Abstract
One of the main challenges in biomedicine is improvement of detection sensitivity to achieve tumor marker recognition at a very low concentration when the disease is not significantly advanced. A pivotal role in cancer defense is played by the p53 tumor suppressor, therefore its detection with high sensitivity may contribute considerably to early diagnosis of cancer. In this work, we present a new analytical method based on surface-enhanced Raman spectroscopy which could significantly increase the sensitivity of traditional bioaffinity techniques. p53 molecules were anchored to gold nanoparticles by means of the bifunctional linker 4-aminothiophenol (4-ATP). The characteristic vibrational bands of the p53-4-ATP nanoparticle system were then used to identify the p53 molecules when they were captured by a recognition substrate comprising a monolayer of azurin in molecules possessing significant affinity for this tumor suppressor. The Raman signal enhancement achieved by 4-ATP-mediated crosslinking of p53 to 50 nm gold nanoparticles enabled detect of this protein at a concentration down to 5 × 10−13 M.
Introduction
Tumor markers play an important role in cancer diagnosis and many efforts are currently devoted to development of ultrasensitive and rapid analytical approaches to identify them.Citation1,Citation2
The human p53 tumor suppressor is considered an indicator of special interest because its tissue and/or serum level has been associated with tumor invasiveness and prognosis in patients with various malignant diseases.Citation3–Citation7 For this reason, several p53 detection procedures have been developed.Citation8–Citation16 Undoubtedly, among these procedures, the most common ones are the traditional fluorescence-labeled immunological methods such as the enzyme-linked immunosorbent assay.Citation8–Citation11 However, these methods require multiple steps, long incubation periods, and provide only a semiquantitative analysis.Citation12 In addition to the more traditional methods, novel label-free detection techniques have paved the way for the realization of protein chip-based immunoassays with enhanced sensitivity and specificity.Citation13–Citation16
On the other hand, nanoparticle applications have received much attention in clinical diagnosis in the last few years.Citation15,Citation17,Citation18 Gold nanocolloids have been widely used to design immunoassay tests for tumor markers, based on their peculiar properties of a high surface-to-volume ratio, the possibility of suitable biomolecular conjugation, their rewarding chemical stability, and the collective electronic behavior at their surface. In particular, their optical and electromagnetic signal enhancement capability, combined with their ability to form hybrid assemblies with biomolecules,Citation2,Citation15 provide the basis for ultrasensitive and molecular specific detection.
Within this context, exploitation of surface-enhanced Raman scattering (SERS)Citation19–Citation22 offers great promise for simplified, sensitive detection of biomolecular interactions and several advantages in early diagnostic over the previously mentioned assay methodologies.Citation23–Citation27 SERS is based on the huge enhancement of the Raman cross-section of molecules when they are placed in the proximity of a nanostructured metal surface,Citation28 due to the contribution of an electromagnetic and chemical or charge transfer effect.Citation19,Citation29,Citation30 Depending on both the chemical nature of the adsorbed molecules and the metal surface features, SERS may reach a 1010-fold increase of conventional Raman detection sensitivity.Citation21,Citation22,Citation31
In previous work we have demonstrated the remarkable potential of this approach in the detection of single molecules and in nanobiodiagnostics.Citation23–Citation26 Proceeding from these studies, we now report a novel SERS-based detection method arising from the synergic combination of Raman signal enhancement and the bioaffinity assay, by which it has been possible to lower the current threshold sensitivity considerably in the detection of p53. In particular, we have exploited the specific interaction of p53 with azurin, a bacterial blue-copper protein. Indeed, cellularCitation32,Citation33 and molecular studiesCitation34,Citation35 have demonstrated that azurin is an exogenous vector able to enter cancer cells preferentially and form a specific and stable complex with human p53, increasing its tumor suppressor activity. It is believed that their interaction involves a portion of the hydrophobic patch surrounding the copper-containing active site of azurin, and the DNA binding domain of p53,Citation32,Citation35 within which the majority of the p53 tumor-derived mutation resides. In our laboratory, the binding and kinetic features of the azurin-p53 complex have been recently investigated by single-molecule atomic force spectroscopy and by surface plasmon resonance.Citation34,Citation36 These studies have demonstrated that this complex is significantly stable and that the azurinp53 interaction is affected neither by the immobilization strategies used nor by the binding of the Mdm2 protein to p53, with the former playing a key physiological role in lowering the intracellular stability of p53.Citation36
In this work we used the 4-aminothiophenol (4-ATP) linker, which has on one side a thiol group able to bind to a gold nanoparticle, and on the other side a diazonium moiety capable of reacting with the electron-rich aromatic lateral chains of p53. p53 conjugated to a gold nanoparticle was transferred onto a capture substrate composed of an azurin monolayer, and the strong intense SERS bands, characteristic of the p53-4-ATP nanoparticle system, were followed to identify the p53 molecules recognized by the azurin partner molecules. This approach was demonstrated to be able to reveal p53 initially present in a solution at 5 × 10−13 M. The proposed methodology couples remarkable sensitivity with high selectivity, even in the presence of other proteins present in the human serum environment.
