Abstract
Background
Recent studies report curcumin nanoformulation(s) based on polylactic-co-glycolic acid (PLGA), β-cyclodextrin, cellulose, nanogel, and dendrimers to have anticancer potential. However, no comparative data are currently available for the interaction of curcumin nanoformulations with blood proteins and erythrocytes. The objective of this study was to examine the interaction of curcumin nanoformulations with cancer cells, serum proteins, and human red blood cells, and to assess their potential application for in vivo preclinical and clinical studies.
Methods
The cellular uptake of curcumin nanoformulations was assessed by measuring curcumin levels in cancer cells using ultraviolet-visible spectrophotometry. Protein interaction studies were conducted using particle size analysis, zeta potential, and Western blot techniques. Curcumin nanoformulations were incubated with human red blood cells to evaluate their acute toxicity and hemocompatibility.
Results
Cellular uptake of curcumin nanoformulations by cancer cells demonstrated preferential uptake versus free curcumin. Particle sizes and zeta potentials of curucumin nanoformulations were varied after human serum albumin adsorption. A remarkable capacity of the dendrimer curcumin nanoformulation to bind to plasma protein was observed, while the other formulations showed minimal binding capacity. Dendrimer curcumin nanoformulations also showed higher toxicity to red blood cells compared with the other curcumin nanoformulations.
Conclusion
PLGA and nanogel curcumin nanoformulations appear to be very compatible with erythrocytes and have low serum protein binding characteristics, which suggests that they may be suitable for application in the treatment of malignancy. These findings advance our understanding of the characteristics of curcumin nanoformulations, a necessary component in harnessing and implementing improved in vivo effects of curcumin.
Video abstract
Point your SmartPhone at the code above. If you have a QR code reader the video abstract will appear. Or use:
Introduction
Curcumin (structure shown in ) is a natural diphenolic compound extracted from the herb Curcuma longa, and is widely used in traditional Indian and Chinese medicine. Curcumin has numerous biological and pharmacological activities, with no major side effects, and is currently being used in several clinical trials for treating several disorders, including Alzheimer’s disease, hypercholesterolemia, atherosclerosis, psoriasis, Crohn’s disease, neurological disorders, and cancer.Citation1,Citation2 Recent studies establish that curcumin can be translated into a therapeutic molecule to treat a variety of cancers.Citation3,Citation4 In addition, studies have demonstrated that curcumin is an efficient molecule for overcoming multidrug resistance phenomena as well as inducing sensitization of tumor cells for chemotherapy and radiation.Citation5–Citation10 Unfortunately, the strong potential of curcumin to improve the effectiveness of cancer treatment is hindered by its poor bioavailability, attributable to its poor aqueous solubility, degradation, and high rate of metabolism.Citation11–Citation13
Figure 1 Structural variations of the curcumin nanoformulations and their uptake in cancer cells. (A) Chemical structure of curcumin. (B) Different types of curcumin nanoformulations and their structures based on chemical composition. Structures do not represent exact size or orientation, and the solid background is intended to distinguish the different formulations. Entire polymer chains, drugs (curcumin), and other structures are defined in the figure itself. (C) Cellular uptake of curcumin nanoformulations in SKBR-3, MDA-MB-231, and HPAF-II cancer cells. Cancer cells were treated with curcumin 20 μg or curcumin nanoformulations (20 μg equivalent to curcumin) for 6 hours. Uptake was determined by recording absorption of acetone cell lysates at λmax 450 nm using UV-vis spectrophotometer.
Notes: Data are reported as the mean of three repeats for each uptake (mean ± standard error of the mean deviation; *P < 0.05, curcumin nanoformulations versus free curcumin).
Abbreviations: CD, β-cyclodextrin; CUR, curcumin; PLGA, polylactic-co-glycolic acid; PVA, polyvinyl alcohol; PLL, poly-L-lysine.
