphiphilic-grafted copolymer, ie, stearic acid-grafted chitosan copolymer (CS-SA). The micelles have a specific spatial structure with multiple hydrophobic domains near their surface, and thus achieve excellent internalization into cancer cells.Citation24 They have been investigated in detail as a gene vector, but their transfection efficiency is not so well understood. Moreover, the cellular uptake of CS-SA/DNA complexes lacks specificity. Therefore, CS-SA copolymers were further modified with folic acid to increase gene transfection efficiency in target cells specifically, and then folate-conjugated CS-SA (FA-CS-SA) was synthesized, and its properties were investigated. A FA-CS-SA/pDNA complex was then prepared by ionic interaction, and physicochemical characterization of the complex was carried out. Cellular uptake in vitro was evaluated in SKOV3 cells (a folate receptor-overexpressing cell line) and A549 cells (a folate receptor-deficient cell line). In addition, the in vitro transfection efficiency of FA-CS-SA/pDNA complexes in terms of green fluorescent protein detection and the luciferase assay was also examined in the two cell lines.
Materials and methods
Materials
Chitosan of low molecular weight (5.0 kDa) was obtained by enzymatic degradation (molecular weight 450.0 kDa, 95% deacetylated, Yuhuan Marine Biochemistry Co Ltd, Zhejiang, China).Citation25 Stearic acid was obtained from Chemical Reagent Co Ltd (Shanghai, China). 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide (EDC), 2,4,6-trinitrobenzene sulfonic acid (TNBS), and 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-tetrazolium bromide (MTT) were purchased from Sigma (St Louis, MO). Fetal bovine serum was purchased from Sijiqing Biologic Co Ltd (Zhejiang, China). Folic acid and fluorescein isothiocyanate (FITC) came from Sigma (St Louis, MO), and LipofectamineTM 2000 was supplied by Invitrogen Corporation (Carlsbad, CA). DNase I, Dulbecco’s Modified Eagle’s Medium, and trypsin-ethylenediamine tetra-acetic acid were purchased from Gibco BRL (Gaithersberg, MD). Green fluorescent protein plasmid DNA (EGFP-C1) was donated by the first Affiliated Hospital of College of Medicine, Zhejiang University, Hangzhou, China. PGL-3, as the luciferase reporter gene, was kindly provided by the Institute of Life Science, Zhejiang University, Hangzhou, China. A luciferase assay kit and bicinchoninic acid protein assay kit were obtained from Beyotime Institute of Biotechnology, China. All other chemicals were of analytical grade and used without further purification.
Synthesis of CS-SA
CS-SA was synthesized via reaction of the carboxyl groups of SA with the amine groups of CS, catalyzed by EDC. In brief, 1.0 g of CS was dissolved in 120 mL of distilled water. SA 0.4 g with EDC (SA:EDC 1:10, mol:mol) was then dissolved in 80 mL of ethanol by sonication, followed by stirring in a water bath at 60°C for 1 hour. The mixture was added to the CS solution and the reaction was conducted with 400 rpm for 5 hours at 80°C in a water bath. After the reaction mixture was cooled to room temperature, the synthesized product was dialyzed using a dialysis membrane (molecular weight cutoff 3.5 kDa, Spectrum Laboratories, Laguna Hills, CA) against distilled water for 48 hours in order to remove byproducts, followed by freeze drying. The lyophilized product was washed twice with ethanol to remove any unreacted SA.
Synthesis of FA-CS-SA copolymer
The free amino groups of CS-SA were conjugated with the carboxylic groups of FA in the presence of EDC. The carboxyl groups of FA were activated by EDC. Briefly, FA dissolved in dimethyl sulfoxide was reacted with 10 molar equivalents of EDC at room temperature for 1 hour under stirring. Thereafter, it was added to the CS-SA solution, and the resulting mixture was stirred at room temperature in the dark for 16 hours. The mixture was dialyzed against distilled water for 3 days and then lyophilized.
Characterization of FA-CS-SA
The critical micelle concentration of FA-CS-SA in aqueous medium was determined by fluorescence measurement using pyrene as a probe.Citation26 Briefly, 6 mg of pyrene was firstly dissolved in 50 mL of acetone as a pyrene stock solution. Before use, the pyrene stock solution was further diluted 100-fold with acetone; 0.5 mL of pyrene solution with a concentration of 0.0012 mg/mL was transferred into test tubes, the acetone was then evaporated at 50°C, and 5 mL of FA-CS-SA solution at different concentrations varying from 1.0 × 10−3 to 1.0 mg/mL was added to the test tubes. The final concentration of pyrene was fixed at 6 × 10−7 M. The fluorescence emission spectrum of pyrene was obtained using a fluorometer (F-2500, Hitachi Corporation, Osaka, Japan). The slit openings were set at 10 nm (excitation) and 2.5 nm (emission), and the excitation wavelength was 337 nm. The intensity ratio of the first highest energy bands (374 nm) to the third (385 nm) in the pyrene emission spectra was then calculated.