Materials and methods
Materials
Gold colloidal nanoparticles with a mean diameter of 48.9 ± 3.9 nm (water solution of 4.5 × 1010 particles/mL; gold chloride concentration 0.01%), were purchased from Ted Pella (Reading, CA). Solid 4-ATP (assay 97%, absorbivity ∈ = 9600 M−1·cm−1), human serum albumin, human serum, and azurin were purchased from Sigma-Aldrich (St Louis, MO). The proteins were dissolved in 50 mM phosphate buffer (pH 7.2). A characteristic spectral absorption ratio of 0.48 at 630 nm and 280 nm was found for azurin solution, indicating a good degree of purity of the protein.Citation37–Citation39
Human glutathione S-transferase (GST)-p53 1.23 mM in 50 mM Tris-acetate, pH 7.5, 1 mM EDTA, 20% glycerol) was purchased from Millegen (Labège, France). Cleavage of the GST portion was achieved by digestion with the thrombin CleaveClean kit (Sigma) according to the manufacturer’s instructions.
The purity of the proteins was assessed using gel electrophoresis experiments. The thrombin-purified proteins were separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and analyzed by densitometry. All thrombin-cleaved preparations were of ≥95% homogeneity. The wild-type conformation of purified p53 was verified by an immunoprecipitation technique, using two conformation-specific antibodies and circular dichroism measurements. The methods used to assess the quality, correct folding, and purity of the proteins have been published elsewhere.Citation40 The water used for the experiments was purified using a Milli-Q Reagent water system (Millipore, Bedford, MA).
Methods
The procedure used to functionalize the gold nanoparticles is briefly described in . In the first step, a gold nanoparticle mother solution was mixed with 4-ATP (0.5 mg/mL in absolute ethanol) in a volume ratio of 1:1. The solution obtained was incubated at 20°C for three hours. The formation of a covalent bond between 4-ATP and the gold nanoparticles was followed by Raman spectroscopyCitation24 and ultraviolet-visible spectrophotometry (see Results section). According to the literature,Citation24 we have estimated that about 104 4-ATP molecules can fully cover a single gold nanoparticle.
Figure 1 Chemical reactions (left side) and schematic sketches (right side) describing the functionalization of (A) gold nanoparticles with p53 and (B) glass slides with azurin.
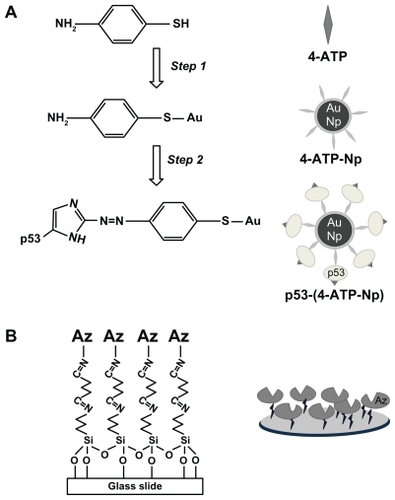
To remove the excess of unbound 4-ATP and promote the ethanol to Milli-Q water solvent exchange, the solution was dialyzed by a membrane (purchased from Spectra/Por®), having a molecular weight cutoff of 100 kDa. Dialysis was stopped when no more 4-ATP was detected by ultraviolet-visible spectroscopy in the fluid beyond the membrane. In the second step, diazo coupling chemistryCitation24,Citation41,Citation42 was employed for the covalent capture of p53 (or azurin in the alternative configuration) onto gold nanoparticles exposing the aromatic amino compound 4-ATP.
To this end, 200 μL of acidified nitrosating agent NaNO2 (0.1 M, pH 3) was added to 200 μL of the dialyzed 4-ATP nanoparticle solution very slowly and under constant stirring for 15 minutes at about 2°C. Only rare bubbles of nitrogen were observed, confirming the stability of the diazotized 4-ATP nanoparticle system. The reaction was monitored by adding phenol to the mixture, since this confers a characteristic red color to the solution when the product appears in the reaction. Excesses of nitrous acid and hydrochloric acid were removed by dialysis against Milli-Q ice water at 2°C, and the pH of the resulting solution was stabilized at 7.2 by phosphate buffer, avoiding the hazard of deamination and unfolding of proteins during the coupling reaction.
The diazonium compound obtained was slowly added to 200 μL of p53 (from 10−9 to 10−13 M, pH 7.2), or alternatively of azurin (1 × 10−5 M, pH 7.2) and kept under gentle stirring at 2°C for two hours. The final solution was further dialyzed to remove unbound proteins. The success of the diazo coupling reaction was assessed by Raman and ultraviolet-visible spectroscopy (see Results section) and the efficiency of the coupling reaction was estimated to be about 0.5. Under the described conditions, proteins were expected to be involved in the diazo coupling with 4-ATP-nanoparticle mainly by means of their exposed histidyl and tyrosyl groupsCitation24,Citation42 (see ).