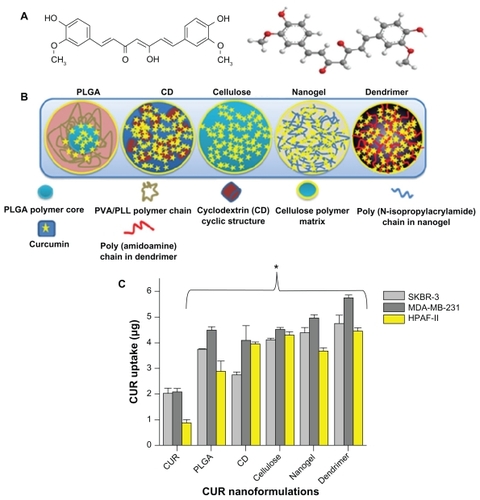
Because of its hydrophobic nature and poor bioavailability, the clinical development of curcumin as a “medicine” is dependent upon developing a nanocarrier for efficient transport in the body.Citation12,Citation14,Citation15 Recently, several curcumin nanoformulations, such as polymer nanoparticles, self- assemblies, nanocrystal dispersions, nanoemulsions, lipid nanoparticles, and protein-based drug delivery systems, have shown improved solubility, stability, and bioavailability of the curcumin molecule.Citation14–Citation20 Moreover, curcumin nanoformulations have demonstrated improved cellular uptake in cancer models that increases the chance of a positive therapeutic outcome. Curcumin nanoformulations have also exhibited superior in vitro anticancer responses compared with free curcumin due to sustained release of the active compound and the enhanced permeation and retention effects of nanoformulations.Citation16,Citation19,Citation21,Citation22 Several of these nanoformulations have been successful in raising the area under the concentration-time curve, half-life, and mean residence time of curcumin in serum and in various organs.Citation16,Citation23–Citation25 In our previous study, we describe a comparative evaluation of β-cyclodextrin, hydroxyl propyl methyl cellulose (cellulose), polylactic-co-glycolic acid (PLGA) and dendrimer curcumin nanoformulations with free curcumin using cytotoxicity, cellular uptake, and apoptosis studies in cancer cells and suggests a close relationship with the therapeutic efficacy of the formulation.Citation26 However, there is no tool to evaluate the behavior of curcumin nanoformulations in vivo. Similarly, while a number of curcumin nanoformulations have been reported in the literature, so far there has been no comparative study to evaluate which formulation(s) is more efficient for in vivo cancer therapeutics.
Preclinical properties of nanoformulations depend upon avoiding rapid clearance from the systemic circulation by the cells of the immune system.Citation27–Citation29 Usually, the in vivo fates of nanoformulations follow many potential routes before reaching their actual target(s).Citation30–Citation32 Therefore, the in vivo outcome of nanoformulations depends on the biological fluid interactions with particle surfaces where particles may aggregate or stabilize, or be subjected to clearance, uptake, and trafficking processes.Citation33–Citation35 Conventional nanoparticle formulations readily interact with plasma proteins, opsonize, and are taken from the blood circulation by phagocytes. Modification of the surface properties of nanoformulations remains a key feature that can enhance their half-life, which is likely to be predictive of therapeutic outcome. Further, hemocompatibility of nanoformulations is an initial check point in understanding their utility in vivo. Therefore, the aim of this study was to examine the interaction of curcumin nanoformulations with plasma proteins and red blood cells and as a result, predict the implications of their biological characteristics for in vivo application.
Materials and methods
Curcumin (≥95% purity), β-cyclodextrin, cellulose (molecular weight 10,000, methoxy 1.8–2.0 and propylene oxide 0.2–0.3, viscosity 2 wt% in water, 5 cps), polyvinyl alcohol (molecular weight 30,000–70,000), poly-L-lysine (molecular weight 30,000–70,000), N-isopropyl acrylamide (NIPAM), polyamidoamine dendrimer generation 4 (10 wt% solution), sodium dodecyl sulfate, fibrinogen, immunoglobulin G, transferrin, human serum albumin (HSA) and acetone (≥99.5, American Chemical Society reagent grade) were purchased from Sigma Chemical Co (St Louis, MO). PLGA, 50:50 lactide-glycolide ratio, inherent viscosity 1.32 dL/g at 30°C) was purchased from Birmingham Polymers (Pelham, AL). All chemicals were used as received without further purification. Ultrapure water was produced by purification with an Ultrapure® water system (Elga, Woodridge, IL). Curcumin nanoformulations () were obtained following our previous protocols using PLGA, β-cyclodextrin, cellulose, nanogel (polyNIPAM), and dendrimer through self-assembly/ complexation/redox free radical solution polymerization or encapsulation techniques.Citation21,Citation22,Citation26,Citation36 For consistency, all curcumin nanoformulations were processed with 20 wt% curcumin loading. A schematic representation of curcumin nanoformulations are provided in .