The degrees of amino substitution of FA-CS-SA were determined by the TNBS method.Citation27 Then 0.3 mL of FA-CS-SA solution at a concentration of 1 mg/mL was incubated with 2 mL of 4% NaHCO3 and 2 mL of 0.1% TNBS at 37°C for 2 hours. The mixture was then added to 2 mL of HCl (2 mol/L) and measured at 344 nm by an ultraviolet spectrophotometer (TU-1800PC, Beijing Purkinje General Instrument Co Ltd, Beijing, China). The degrees of amino substitution for CS-SA were measured in a similar way to that described above. The FA to amino group molar ratio in the FA-CS-SA copolymer was estimated indirectly.
The size and zeta potential of the micelles with 1 mg/mL of FA-CS-SA were measured by dynamic light scattering using a Zetasizer (3000 HS; Malvern Instruments Ltd, Worcester, UK). Each sample determination was done in triplicate.
Preparation of FA-CS-SA/pDNA nanoparticles
Briefly, FA-CS-SA was dissolved in deionized water at a concentration of 1 mg/mL. The solution was then filtered using a 0.22 μm Millipore filter. The plasmid DNA was diluted with phosphate-buffered saline to give a concentration of 0.5 mg/mL. Nanoparticles were prepared by mixing pDNA with appropriate polymer solution at various charge ratios. The charge ratio (N/P) of FA-CS-SA/pDNA was expressed as the molar ratio of the primary amino group of FA-CS-SA to the phosphate groups of DNA. The complex was then vortexed for 30 seconds and incubated for 30 minutes at room temperature for complete complex formation.
Determination of particle size and zeta potential
Particle size and zeta potential of the FA-CS-SA/pDNA nanoparticles was determined by dynamic light scattering (Zetasizer 3000HS). Each sample determination was done in triplicate.
Gel retardation assay
The DNA binding ability of FA-CS-SA was evaluated by agarose gel electrophoresis. The FA-CS-SA/DNA nanoparticles containing 1.0 μg of DNA were prepared at different N/P ratios (0.1, 0.2, 0.4, 0.8, 1.6, 3.2, and 6.4, respectively), by varying the concentration of FA-CS-SA. The nanoparticles and the naked plasmid were loaded into individual wells with 1.0% agarose gel in Tris-acetate-ethylenediamine tetra-acetic acid buffer. The samples were run on the gel at 100 V for 40 minutes. Finally, the gel was stained with 0.5 mg/mL ethidium bromide and photographed using an ultraviolet spectrophotometer (UV-1601; Shimadzu, Kyoto, Japan). As a control, the DNA binding ability of CS-SA was also evaluated by the same process as described for FA-CS-SA.
DNase I protection assay
Protection and release of DNA in FA-CS-SA/pDNA nanoparticles were carried out by electrophoresis according to the method described by Park et al.Citation28 Briefly, 4 μL of DNase I (2 units) or phosphate-buffered saline in DNase/Mg2+ digestion buffer (50 mM, Tris-Cl, pH 7.6 and 10 mM MgCl2) was added to 0.5 μg of naked plasmid DNA or 4 μL of nanoparticle solution and incubated at 37°C with shaking at 100 rpm for 30 minutes. For DNase inactivation, all the samples were treated with 4 μL of ethylenediamine tetra-acetic acid (250 mM) for 10 minutes, and 1% sodium dodecyl sulfate dissolved in 0.1 M NaOH (pH 7.2) was then added to a final volume of 18 μL. The final samples were further incubated for 2 hours to allow complete dissociation of complexes, and electrophoresis was performed with 1% agarose gel in Tris-acetate-ethylenediamine tetra-acetic running buffer for 1 hour at 100 V.
Cell culture
SKOV3 and A549 cells were maintained in Dulbecco’s Modified Eagle’s Medium or RPMI 1640 media supplemented with 10% fetal bovine serum, penicillin 100 U/mL, streptomycin 100 U/mL. Both cell lines were cultured at 37°C in a humidified 5% CO2 atmosphere.