Glass substrates were cleaned by sonication in acetone for two minutes and subjected to Piranha etch (H2O2:H2SO4, 1:3 v/v) which strongly increases the number of silanol groups on the surface. Successively, they were treated with 3-amino- propyl triethoxy-silane dissolved in 2-propanol (8% solution) for four hours to form a self-assembled monolayer.Citation43 The modified plates were thoroughly rinsed with 2-propanol, baked at 110°C for 10 minutes, and then reacted with 1% glutaraldehyde solution for 30 minutes at room temperature.Citation43,Citation44 After thoroughly rinsing with Milli-Q water, the aldehyde-modified surfaces were incubated with azurin (1 × 10−5 M) or with different molar ratios of p53/human serum albumin (at a minimum concentration of 5 pmol) to form a self-assembled protein monolayer through their solvent-exposed amino groupsCitation36 (see ). Additional control experiments were carried out by incubating the functionalized substrate with human serum solution.
Tapping-mode atomic force microscopy (TM-AFM) images in buffer solution (pH 7.2), using a cantilever force constant, k, of 0.5 N/m, a scan rate of 1 Hz, and a 1 μm × 1 μm scan size, were recorded by a Park XE-100 multimode scanning probe microscope during the different steps of the sample preparation.
The protein-covered substrates were incubated for three hours at room temperature with the corresponding protein partner previously conjugated to 4-ATP nanoparticles in phosphate buffer solution (pH 7.2), allowing the azurin-p53 interaction. Then the substrates were washed with phosphate buffer to remove the unbound p53-4-ATP nanoparticles. For each protein concentration, three samples were prepared to repeat independent experiments.
Raman and SERS spectra were recorded in air at 20°C by a Labram confocal setup (Jobin-Yvon, Edison, NJ) equipped with a Peltier-cooled CCD detector and a single-grating spectrograph (1800 grooves/mm), allowing a resolution of 5 cm−1 and a 16-bit dynamic range. The microscope objective was 100× with a numerical aperture of 0.9 producing a laser spot size of about 1 μm in diameter. The source was a He-Ne laser (Melles Griot) providing 632.8 nm radiation with power emerging from the objective of 6.5 ± 0.5 mW. SERS spectra were acquired by scanning different sample areas per time unit, at the same confocal section. This allowed us to increase the irradiated number of SERS “hot spots” and, at the same time, to reduce the bleaching effects on the irradiated sample. The typical Raman acquisition parameter was an integration time of 20 seconds with five scan repetitions. Optical absorbance was recorded at room temperature using a double-beam Jasco V-550 ultraviolet-visible spectrophotometer with 1 cm path length cuvettes and a 0.5 nm bandwidth.
Results
The approach developed for SERS detection of p53 at very low concentrations is shown in . The 4-ATP molecules are linked to gold nanoparticles by means of a covalent S-Au bond, and the resulting 4-ATP nanoparticle system is successively conjugated to p53 molecules via a diazo coupling reaction to form the p53-4-ATP nanoparticle system (). Glass slides are instead treated in sequence with silane and glutaraldehyde and then reacted with azurin to form the capture substrate (). The p53-4-ATP nanoparticle system is then incubated onto the azurin platform to allow p53 recognition, capture, and successive Raman interrogation.
Each step of the p53-4-ATP nanoparticle system assembly was investigated by Raman spectroscopy and the resulting spectra are shown in . The vibrational bands found in the selected wave number region where the main spectral changes occur are listed in , together with the literature-based assignments.Citation45–Citation48
Table 1 Selected Raman and SERS bands of 4-ATP, 4-ATP nanoparticle and p53-4-ATP nanoparticles with their vibrational assignments
Figure 2 Surface-enhanced Raman scattering spectra of water solutions of (A) 4-ATP, (B) 4-ATP self-assembled on 50 nm gold nanoparticles, and (C) p53 conjugated via diazotization to 4-ATP functionalized gold nanoparticles. The spectral range corresponding to the S-H stretching mode is shown in the insets. The measurements were performed by a 633 nm laser line, objective 100× and 50× (no smoothed spectra).
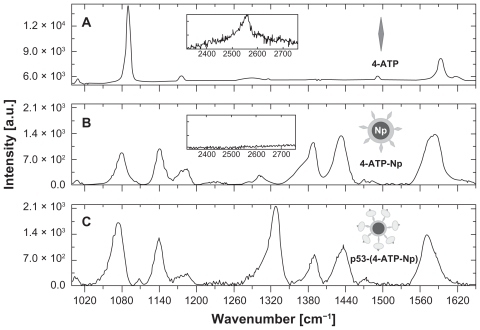
We can note striking differences between the spectra of the 4-ATP linker before () and after () conjugation with the gold nanoparticles. Indeed, the formation of an S-Au covalent bond, indicative of conjugation of 4-ATP with gold nanoparticles, is evident from the disappearance of the band centered at 2550 cm−1, inherent to the stretching vibration of S-H (see the insets of ).Citation24,Citation48 Moreover, most of the peaks in the 1000–1700 cm−1 region of 4-ATP show a strong intensity increase upon binding to the nanoparticles, together with some changes in the relative intensity, with an appreciable frequency red-shift also observed.