In vitro cellular uptake
In vitro cellular uptake studies were performed as described in our previous reports.Citation21,Citation22 MDA-MB-231, SKBR-3 (breast), and HPAF-II (pancreatic) cancer cells (1 × 105) in Dulbecco’s modified Eagle’s medium or F12 medium were incubated in either 20 μg of free curcumin (dissolved in dimethylsulfoxide) or an equivalent amount of curcumin nanoparticles. After 6 hours of incubation, the cells were washed with phosphate-buffered solution (10 mM, pH 7.4), trypsinized, and centrifuged at 1000 rpm. The cell pellet containing curcumin in cancer cells was resuspended in 1 mL of acetone and sonicated for 5 minutes. The lysates were centrifuged at 12,000 rpm for 15 minutes and the curcumin levels were recorded for the supernatant curcumin solution.Citation21 A standard plot of curcumin in acetone (1–10 μg) was prepared identically to calculate curcumin uptake by the cells.
Protein adsorption
Particle size, shape, and zeta potential
The particle size, shape, and zeta potential of nanoparticles can vary after serum protein adsorption/interaction. To evaluate this phenomenon, curcumin nanoformulations (1 mg/mL) were dispersed in 100 μg HSA for 2 hours at 25°C. The nanoformulations were centrifuged at 12,000 rpm for 15 minutes to obtain HSA-bound nanoparticles. The particle size and zeta potential of HSA-bound curcumin nanoformulations were measured using a Delsa™ Nano C particle size analyzer (Beckman Coulter, Miami, FL). Particle size and distribution was measured for 3 minutes, and the zeta potential was measured for 90 runs (9 minutes). The surface morphology of curcumin formulations and HSA- bound curcumin nanoformulations was observed under a JEOL-1210 transmission electron microscope (JEOL, Tokyo, Japan) operating at 80 kV. The nanoformulation samples were stained with uranyl acetate solution to accentuate fine structures under transmission electron microscopy.
Western blot
Curcumin nanoformulations (20 μM) were incubated in 100 μg of various plasma protein solutions at 37°C. During incubation, adsorption of proteins occurred on the nanoparticle surfaces. Following 2 hours of incubation, the samples were centrifuged at 12,000 rpm for 15 minutes to obtain pellets of protein-adsorbed curcumin nanoparticles. The adsorbed proteins were removed by adding an electrophoresis sample buffer (Santa Cruz Biotechnology, Santa Cruz, CA) and heating to 95°C for 5 minutes. These protein samples were loaded in 4%–20% polyacrylamide gel and then underwent sodium dodecyl sulfate-polyacrylamide gel electrophoresis at 150 V for 60 minutes. The gels were placed in 100 mL ultrapure water and microwaved (Emerson ® microwave oven, MW8777 W, 700 W) on high for 3 minutes and allowed to cool down for 5 minutes. After the water was discarded, 20 mL of SimplyBlue™ SafeStain solution (Coomassie® G-250 stain, Invitrogen, Carlsbad, CA) was added and the gel was microwaved on high for 45 seconds. The gel was allowed to cool down, washed with ultrapure water and incubated in 20 mL of 20% sodium chloride solution for 5 minutes. The gels were scanned using a BioRad Gel Doc (BioRad, Hercules, CA) and the intensity quantification of bands was acquired using AlphaEase Fc software.Citation10
Flow cytometry
To determine the adhesion properties of curcumin nanoformulations in human red blood cells, a flow cytometric assay was employed. For this study, 100 μL of healthy male human blood (Biological Specialty Corporation, Colmar, PA) was incubated with 10–40 μM curcumin nanoformulations for 2 hours in 1 mL of RPMI medium at 37°C. During incubation, the curcumin nanoformulations readily established interaction with plasma proteins while the free curcumin nanoparticles bound with red blood cells.