Cellular uptake studies
In order to investigate cellular uptake, FA-CS-SA copolymer was labeled with FITC via reaction of the amino groups of chitosan and the isothiocyanate groups of FITC. The molar ratio of FA-CS-SA to FITC was fixed at 2:1. The solution was then stirred for 12 hours with 400 rpm at room temperature in aphotic environment. The reaction product was dialyzed against distilled water using a dialysis membrane (molecular weight cutoff 3.5 kDa, Spectrum Laboratories, Laguna Hills, CA) for 24 hours to remove the unreacted FITC. SKOV3 and A549 cells were seeded at 3 × 104 cells/well in a 24-well plate, and grown for 24 hours. After removing the growth medium, 1.0 mL RPMI 1640 medium containing 100 μg FITC-labeled FA-CS-SA was added. The cells were further incubated for 1, 2, 3, 6, 12, and 24 hours, respectively. The cells were then washed twice with phosphate-buffered saline (pH 7.4) and directly viewed under a fluorescence microscope (Leica DM4000 B; Leica, Solms, Germany). The fluorescence images were obtained at 20× magnification. As a control, FITC-labeled CS-SA was used to observe cellular uptake as described above.
In vitro cytotoxicity assay
The cytotoxicity of the copolymers was measured by MTT assay. SKOV3 and A549 cells were seeded at a density of 1 × 104 cells/well in 200 μL complete medium in 96-well plates and incubated for 24 hours at 37°C. The growth medium was then removed and replaced with fresh medium containing copolymer (CS-SA or FA-CS-SA) or Lipofectamine 2000. The cells were then incubated further for 48 hours. After incubation, 20 μL of MTT (5.0 mg/mL) was added and incubated for 4 hours under normal growing conditions. After the medium with the unreacted MTT was removed, 200 μL of dimethyl sulfoxide was added to each well. The well plates were shaken for 20 minutes and the absorbance of formazan product was measured at 570 nm in a microplate reader (Model 680; BioRad, Mountain View, CA). Cells without polymers were used as a control.
In vitro transfection studies
SKOV3 and A549 cells were seeded in 24-well plates at a density of 1 × 105 cells per well in 1 mL of complete medium and incubated for 24 hours prior to transfection. The media was then replaced with fresh growth medium in each well. pDNA was diluted with phosphate-buffered saline (pH 7.4) to 0.5 mg/mL, and then added to an appropriate amount of the copolymer in the same buffer with an equal volume to obtain the desired N/P ratios. The complexes were mixed using a vortex mixer for 30 seconds and then allowed to stand for 30 minutes at room temperature. Complexes containing 2 μg EGFP plasmid were subsequently added to the wells and incubated with the cells initially for 6 hours. The medium was then replaced with 1 mL of fresh complete medium, and the cells were incubated for an additional 24–72 hours at 37°C. As a positive control, transfection with Lipofectamine 2000/DNA complexes was performed according to the manufacturer’s protocol. Briefly, Lipofectamine 2000/DNA complexes were incubated with cells in 1 mL of serum-free medium for 6 hours, and further incubated with fresh complete medium for 24–72 hours at 37°C. Naked DNA was used on the cell cultures and examined as described above. All transfection experiments were performed in triplicate. The cells were transferred to an inverted fluorescence microscope (Leica DMI 4000B) for imaging.
To evaluate transfection efficiency, FA-CS-SA/DNA complexes (2 μg pGL3 plasmid) were prepared for transfection into SKOV3 and A549 cells. The transfection procedure was carried out using methods similar to those described above. Transfection efficiency was detected by assay of luciferase activity. After incubation for 72 hours with the cells, the growth medium was removed, and the cells were lysed with cell lysate buffer (Beyotime) and shaken for 30 minutes at room temperature. The luciferase assay was carried out according to the manufacturer’s instructions. Relative light units were measured using a luminometer (Turner Designs Luminometer Model TD-20/20, Promega), and the protein assay was performed using a bicinchoninic acid protein assay reagent kit (Beyotime). The final results were reported in terms of relative light units/mg cellular protein. All transfection experiments were performed in triplicate.
FA competition studies
SKOV3 and A549 cells were cultured in the same conditions as those used for the transfection tests. The cells were seeded in 24-well plates at 1 × 105 cells/well and incubated for 24 hours. One hour before the transfection test, the medium was replaced with RPMI 1640 containing FA 5.0 mM. The cells were then treated with FA-CS-SA/pDNA complex containing 2 μg of plasmid DNA at predetermined N/P ratios for 6 hours at 37°C in 5% CO2. After an exchange of fresh medium, the cells were further incubated for 24–72 hours at 37°C. Transfection efficiency was assessed using the same method as described for the luciferase assay.
Results and discussion
Synthesis of FA-CS-SA
CS-SA was synthesized via reaction of the amino groups of CS with the carboxyl groups of SA in the presence of EDC. The degree of amino substitution in the prepared CS-SA was about 19.9%, as determined by the TNBS method. CS-SA micelles were easily prepared by dispersing CS-SA in distilled water under ultrasound, due to the inherent self-aggregation of CS-FA in an aqueous environment. FA-CS-SA could be synthesized by chemically linking FA to CS-SA via the reaction of the carboxyl groups of FA and the amino groups on the CS-SA micelle surface, in the presence of EDC. The degree of amino substitution of FA-CS-SA was 25.2%, as determined by the TNBS methods. The molar ratio of folic acid/NH2 in FA-CS-SA copolymer was 5.3%.