After the diazotization reaction, which involves the conjugation of p53 through its exposed histidine and/or tyrosine residues, the SERS spectrum of the whole p53-4-ATP nanoparticle system displays an additional band centered at about 1328 cm−1 (). The new band is assigned to the stretching vibration modes of the diazo bond (–N=N–) which keeps the protein tightly linked to the 4-ATP nanoparticle system.Citation46,Citation49,Citation50 The intensity of this band increases with the p53 concentration used for the diazotization reaction (not shown), indicating that each 4-ATP nanoparticle system is able to conjugate a progressively increasing number of p53 molecules as the concentration of the latter is raised. Indeed, we estimated that up to about 250 p53 molecules can be effectively bound onto a single 50 nm 4-ATP nanoparticle. Citation24 Accordingly, the band at 1328 cm−1 constitutes the spectroscopic fingerprint of the covalent conjugation of p53 to a 4-ATP nanoparticle, and its presence then reflects the capture of p53 by the azurin-containing recognition element. Other Raman bands at around 1390 and 1435 cm−1, characteristic of the 4-ATP nanoparticle system, were also followed to check the conjugation of 4-ATP to the nanoparticles.
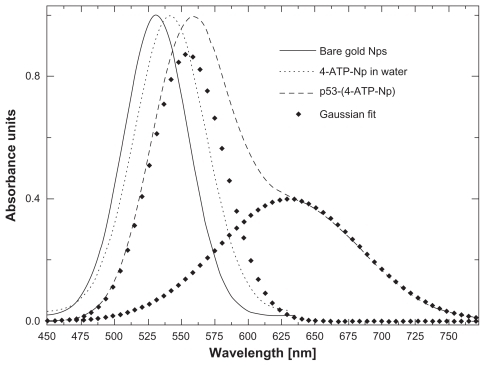
The stepwise molecular assembly of p53-4-ATP nanoparticles was also investigated using ultraviolet-visible spectroscopy, by monitoring the resonant absorption bands due to the collective oscillations of surface electrons of the gold nanoparticles, called localized surface plasmons. shows that the bare nanoparticles are characterized by an absorption band centered at about 530 nm (full line). Upon binding of 4-ATP to the nanoparticles, a shift of the plasmonic resonance band from 530 nm up to a maximum of 541 nm (, light dots) is observed. Since the red-shift is proportional to the number of molecules adsorbed onto the gold nanoparticles,Citation19,Citation20,Citation22,Citation51 we can reasonably assume that, at the maximum shift, the nanoparticles surface is fully covered by 4-ATP molecules. After conjugation of the p53 molecules, a further wavelength shift from 541 nm up to a maximum of 555 nm is registered (, dashed line). At this maximum shift, it is reasonable to assume that each 4-ATP nanoparticle is fully covered by p53 molecules. Moreover, the absorption peak also displays a marked shoulder on the red side (, dashed profile) with a Gaussian deconvolution, indicating a contribution from a further plasmonic band centered at 630 nm (, heavy dots).Citation47
Figure 3 Optical absorption of water solutions of: 50 nm gold nanoparticles (full line), 4-ATP nanoparticles (light dots), and p53-4-ATP nanoparticles (dashed line). The localized plasmon absorption band of the latter is fitted by two components centred at 555 nm and 630 nm (heavy dots).
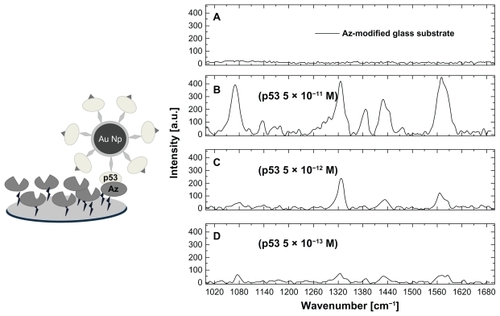
To detect p53 we flowed the p53-4-ATP nanoparticle solution onto a capture platform made up of an immobilized azurin monolayer. After brief incubation, the system was rinsed several times to remove the unbound p53-4-ATP nanoparticle molecules and was then interrogated by SERS spectroscopy. shows the SERS spectra of such an azurin substrate, before () and after () the addition of progressively decreasing concentrations of p53 bound to 4-ATP nanoparticles. While the characteristic Raman fingerprint is absent in , it can be clearly observed in , confirming the presence of p53 molecules which were recognized and captured by azurin. The Raman signal to noise ratio is progressively reduced upon decreasing the p53 concentration. Below 5 × 10−13 M (), this ratio becomes too low (signal to noise ratio <3) and therefore we can assume that such a concentration is the lowest detection limit for p53 by our method. A control experiment in which we dropped 4-ATP nanoparticle molecules (1 pmol of gold nanoparticles) without p53 onto the azurin-coated platform, resulted in a spectrum similar to that shown in , thus revealing no recognition events. These SERS experiments were repeated on three independent samples for each p53 concentration, and obtained good reproducibility. The Raman fingerprint intensity showed a variability of about 5% within samples at the same concentration. In addition, this intensity scaled down in a good correlation with the concentration decrease of p53.