After incubation, the entire solution was filtered through a polystyrene round-bottom tube with cell-strainer cap and injected into an Acuri C6 Flow Cytometer (Accuri™ Cytometers Inc, Ann Arbor, MI) to determine the fluorescence levels of curcumin nanoformulations attached to red blood cells in flow channel 1 absorbance (FL1A, 488 excitation, blue laser, 530 ± 15 nm, fluorescein isothiocyanate/green fluorescent protein). Red blood cells were first gated using side and forward scatter in the Accuri software and nanocurcumin particles bound to red blood cells in this range only were analyzed by flow cytometry. Because unbound nanoparticles were way below the range, they did not interfere with the data.
Hemolysis test
Hemolysis causes damage to red blood cells via the release of iron-containing protein hemoglobin into plasma. To evaluate curcumin nanoformulations for human use, formulations must first be checked for hemolytic compatibility. In this study, 8 mL of whole blood from a healthy male was centrifuged at 2000 rpm for 10 minutes to obtain red blood cells, which were then resuspended in 8 mL RPMI 1640 growth media. Different concentrations of curcumin nanoformulations (0–200 μM) were incubated at 37°C with 100 μL red blood cells in Eppendorf tubes. After 2 hours of incubation at 37°C, the extent of hemolysis caused by curcumin nanoformulations was observed macroscopically, the tubes were centrifuged, and the supernatant fluid was measured for optical density at λmax 570 nm using ultraviolet-visible spectrophotometry (BioMate 3 spectrophotometer, Thermo Electron Corporation, Waltham, MA). Phosphate- buffered solution and sodium dodecyl sulfate were used as the negative and positive controls, respectively. To observe the visual effect of red blood cell membrane damage, the red blood cell pellets were redispersed in phosphate-buffered solution after treatment, and a drop of solution was placed on a glass slide for imaging using an Olympus BX 41 phase contrast microscope (Olympus, Center Valley, PA). A transmission electron microscopy study was performed according to our previously reported method to identify further nanoparticle association with the red blood cells.Citation21
Statistical analysis
The data were processed using Microsoft Excel 2007 software and presented as the mean ± standard error of the mean. Statistical analyses were performed for the overall comparison between groups using analysis of variance models. If the analysis of variance model F-test was significant at the 0.05 level, the difference between groups was examined using a pairwise t-test and each group was compared with the control. The analysis was performed using SAS 9.2 (SAS Institute, Inc, Cary, NC). All graphs were plotted using Origin 6.1 software.
Results and discussion
Many reports confirm that nanoparticles with a size of less than 200 nm can be taken up by cancer cells through endo-cytosis or diffusion.Citation37–Citation39 In addition to particle size, the type and surface characteristics of nanoformulations contribute to the regulation of uptake.Citation40–Citation42 Some nanoformulations are toxic due to the surfactants used in their preparation. Therefore, in this study we explored the feasibility of five different curcumin nanoformulations for cancer therapy based on cancer cell uptake, as well as their interaction with human plasma proteins and red blood cells.
Cellular uptake
First, all five curcumin nanoparticle formulations were studied for uptake into SKBR-3, MDA-MB-231 (breast), and HPAF-II (pancreatic) cancer cells to evaluate their feasibility as drug (curcumin) delivery vehicles. This uptake study will be useful to identify which curcumin nanoformulations are efficient compared with free curcumin. Uptake was achieved by incubating the cancer cells with 20 μg of curcumin or 20 μg equivalent curcumin nanoparticles for 6 hours. Curcumin uptake was determined by the absorbance measurement of acetone-extracted cell lysates. Preferential uptake of curcumin was observed in all three cancer cells by the curcumin nanoformulations in comparison with free curcumin (). This enhanced uptake behavior of various curcumin nanoformulations has been observed previously in many cancer cells.Citation16,Citation21,Citation22,Citation43–Citation45 Curcumin uptake in cancer cells, in descending order, was found to be MDA-MB-231 > SKBR-3 > HPAF-II. Overall, the dendrimer curcumin formulation showed noticeably higher uptake by cancer cells than the PLGA, β-cyclodextrin, cellulose, and nanogel curcumin formulations. This significantly higher uptake could be due to the positive zeta potential or higher penetration capacity via functional amino groups on the dendrimer. These results suggest that nanoparticle-mediated delivery of curcumin is more efficient and better able to reach cancer cells than free curcumin, and may effectively improve therapeutics. Although all five curcumin nanoformulations showed enhanced uptake, their in vivo properties may be different, depending on several factors, including stability of the curcumin nanoformulations in the presence of blood proteins and interaction with erythrocytes. Therefore, we investigated interaction of blood proteins and erythrocytes for all five curcumin formulations.