Characteristics of FA-CS-SA
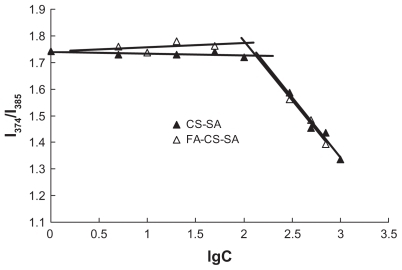
The aggregation behavior of FA-CS-SA and CS-SA in aqueous media was measured by fluorometry using pyrene as a fluorescent probe. The critical micelle concentration refers to the threshold self-aggregation concentration of amphiphilic polymer. The I1/I3 ratio, ie, the first to third highest energy bands in the pyrene emission spectra (I1, em = 374 nm; I3, em = 385 nm) was used to determined the critical micelle concentration value of the polymer. As the concentration increased to form micelles, the total emission fluorescence intensity increased, especially the intensity of the third highest energy band. As a result, the fluorescence intensity ratio of I1/I3 decreased. Variation in the fluorescence intensity ratio of I1/I3 against the logarithm of FA-CS-SA and CS-SA concentration is shown in . The critical micelle concentration values of FA-CS- SA and CS-SA in distilled water were determined to be about 105 μg/mL and 147 μg/mL, respectively, which showed that the modification of FA improved the ability of CS-SA to self-assemble in water.
Figure 1 Variation of intensity ratio (I1/I3) vs the logarithm of concentration of CSSA and FA-CS-SA. Unit of concentration was μg/mL.
Abbreviations: CS-SA, stearic acid-grafted chitosan copolymer; FA-CS-SA, folic acid-conjugated stearic acid-grafted chitosan copolymer.
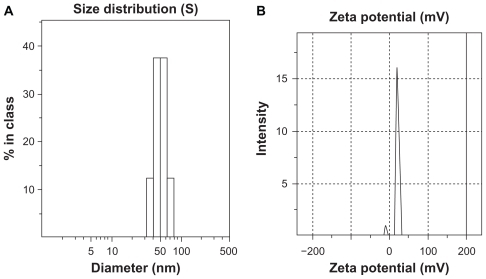
The distributions of hydrodynamic diameter and zeta potential of FA-CS-SA micelles with 1 mg/mL of FA-CS-SA are shown in . The average hydrodynamic diameter and zeta potential of the FA-CS-SA micelles was 51.1 nm and 13.7 ± 4.7 mV, respectively.
Characteristics of FA-CS-SA/DNA nanoparticles
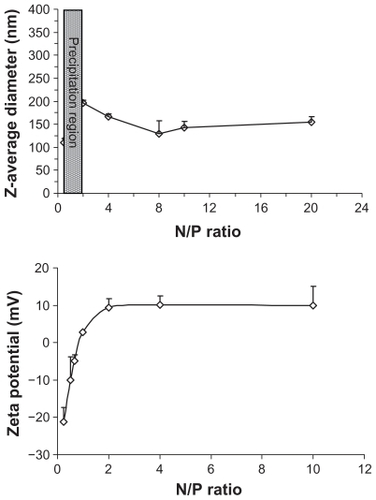
Due to its positive charge, the FA-CS-SA micelle was used to condense DNA, forming FA-CS-SA/DNA complex nanoparticles. The FA-CS-SA/DNA complex nanoparticles with N/P ratios from 0.25 to 20 were prepared by mixing the appropriate volume of FA-CS-SA micelle solution and DNA solution. The size and zeta potential of the nanoparticle complexes were measured using a dynamic light scattering technique. showed the Z-average diameter and zeta potential of FA-CS-SA/DNA complexes at various charge ratios. The size of the FA-CS-SA/DNA complexes increased as the N/P ratio increased from 0.25 to 1. When the N/P ratio was about 1, precipitation occurred and the size increased to microlevels due to the charge neutrality. Again, on increasing the N/P ratio, the size was found to decrease below 200 nm. The zeta potential of the FA-CS-SA/DNA complexes () showed that the zeta potential changed from negative to positive when the N/P ratio increased from 0.5 to 20. The complexes had the highest zeta potential when the N/P ratio was about 2. With increasing N/P ratios, the zeta potential was found to approach a plateau, with no significant variation in the zeta potential.
Figure 3 Variations in Z-average diameter and zeta potential of folic acid-conjugated stearic acid-grafted chitosan copolymer/plasmid DNA nanoparticle complexes versus N/P ratio.