Figure 4 Sketch of a p53-azurin recognition event (left) and surface-enhanced Raman scattering spectra (right) of: the azurin-coated substrate before (A) and after incubation with the p53-4-ATP nanoparticle system, at different p53 concentrations, 5 × 10−11 M (B), 5 × 10−12 M (C), and 5 × 10−13 M (D). Spectra were collected by an objective 100× immersed in phosphate buffer.
Interestingly, we were able to detect p53-azurin biorecognition events even when the SERS bands became hard to distinguish over the noise (at subpicomolar concentrations of p53). To this end, we used a TM-AFM topographical investigation able to detect the formation of biorecognition complexes (p53-azurin) at the level of a single molecule.Citation23 shows representative TM-AFM images of the azurin-coated substrate before () and after incubation with p53-4-ATP nanoparticles (). In , the vertical profile reveals isotropic molecule packing with a height of about 3.1 ± 0.5 nm and roughness (Rq) of 1.7 ± 0.5 nm, which are consistent with those of an azurin monolayer.Citation52 shows instead large and rather regular spots (height 55 ± 8 nm) appearing on the substrate, consistent with the nanoparticles (diameter about 50 nm) bound to p53 (diameter about 5.5 nm).Citation34 Images similar to that shown in were also recorded for the substrate after incubation (and successive rinsing) with 4-ATP-nanoparticles lacking p53. These results indicate that the single atomic force microscopic spots correspond to single capture events between azurin and p53. We also occasionally observed dimers, indicating the presence of a few aggregates in our sample.
Figure 5 Tapping-mode atomic force microscopy images and vertical section analysis of the azurin-functionalized substrate before (A) and after (B) incubation with a solution 0.5 × 10−13 M of p53 linked to 50 nm gold nanoparticles. The vertical profile analysis of panel B indicates the presence of single p53 functionalized nanoparticles (height 55 ± 8 nm) recognized by azurin.
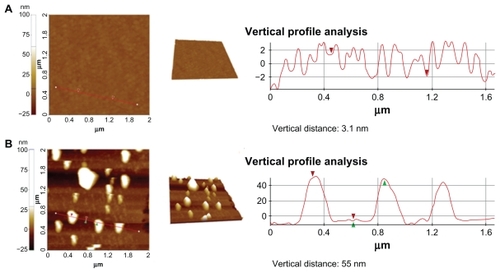
Although this atomic force microscopic approach is quite promising for very ultrasensitive detection, it requires further investigation to gain some ground from a quantitative point of view and reproducibility. However, it did allow us to estimate the amount of complexes formed on the substrate. Indeed, a statistical analysis of recorded images revealed that the complexes formed corresponded to about 20% of the total amount of p53-4-ATP nanoparticle molecules incubated with azurin. This indicates that only 20% of the functionalized nanoparticles give rise to successful interactions with the substrate.
To check if the method was able to reveal the tumor marker p53 among other proteins selectively, we first analyzed if the capture of p53 from azurin was affected by the presence of a molar excess of human serum albumin. The latter is the most abundant protein in human blood serum. We used an alternative configuration in which azurin was conjugated with 4-ATP nanoparticles by diazotization, and then used the SERS marker to screen the various samples. We incubated different samples containing p53 and human serum albumin at various concentrations on glass slides bearing aldehyde moieties on their surface, to allow immobilization of both human serum albumin and p53.Citation36 Successively, we flowed the azurin-4-ATP nanoparticle systems onto these substrates and performed SERS measurements after extensive rinsing to remove what had not been captured by the protein-coated substrates. shows representative SERS spectra of samples with human serum albumin/p53 at a molar ratio of 0, 10, 100, and 1000, and samples containing only human serum albumin, with all of them treated with our azurin-modified nanoparticles. It is worthy of note that the intensity of the SERS bands characteristic of the azurin-4-ATP nanoparticle system undergo a progressive decrease with increasing of the human serum albumin/p53 molar ratio (). These bands are absent on the substrate containing only human serum albumin (). Therefore, it can be inferred that the fingerprint SERS signal arose only from the azurin-4-ATP nanoparticle molecules which formed a complex with p53 immobilized onto the substrate.
Figure 6 Sketch of an azurin-p53 recognition event in presence of an excess of human serum albumin (left) and surface-enhanced scattering spectra (right) of the capture substrates incubated with the azurin-4-ATP nanoparticle system (5 × 10−9 M azurin), and previously functionalized with (A) p53 alone, (B) human serum albumin/p53 at molar ratios equal to 10, (C) human serum albumin/p53 at molar ratios equal to 100, (D) human serum albumin/p53 at molar ratios equal to 1000, and (E) human serum albumin alone. Surface-enhanced Raman scattering spectra were acquired with a laser source at 633 nm in air using a 100× objective.