Association of curcumin nanoparticles with HSA
Newly developed drug nanoformulations may have a direct interaction with tissues and cells after administration to animals and humans.Citation46 The ability of nanoformulations to achieve a higher therapeutic value relies on their biocompatibility and lower serum protein binding capacity which, in turn, is important for industrialization. In general, serum proteins readily bind onto the surfaces of various nanoparticles within one hour.Citation47,Citation48 Proteins adsorb on the surfaces of nanoparticles quickly and their characteristics may change before reaching the intended target(s). The degree of protein interaction may depend on particle size, surface stabilization, functional or chemical groups on the surface, and composition of the particles.Citation48–Citation52 Therefore, we first addressed how the curcumin nanoparticles surface is adsorbed by HSA. We chose HSA because it is the most abundant protein in human plasma (3.4–5.4 g/dL).
Different curcumin nanoparticles have variable particle sizes and morphology (, top panel). PLGA, β-cyclodextrin, cellulose, nanogel, and dendrimer curcumin nanoformulations had particle sizes of about 52, 41, 36, 48, and 42 nm, respectively. After incubation with HSA, most of the nanoparticles had a similar particle size, but their morphology was distinctive, ie, they were either coated with HSA corona or showed a change in their aggregation behavior (, lower panel). This variation in the morphology of nanoparticles is similar to that caused by the formation of soft corona or loosely bound proteins on metal or polymer nanoparticle surfaces.Citation50,Citation53,Citation54 High accumulation of dark spots in the β-cyclodextrin and cellulose curcumin nanoformulations after HSA incubation is related to the stain (uranyl acetate, , red arrows) used to assist detection under transmission electron microscopy, not to actual nanoparticle aggregation. This may be because a higher rate of adsorption of metal ions to β-cyclodextrin and/or cellulose exists through an inclusion complex or ionic bonding. This behavior can be seen in β-cyclodextrin formulation before and after incubation with HSA (red arrows), but not in the cellulose formulation before incubation with HSA. A change in the overall particle morphology or structure after incubation with HSA may occur, and thus more dark staining is present in the case of the cellulose nanoformulation. In the case of the dendrimer curcumin nanoformulation, an aligned network was seen throughout the sample due to the HSA layers (, blue arrows).
Figure 2 Human serum albumin binding alters the physicochemical properties of the curcumin nanoformulations, ie, PLGA, β-cyclodextrin, cellulose, nanogel, and dendrimecurcumin nanoparticles. (A) Transmission electron microscopic images of the curcumin nanoformulations (1 mg/mL) before and after incubation with human serum albumin 100 μg. Incubation with human serum albumin was performed for 2 hours, after which the nanoparticles were deposited on transmission electron microscopy grids, stained with uranyl acetate, and imaged under transmission electron microscopy. Red arrows represent uranyl acetate over stain. Blue arrows indicate aliened HSA networks. (B–C) Change in particle size and zeta potential of curcumin nanoformulations before and after incubation with human serum albumin. Particle size was measured for 3 minutes and the zeta potential was measured for 90 runs (9 minutes).
Note: Data are reported as the mean ± standard error of the mean for three repeats for each incubation.
Abbreviations: CD, β-cyclodextrin; CUR, curcumin; PLGA, polylactic-co-glycolic acid; HSA, human serum albumin.