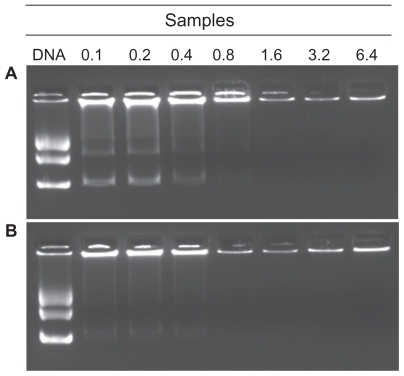
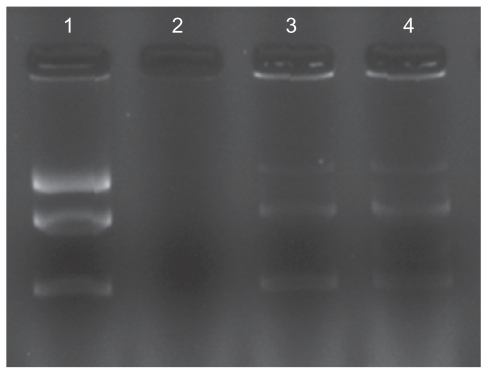
In order to determine the ability of FA-CS-SA to condense DNA, solutions of FA-CS-SA/DNA with different N/P ratios were electrophoresed on agarose gel containing ethidium bromide. Complete retardation of FA-CS-SA/DNA complexes was observed at N/P ratios of 0.8 and onwards (). As a control, the migration of CS-SA/DNA complexes was performed when the N/P ratio was above 0.8 (). The results indicate that FA-CS-SA had a stronger ability than CS-SA to condense DNA into nanoparticles.
Figure 4 Gel retarding analysis of (A) stearic acid-grafted chitosan copolymer/plasmid DNA and (B) folic acid-conjugated stearic acid-grafted chitosan copolymer/plasmid DNA complex nanoparticles. Lane 1 represents naked DNA; lanes 2–8 represent complex nanoparticles with N/P ratios of 0.1, 0.2, 0.4, 0.8, 1.6, 3.2, and 6.4, respectively.
DNase I protection assay
Protection of plasmid DNA from nucleases is a primary requisite for efficient gene delivery in vivo and in vitro.Citation29 As shown in , dissociation of DNA from copolymers was achieved although some still remained as a complex. Unlike the naked plasmid DNA control, the copolymer protected DNA. When naked pDNA was treated with two units of DNase I, it was completely degraded without any smear (lane 2). After adding 1% sodium dodecyl sulfate, the pDNA could be released from the FA-CSO-SA/pDNA complexes (lane 3 and lane 4). The Figure shows that at the same concentration of DNase I, the pDNA was protected from degradation by FA-CSO-SA micelles effectively when the N/P ratio was 2.
Figure 5 Protection and release assay of DNA. Lane 1 represents naked DNA(pEGFP-C1); lane 2 represents naked DNA treated with DNase I; lane 3 represents folic acid-conjugated stearic acid-grafted chitosan copolymer/DNA complex; lane 4 represents folic acid-conjugated stearic acid-grafted chitosan copolymer/DNA complex treated with DNase I.
Cellular uptake
It has been demonstrated that folate-mediated biomolecular uptake is roughly proportional to the folate receptor density on the target cell membrane. To investigate the targeting efficiency of FA-CS-SA micelles, the cellular uptake of FA-CS-SA labeled with FITC was evaluated in SKOV3 cells and visualized using fluorescence microscopy (). After 0.5 hours of incubation, FITC-labeled FA-CS-SA was distributed in the SKOV3 cells, with green fluorescence throughout the entire cytoplasm. However, CS-SA was detected only as spot-like fluorescence. With a longer incubation time, fluorescence was enhanced, with a significant difference in fluorescence intensity between FA-CS-SA and CS-SA in the SKOV3 cells.
Figure 6 Fluorescence images of (A) SKOV3 cells and (B) A549 cells incubated with FITC-labeled stearic acid-grafted chitosan copolymer and folic acid-conjugated stearic acid-grafted chitosan copolymer for 0.5, 3.0, and 8.0 hours.
Abbreviations: CS-SA, stearic acid-grafted chitosan copolymer; FA-CS-SA, folic acid-conjugated stearic acid-grafted chitosan copolymer.