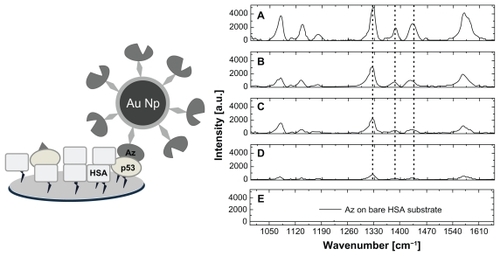
Notably, the Raman marker derived from conjugation of azurin to the 4-ATP nanoparticle gives a more intense SERS signal than that corresponding to the previous configuration, due to the higher number of azurin molecules surrounding each nanoparticle. Indeed, by assuming that azurin and p53 can be represented as spheres with a diameter of about 3 nm and 6 nm, respectively, we can estimate that about 980 molecules of azurin and 250 molecules of p53 can be, at maximum, closely packed around each nanoparticle, the diameter of which is about 50 nm.
A competitive experiment was also performed using human serum which is known to contain other molecules besides albumin. For this purpose, glass slides bearing aldehyde moieties were incubated with human serum. Subsequently, the azurin-4-ATP nanoparticle solution was flowed on these substrates to check for any interaction between azurin and the molecules contained in the serum. After rinsing, no appreciable SERS signal over the noise was detected, and the resulting spectrum was indeed similar to that shown in . This indicates that the azurin-modified nanoparticles do not interact with human serum molecules, so no interference with the p53-azurin complex formation can be reasonably inferred.
Discussion
Our Raman-SERS results show that the p53-4-ATP nanoparticle system exhibits remarkable enhancement of the fingerprint Raman signal, with vibrational features well distinguishable and stable in time. Generally, the huge enhancement of the Raman cross-section when the molecules are bound to a metal surface (SERS effect) is attributed to two main SERS mechanisms, ie, an electromagnetic mechanism associated with a large local field caused by surface plasmon resonance, and a charge transfer from the metal to the adsorbed molecules.Citation19,Citation29,Citation30 For 4-ATP molecules bound to gold nanoparticles, the enhancement of bands at about 1141 cm−1 and 1435 cm−1 seen in can be ascribed to the charge transfer mechanism, while the other bands reported in for 4-ATP nanoparticles are thought to be selectively enhanced mainly via electromagnetic mechanisms.Citation45–Citation48 From the signal to noise intensity ratio of the C-S stretching mode (around 1089 cm−1 and 1078 cm−1 for the solid 4-ATP and the 4-ATP nanoparticle system, respectively), enhancement due to a SERS effect of about seven orders of magnitude was estimated.
Such a huge enhancement allowed us to demonstrate p53 at extremely low concentrations (down to about 5 × 10−13 M) which should be compared with those obtained by the other methods reported in the literature. Recent research reports novel bioaffinity-based methods, employing consensus DNA sequences and monoclonal antibodies adsorbed onto surface plasmon resonance sensor discs, gold electrodes, and metal nanostructures, which are able to improve the tumor marker detection limits of the enzyme-linked immunosorbent assay test. These new techniques can demonstrate p53 at picomolar concentrations,Citation53,Citation54 indicating that our detection value is comparable with or even better than the best results obtained by these biosensors.Citation53 Nevertheless, our method has the advantage of low concentrations which are easy to detect and a relatively short assay time. In addition, it does not require multiple steps, as does the classical sandwich enzyme immunoassay. Notably, our SERS-based detection approach is amenable to further improvements of the enhancement factor by optimizing the dimensions of the gold nanoparticles, the distance between the Raman marker and the nanoparticle surface, and perhaps even the arrangement of azurin molecules on the substrate.
The presence of very few larger spots in the TM-AFM images, reasonably attributed to the formation dimers, finds a correspondence with the observation of plasmonic modes centered at around 630 nm in the ultraviolet-visible spectra, which can likely be attributed to some interparticle coupling effects.Citation47 Accordingly, we can hypothesize that, during the diazo coupling reaction, some p53 molecules may form multiple bonds with different azotated 4-ATP nanoparticles, thus promoting interparticle modulation of the common localized plasmonic band. Such an effect could be responsible for some colloid polydispersivity.
A further lowering of the detection level could also be achieved by coupling SERS with the single molecule capability of atomic force microscopy imaging which allows monitoring of formation of an individual complex. Indeed, the observation by TM-AFM of large spots on the azurin-coated substrate after incubation with the p53-4-ATP nanoparticle solution showed the effective deposition of nanoparticles on the substrate, with concomitant formation of specific complexes between azurin and p53.
The ability of our method to detect p53 specifically among the other molecules present in human serum provides an indication of the selectivity of this method. Such a property, coupled with high reproducibility, accuracy and speed, represents a starting point for further implementation in real biosensors. In this respect, we suggest that monitoring of p53 in serum could be extremely helpful as an indicator of prognosis and response to chemotherapy and radiotherapy in various malignant diseases,Citation3–Citation7 and even to relate their insurgence to genotoxic environmental carcinogenic exposure.Citation11,Citation55–Citation57
Conclusion
The proposed azurin-based SERS approach for detection of the p53 tumor suppressor combines high sensitivity and selectivity. It is able to detect p53 at a concentration of 5 × 10−13 M, with the further ability to reveal the target biomolecule selectively in the presence of human serum albumin, and even in the serum environment in which other biomolecules are present. Accordingly, it can constitute a basis to design novel protocols for early clinical screening of human and environmental pathologies. This strategy can be easily extended to the detection of multiple specific analytes, thus providing a multiplex assay for ultrasensitive and targeted screening. Additionally, a combination of this SERS-based detection method with atomic force microscopic topography offers the opportunity to improve further the detection sensitivity towards the single molecule regime.