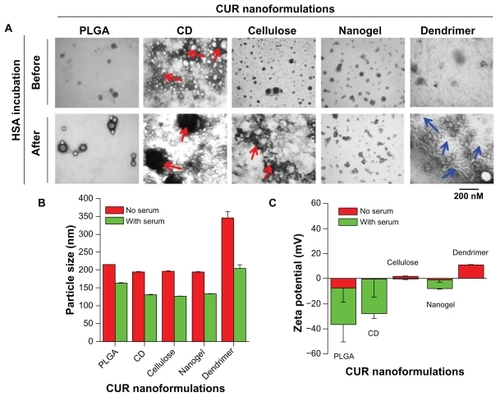
Since most of the HSA protein adsorption is loosely coated to all curcumin nanoparticle surfaces, their overall particle size aggregation decreased (). This indicates that the adsorbed HSA helps to reduce particle–particle interaction but does not change in the individual particle grain size, as seen in the transmission electron microscopy study. A similar phenomenon was observed when nanoparticles were incubated with HSA, bovine serum albumin, or Tween 80 dispersions.Citation53 However, we observed little change in the zeta potential of the curcumin nanoformulations after adsorption of HSA (). The zeta potential values for the curcumin PLGA, β-cyclodextrin, and nanogel formulations were −7.71, −0.41, and −1.46 mV, and the values for their HSA complexes further decreased to −28.86, −27.63, and −7.28 mV, respectively. HSA-adsorbed molecules carry a net negative charge in aqueous solution, resulting in a lowered negative zeta potential.Citation55 No significant change was observed in the zeta potential values of the cellulose and dendrimer curcumin nanoformulations after HSA complexation. This could be possible because a HSA corona was not formed, HSA was loosely bound on the surface of the nanoparticles, or because HSA polymer chains were dispersed throughout the nanoformulations.
Interaction of curcumin nanoparticles with human serum proteins
Chemotherapeutic drug nanoformulations intended for human applications need to be evaluated for their impact on the immune system.Citation56–Citation58 Some nanoparticles can escape the phagocytosis process but may have a tendency to bind to serum proteins which disturb native immunity. To avoid this problem, selection of nanoformulations must be based on minimal interaction with serum proteins. Low protein-binding nanoformulations may carry and deliver drug molecules to target site(s) for an extended period. On the other hand, higher protein binding may lead to aggregation of nanoformulations over time, that allows elimination from the body. In this study, we evaluated the adsorption of protein onto curcumin nanoparticles, which varies according to the type of plasma protein (). Fibrinogen and immunoglobulin G adsorption is very low, while transferrin and HSA adsorption is very high. The decreasing degree of protein adsorption to nanoparticles is HSA > transferrin > immunoglobulin G > fibrinogen. However, it is evident that dendrimer curcumin nanoparticles adsorbed more proteins compared with other types of curcumin nanoparticles. This suggests a higher degree of association/interaction of plasma proteins with the dendrimer curcumin nanoformulation which may not be appropriate for in vivo therapeutics. Densitometry quantification revealed that the order of adsorption of proteins to the different curcumin nanoformulations was dendrimer > β-cyclodextrin > cellulose > nanogel > PLGA (). A 3.0–3.5-fold increase in protein adsorption was found with dendrimer curcumin nanoparticles compared with PLGA curcumin nanoparticles. A similar pattern of binding was observed for all curcumin nanoformulations incubated with bovine serum albumin (data not shown).
Figure 3 Human serum proteins binding to curcumin nanoformulations (PLGA, β-cyclodextrin, cellulose, nanogel, and dendrimer). (A) Sodium dodecyl sulfate-polyacrylamide gel electrophoresis of curcumin nanoformulations (20 μM) incubated in 100 μg human plasma proteins (fibrinogen, immunoglobulin G, transferrin, and serum albumin). After 2 hours of incubation, bound or adsorbed proteins were separated, and sodium dodecyl sulfate-polyacrylamide gel electrophoresis was run at 150 V for 60 minutes and stained with Coomassie® G-250 stain. (B) Adsorbed protein bands were quantified by densitometry using AlphaEase Fc software. Data are reported as the mean of three repeats for each incubation (mean ± standard error of the mean, *P < 0.05, compared with PLGA curcumin nanoformulation). (C) Adsorption of curcumin nanoparticles on red blood cells using different concentrations of curcumin nanoformulations (10–40 μM).
Notes: Data are reported as the mean of three repeats for each adsorption (mean ± standard error of the mean deviation, *P < 0.05, compared with free curcumin or PLGA curcumin nanoformulation).
Abbreviations: CD, β-cyclodextrin; CUR, curcumin; PLGA, polylactic-co-glycolic acid; HSA, human serum albumin.