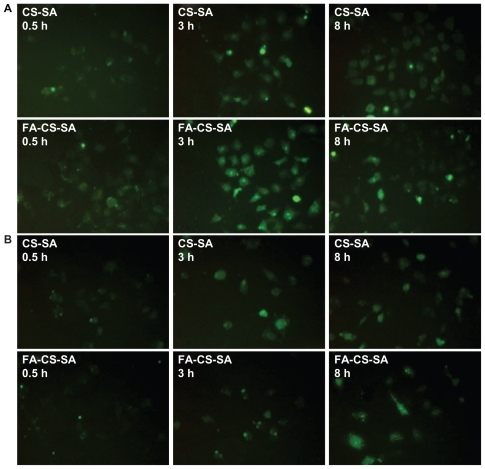
When the CS-SA copolymers were conjugated with FA, cellular uptake was mainly mediated by FA during a short incubation time, which indicated that shortening the incubation time was helpful to see the difference in cellular uptake between the two micelles. After 8 hours of incubation, with further enhanced fluorescence of CS-SA, the difference between the two micelles disappeared. This is because of the increasing concentration of micelle complexes in the cells, with the internalization ability attributed to the cellular components and their special spatial structure.Citation30
To verify further the effect of the folate receptor on the intracellular uptake of FA-CS-SA, A549 cells were used as a negative control for the intracellular uptake experiment. showed that there was no significant difference between intracellular uptake of FA-CS-SA and that of CS-SA, regardless of the incubation time.
Evaluation of cytotoxicity
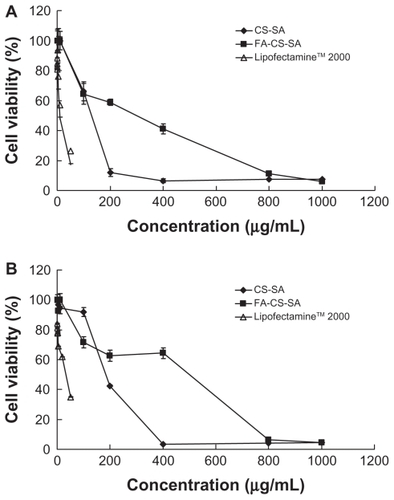
The cytotoxicity of the FA-CS-SA and CS-SA copolymers were estimated by MTT assay. The IC50 value of CS-SA and FA-CS-SA were higher than that of Lipofectamine 2000, suggesting that both CS-SA and FA-CS-SA had lower cytotoxicity compared with Lipofectamine 2000 regardless of the cell lines used, and the difference in cytotoxicity between FA-CS-SA and CS-SA was not significant. As shows, the IC50 values of the CS-SA and FA-CS-SA micelles against SKOV3 cells were 115.15 μg/mL and 217.18 μg/mL, respectively, and against A549 cells were 201.91 μg/mL and 298.46 μg/mL, while the IC50 values of Lipofectamine 2000 were about 9.91 μg/mL and 29.50 μg/mL. The cytotoxicity of CS-SA and FA-CS-SA micelles at a higher concentration might originate from the large amounts of exogenous substances internalized into the cells. Thereafter, a lower concentration of micelles was used. The highest concentration used in gene delivery was 40 mg/mL when the N/P was 20, which was markedly lower than that of the IC50 value.
Figure 7 Cytotoxicity of folic acid-conjugated stearic acid-grafted chitosan copolymer and stearic acid-grafted chitosan copolymer in comparison with LipofectamineTM 2000 in (A) SKOV3 cells and (B) A549 cells.
Abbreviations: CS-SA, stearic acid-grafted chitosan copolymer; FA-CS-SA, folic acid-conjugated stearic acid-grafted chitosan copolymer.
Transfection experiments
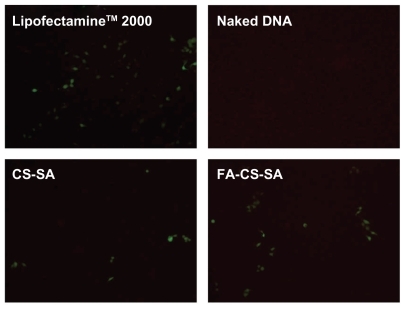
SKOV3 cells expressing folate receptors were used to determine whether gene delivery with FA-CS-SA is folate receptor-mediated. Transfection efficiencies of the FA-CS- SA/DNA and CS-SA/DNA complexes at different N/P ratios varying from 1 to 20 were determined in our preliminary experiments. The maximum transfection efficiency of the FA-CS-SA/DNA complex was observed at an N/P ratio of 2, and this tended to decrease with further increases in the N/P ratio. The transfection efficiency of the copolymer/DNA complexes was visualized by fluorescence microscopy. The images in show that SKOV3 cells were transfected with copolymer/pEGFP-C1 complexes at optimal N/P ratios. The transfection efficiency of FA-CS-SA/DNA could not reach the high level of Lipofectamine 2000, but it was much higher than that of CS-SA/DNA.
Figure 8 Green fluorescent protein detection of transfected SKOV3 cells observed by inverted microscopy.
Abbreviations: CS-SA, stearic acid-grafted chitosan copolymer; FA-CS-SA, folic acid-conjugated stearic acid-grafted chitosan copolymer.