Acknowledgment
This work was partly supported by a grant from the Italian Association for Cancer Research (AIRC No. IG 10412).
Disclosure
The authors report no conflicts of interest in this work.
References
- StefanekMEAndrykowskiMALermanCManneSGlanzKBehavioral oncology and the war on cancer: Partnering with biomedicineCancer Res2009697151715619706775
- SoperSABrownKEllingtonAPoint-of-care biosensor systems for cancer diagnostics/prognosticsBiosens Bioelectron2006211932194216473506
- ZusmanISandlerBGurevichPComparative study of the role of serum levels of p53 antigen and its tumor cell concentration in colon cancer detectionHum Antibodies Hybridomas199671231289057060
- ShurbajiMSKalbfleischJHThurmondTSImmunohistochemical detection of p53 protein as a prognostic indicator in prostate cancerHum Pathol1995261061097821906
- PsyrriAKountourakisPYuZAnalysis of p53 protein expression levels on ovarian cancer tissue microarray using automated quantitative analysis elucidates prognostic patient subsetsAnn Oncol20071870971517220511
- MalviyaVSinghHUshaUSDSinghPBSerum p53 and bladder cancer: Can serum p53 be used as a tumor markerUrol Res20043239139415503000
- ShimadaHNakajimaKSakamotoKExistence of serum p53 antibodies in cyclosporine A-treated transplant patients: Possible detection of p53 protein over-expressionTransplant Proc200032177911119931
- PortefaixJMFanuttiCGranierCDetection of anti-p53 antibodies by ELISA using p53 synthetic or phage-displayed peptidesJ Immunol Methods2002259657511730842
- LevesqueMAKatsarosDYuHImmunofluorometrically determined p53 accumulation as a prognostic indicator in Italian breast cancer patientsInt J Cancer1998791471529583729
- ThomasMDMcIntoshGGAndersonJJA novel quantitative immunoassay system for p53 using antibodies selected for optimum designation of p53 statusJ Clin Pathol1997501431479155696
- BeniniECostaAAbolafioGSilvestriniRp53 Expression in human carcinomas: Could flow cytometry be an alternative to immunohistochemistryJ Histochem Cytochem19984641479405493
- GuihongYDaXShiciTQunCRapid and sensitive immunomagnetic-electrochemiluminescent detection of p53 antibodies in human serumJ Immunol Methods2004288475415183084
- CloarecJPChevolotYLaurenceauEPhaner-GoutorbeMSouteyrandEA multidisciplinary approach for molecular diagnostics based on biosensors and microarraysIRBM200829105127
- ChungJWKimSDBernhardtRPyunJCApplication of SPR biosensor for medical diagnostics of human hepatitis B virus (hHBV)Sensors and Actuators B2005111–112416422
- ChenHJiangCYuCZhangSLiuBKongJProtein chips and nanomaterials for application in tumor marker immunoassaysBiosens Bioelectron2009243399341119398322
- YeoJParkJYBaeWJLabel-free electrochemical detection of the p53 core domain protein on its antibody immobilized electrodeAnal Chem2009814770477719435284
- El-SayedIHHuangXEl-SayedMASurface plasmon resonance scattering and absorption of anti-EGFR antibody conjugated gold nanoparticles in cancer diagnostics: Applications in oral cancerNano Lett2005582983415884879
- BoisselierEAstrucDGold nanoparticles in nanomedicine: Preparations, imaging, diagnostics, therapies and toxicityChem Soc Rev2009381759178219587967
- CampionAKambhampatiPSurface enhanced Raman scatteringChem Soc Rev1998127241250
- WeiAKimBSadtlerBTrippSLTunable surface-enhanced Raman scattering from large gold nanoparticles arraysChemphyschem200112743745
- KneippKWangYKneippHSingle molecule detection using surface enhanced Raman scatteringPhys Rev Lett19967016671670
- NieSEmorySRProbing single molecules and single nanoparticles by surface-enhanced Raman scatteringScience1997275110211069027306
- BizzarriARCannistraroSSurface-enhanced Raman spectroscopy combined with atomic force microscopy for ultrasensitive detection of thrombinAnal Biochem200939314915419563767
- BizzarriARCannistraroSSERS detection of thrombin by protein recognition using functionalized gold nanoparticlesNanomedicine: Nanotechnology, Biology, and Medicine20073306310
- BizzarriARCannistraroSLévy statistics of vibrational mode fluctuations of single molecules from surface-enhanced Raman scatteringPhys Rev Lett20059406830306830615783781
- BizzarriARCannistraroSStatistical analysis of intensity fluctuations in single molecule SERS spectraPhys Chem Chem Phys2009953145319
- WangJKongLTGuoZXuJYLiuJHSynthesis of novel decorated one-dimensional gold nanoparticle and its application in ultrasensitive detection of insecticideJ Mater Chem20102052715279
- JacksonJBHalasNJSurface-enhanced Raman scattering on tunable plasmonic nanoparticle substratesProc Natl Acad Sci U S A2004101179301793515608058