To quantify transfection efficiency, the plasmid pGL-3 was used as the luciferase reporter gene, and a luciferase expression assay induced by the copolymers/pGL-3 complexes was carried out. shows the transfection efficiencies of FA-CS-SA/DNA, CS-SA/DNA, and Lipofectamine 2000/DNA complexes at different N/P ratios in terms of luciferase activity of the protein expressed. showed the transfection efficiencies of FA-CS-SA/DNA and CS-SA/DNA complexes at optimal N/P ratios. It was found that the transfection efficiency of FA-CS-SA/DNA complexes was 2.3-fold higher than that of the CS-SA/DNA complexes.
Figure 9 (A) Transfection efficiency of folic acid-conjugated stearic acid-grafted chitosan copolymer/DNA and stearic acid-grafted chitosan copolymer/DNA complexes at various N/P ratios in SKOV3 cells. (B) Transfection efficiency of folic acid-conjugated stearic acid-grafted chitosan copolymer DNA and stearic acid-grafted chitosan copolymer/DNA complexes at functional N/P ratios in SKOV3 cells. (C) Transfection efficiency of folic acid-conjugated stearic acid-grafted chitosan copolymer/DNA and stearic acid-grafted chitosan copolymer/DNA complexes at various N/P ratios in A549 cells. (D) Competitive assay of folic acid-conjugated stearic acid-grafted chitosan copolymer/DNA and stearic acid-grafted chitosan copolymer/DNA complexes by adding free folic acid 5.0 mM.
Notes: Values are means ± standard deviation. *P < 0.05 and **P < 0.01.
Abbreviations: CS-SA, stearic acid-grafted chitosan copolymer; FA-CS-SA, folic acid-conjugated stearic acid-grafted chitosan copolymer.
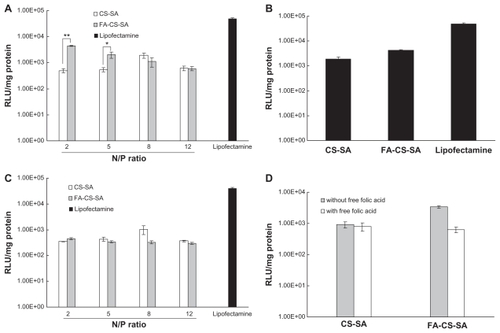
In order to demonstrate further the effect of the folate receptor on the transfection efficiencies of FA-CS-SA/DNA complexes, a transfection experiment was conducted using A549 cells deficient in the folate receptor. shows that the two complexes exhibited no difference in transfection efficiency in A549 cells, indicating that the folate ligand on FA-CS-SA plays a significant role in folate receptor recognition and enhanced transfection efficiency in SKOV3 cells.
From , it can be observed that, due to the competitive displacement of free FA, the transfection efficiency of the FA-CS-SA/DNA complexes in SKOV3 cells was greatly reduced (by 5.4-fold) in the presence of excess FA (5.0 mM) compared with the medium free of FA, whereas the free FA in the medium did not affect the transfection efficiency of the CS-SA/DNA complexes. The results suggest that free FA molecules could prevent cellular uptake of the complexes by competitive binding to folate receptors on the cell surface. In other words, conjugation of FA indeed improved the transfection efficiency of FA-CS-SA for gene delivery to folate receptor-positive cells.
Conclusion
FA-CS-SA was synthesized in this study by the reaction between the carboxyl groups of folate and the amine groups of CS-SA in the presence of EDC, which could self-aggregate to form micelle-like structures in aqueous solution. FA-CS-SA showed a good ability to form complexes with plasmid DNA and could efficiently protect condensed DNA from enzymatic degradation by DNase I. It had much lower cytotoxicity compared with Lipofectamine 2000. Folate conjugation increased the intracellular uptake of FA-CS-SA in SKOV3 cells via folate receptor-mediated endocytosis at shorter incubation times. The FA-CS-SA/DNA complexes displayed higher transfection efficiency at certain N/P ratios in comparison with that of CS-SA/DNA complexes in SKOV3 cells, whereas no difference in folate receptor-negative A549 cells was found. The results suggest that FA-CS-SA has the potential to be a safe and effective nonviral vector for gene delivery.
Acknowledgments
We appreciate the financial support of the National Nature Science Foundation of China (30873174 and 81072583) and the Nature Science Foundation of Zhejiang Province (z207489 and Y2090336).
Disclosure
The authors report no conflicts of interest in this work.