- MoskovitsMSurface-enhanced spectroscopyRev Mod Phys198557783826
- LombardiJRBirkeRLLuTXuJCharge-transfer theory of surface enhanced Raman spectroscopy: Herzberg-Teller contributionsJ Chem Phys19868441744180
- RuECMeyerMEtchegoinPGProof of single-molecule sensitivity in surface enhanced Raman scattering (SERS) by means of a two-analyte techniqueJ Phys Chem B20061101944194816471765
- YamadaTGotoMPunjVBacterial redox protein azurin, tumor suppressor protein p53, and regression of cancerProc Natl Acad Sci U S A200299140981410312393814
- YamadaTHiraokaYIkehataMApoptosis or growth arrest: Modulation of tumor suppressor p53’s specificity by bacterial redox protein azurinProc Natl Acad Sci U S A20041014770477515044691
- TarantaMBizzarriARCannistraroSProbing the interaction between p53 and the bacterial protein azurin by single molecule force spectroscopyJ Mol Recognit20082637018247358
- De GrandisVBizzarriARCannistraroSDocking study and free energy simulation of the complex between p53 DNA-binding domain and azurinJ Mol Recognit20072021522617703463
- DomeniciFFrasconiMMazzeiFD’OraziGBizzarriARCannistraroSAzurin modulates the association of Mdm2 with p53: SPR evidence from interaction of the full-length proteinsJ Mol Recognit20112470771421584881
- PunjVDas GuptaKTChakrabartyMABacterial cupredoxin azurin and its interactions with the tumor suppression protein p53Biochem Biophys Res Commun200331210911414630027
- AdmanETCopper protein structuresAdv Protein Chem1991421451971793005
- GuzziRSportelliLInteraction of azurin with alcohols: An ESR, optical absorption, and fluorescence emission investigationJ Inorg Biochem1992453945
- FunariGDomeniciFNardinocchiLInteraction of p53 with MDM2 and azurin as studied by atomic force spectroscopyJ Mol Recognit20092334335119941302
- MorrisonRTBoydRNOrganic Chemistry6th EditionNew DelhiPrentice Hall1999
- PhillipsJHRobrishSABatesCHigh efficiency coupling of diazonium ion to proteins and amino acidsJ Biol Chem196524069970414275124
- ParkSHImJHChunBHKimJHAdsorption kinetics of Au and Ag nanoparticles on functionalized glass surfacesMicrochem J1999637191
- HalliwellCMCassAEGA factorial analysis of silanization conditions for the immobilization of oligonucleotides on glass surfacesAnal Chem2001732476248311403288
- BaiaMToderasFBaiaLPoppJAstileanSProbing the enhancement mechanisms of SERS with p-aminothiophenol molecules adsorbed on self-assembled gold colloidal nanoparticlesChem Phys Lett2006422127132
- JiaoLSNiuLShenJYouTDongSIvaskaASimple Azo derivation on 4-aminothiophenol/Au monolayerElectrochem Commun20057219222
- ZhuZZhuTLiuZRaman scattering enhancement contributed from individual gold nanoparticles and interparticle couplingNanotechnology200415357364
- OsawaMMatsudaNYoshiiKUchidaICharge transfer resonance raman process in surface-enhanced Raman scattering from p-aminothiophenol adsorbed on silver: Herzberg-Teller contributionJ Phys Chem1994981270212707
- WuYBurandaTMetzenbergRLSklarLALopezGDiazo coupling method for covalent attachment of proteins to solid substratesBioconjug Chem20061735936516536466
- AndrikopoulosPCMcCarneyKMArmstrongDRLittlefordREGrahamDSmithWEADensity functional theory and resonance Raman study of a benzotriazole dye used in surface enhanced resonance Raman scatteringJ Mol Struct20067895970
- SarahLWStevenJOLeeTRHalasNJConstruction of simple gold nanoparticle aggregates with controlled plasmon-plasmon interactionsChem Phys Lett1999300651655
- WangYZhuXWuMXiaNWangJZhouFSimultaneous and label-free determination of wild-type and mutant p53 at a single surface plasmon resonance chip preimmobilized with consensus DNA and monoclonal antibodyAnal Chem2009818441844619772286
- JiaCPZhongXQHuaBNano-ELISA for highly sensitive protein detectionBiosens Bioelectron2009242836284119339168
- AndolfiLBizzarriARCannistraroSAssembling of redox proteins on Au(111) surfaces: A scanning probe microscopy investigation for application in bio-nanodevicesThin Solid Films2006515212219
- LiJDetection of ras p21 and p53 proteins in serum of the lung cancer patients exposed to indoor coal-burning pollution with Western blottingFuel and Energy Abstracts199738350
- HofsethLJSaitoSHussainSPNitric oxide-induced cellular stress and p53 activation in chronic inflammationProc Natl Acad Sci U S A200210014314812518062
- YangJDuerksen-HughesPA new approach to identifying genotoxic carcinogens: p53 induction as an indicator of genotoxic damageCarcinogenesis199819111711259667752