References
- KabanovAVKabanovVADNA complexes with polycations for the delivery of genetic material into cellsBioconjug Chem199567207711106
- SmithAViral vectors in gene therapyAnnu Rev Microbiol1995498078388561480
- BediDMusacchioTFagbohunOADelivery of siRNA into breast cancer cells via phage fusion protein-targeted liposomesNanomedicine2011731532321050894
- ChenXAZhangLJHeZJPlasmid-encapsulated polyethylene glycol-grafted polyethylenimine nanoparticles for gene delivery into rat mesenchymal stem cellsInt J Nanomedicine2011684385321589652
- GaoYLiuXLLiXRResearch progress on siRNA delivery with nonviral carriersInt J Nanomedicine201161017102521720513
- BorchardGChitosans for gene deliveryAdv Drug Deliv Rev20015214515011718938
- MansouriSLavignePCorsiKChitosan-DNA nanoparticles as non-viral vectors in gene therapy: strategies to improve transfection efficacyEur J Pharm Biopharm2004571814729076
- CuiZMumperRJChitosan-based nanoparticles for topical genetic immunizationJ Control Release20017540941911489327
- IllumLJabbal-GillIHinchcliffeMFisherANDavisSSChitosan as a novel nasal delivery system for vaccinesAdv Drug Deliv Rev200151819611516781
- LeeMNahJWKwonYKohJJKoKSKimSWWater-soluble and low molecular weight chitosan-based plasmid DNA deliveryPharm Res20011842743111451027
- Köping-HöggårdMVårumKMIssaMImproved chitosan-mediated gene delivery based on easily dissociated chitosan polyplexes of highly defined chitosan oligomersGene Ther2004111441145215269712
- PengJXingXWangKTanWHeXHuangSInfluence of anions on the formation and properties of chitosan-DNA nanoparticlesJ Nanosci Nanotechnol2005571371716010926
- YoksanRMatsusakiMAkashiMChirachanchaiSControlled hydrophobic/hydrophilic chitosan: colloidal phenomena and nanosphere formationColloid Polym Sci2004282337342
- KimKYKwonSLParkJHChungHJeongSYKwonICPhysicochemical characterizations of self-assembled nanoparticles of glycol chitosan-deoxycholic acid conjugatesBiomacromolecules200561154115815762689
- KurisawaMYokoyamaMOkanoTTransfection efficiency increases by incorporating hydrophobic monomer units into polymeric gene carriersJ Control Release2000681810884574
- LutenJSteenbergenMJvan LokMCDegradable PEG-folate coated poly(DMAEA-co-BA)phosphazene-based polyplexes exhibit receptor-specific gene expressionEur J Pharm Sci20083324125118207707
- LeeHKimTHParkTGA receptor-mediated gene delivery system using streptavidin and biotin-derivatized, pegylated epidermal growth factorJ Control Release20028310911912220843
- HashimotoMMorimotoMSaimotoHSatoTLactosylated chitosan for DNA delivery into hepatocytes: the effect of lactosylation on the physicochemical properties and intracellular trafficking of pDNA/chitosan complexesBioconjug Chem20061730931616536460
- KimYChoiJYYooMReceptor-mediated gene delivery by folate-PEG-baculovirus in vitroJ Biotechnol200713135336117727999
- SudimackJLeeRJTargeted drug delivery via the folate receptorAdv Drug Deliv Rev20004114716210699311
- MislickKABaldeschwielerJDKayyemJFMeadeTJTransfection of folate-polylysine DNA complexes: evidence for lysosomal deliveryBioconjug Chem199565125158974447
- GuoWJLeeRJReceptor-targeted gene delivery via folate-conjugated polyethyleneimineAAPS Pharmsci19991E1911741215
- GuoWJLeeRJEfficient gene delivery via non-covalent complexes of folic acid and polyethylenimineJ Control Release20017713113811689266
- YouJHuFQDuYZYuanHPolymeric micelles with glycolipid-like structure and multiple hydrophobic domains for mediating-target delivery of paclitaxelBiomacromolecules200782450245617661518
- HuFQZhaoMDYuanHYouJDuYZZengSA novel chitosan oligosaccharide-stearic acid micelles for gene delivery: properties and in vitro transfection studiesInt J Pharm200631515816616632285
- MoromotoHHashidzumeAMorishimaYFluorescence studies of associative behavior of cationic surfactant moieties covalently linked to poly(acrylamide) at the surfactant head or tailPolymer200344943952
- SchnurchABKrajicekMEMucoadhesive polymers as platforms for peroral peptide delivery and absorption: synthesis and evaluation of different chitosan-EDTA conjugatesJ Control Release1998502152239685888
- ParkMRHanKOHanIKDegradable polyethylenimine-alt-poly(ethylene glycol) copolymers as novel gene carriersJ Control Release200510536738015936108
- KatayoseSKataokaKRemarkable increase in nuclease resistance of plasmid DNA through supramolecular assembly assembly with poly(ethylene glycol)-poly(L-lysine) block copolymerJ Pharm Sci1998871601639519147
- YouJLiXCuiFDDuYZYuanHHuFQFolate-conjugated polymer micelles for active targeting to cancer cells: preparation, in vitro evaluation of targeting ability and cytotoxicityNanotechnology2008191919436766