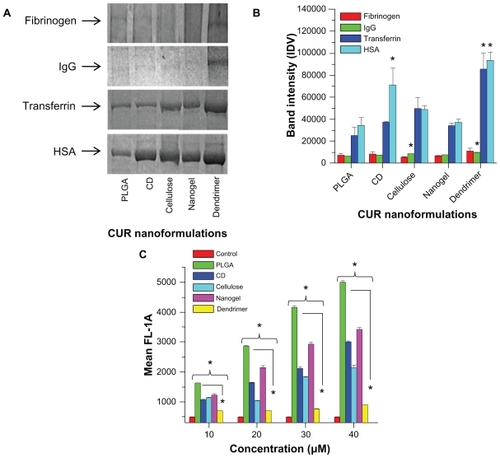
Chambers and Mitragotri reported that nanoparticles attached to the surface of red blood cells can dramatically improve their in vivo circulation characteristics.Citation59 Unlike particle-protein complexes, particles adhered on red blood cells or red blood cell mimic nanoparticles remain in the blood circulation unless there are large shear forces and cell–cell interactions. Therefore, we investigated the adhesion properties of curcumin particles in whole human blood. The adhesion process occurred after nanoparticles had interacted with plasma proteins in the blood. Adhesion of curcumin nanoparticles on red blood cells was determined by curcumin fluorescence using flow cytometry in channel 1 (488 excitation, blue laser, 530 ± 15 nm, fluorescein isothiocyanate/ green fluorescent protein, ). The order of adhered curcumin nanoparticles on red blood cells was found to be PLGA > nanogel > β-cyclodextrin > cellulose > dendrimer curcumin nanoformulations. The dendrimer exhibited very low adherence on red blood cells due to strong binding with plasma proteins, findings which are consistent with the above protein study.
From this study, our understanding is that both PLGA and nanogel curcumin nanoparticle interactions are minimal with both plasma proteins alone and in the presence of whole blood. Therefore, these formulations may be used for future in vivo and human clinical trials after detailed toxicological studies.
Hemolysis
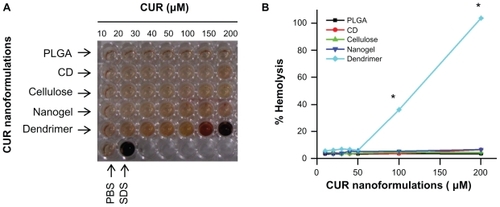
Hemolysis is a type of acute toxicity assay used to evaluate the hemocompatibility of the nanomaterials and to detect hemolyzation of erythrocytes.Citation60–Citation63 To assess the in vivo utility of a nanoformulation as a carrier for curcumin, the hemolytic potential in human blood needs to be tested.Citation64,Citation65 Therefore, we evaluated a direct nanoparticle-erythrocyte membrane interaction in which the extent of disruption of the erythrocyte membrane was a direct measure of nanoparticle toxicity (). The dark brown/red color () signifies toxicity to red blood cells at those concentrations. The mechanism of red blood cell disruption depends entirely on the physical and chemical nature of the nanoformulations. Red blood cells treated with PLGA, β-cyclodextrin, cellulose, and nanogel curcumin nanoformulations showed no signs of hemolysis at any concentration tested (). This behavior is very similar to that of the negative control treatment (). The dendrimer curcumin nanoformulation did not show signs of hemolysis up to 100 μM, but after that, extreme dose-dependent hemolytic activity was observed (). Hemolysis is a rapid process and is likely due to binding with the negatively charged red blood cell membrane and its disturbance. This formulation demonstrated hemolysis equivalent to that of the positive control, suggesting that serious toxicity would occur in vivo. The extensive hemolysis of the dendrimer curcumin formulation results from the positively charged amino surface groups which are highly likely to destabilize the cell membrane and cause cell lysis.Citation66 This type of lytic effect on red blood cells is extremely dangerous when administered in vivo, and therefore polyethylene glycol (PEG) conjugation or PEGylation of dendrimer formulations may be required to decrease this activity.Citation67
Figure 4 Hemolysis of curcumin nanoformulations incubated in red blood cells. (A) Optical image of supernatants from hemolysis study. Hemolysis was performed by incubating curcumin nanoformulations in red blood cells for two hours, centrifuging and collecting the supernatant for analysis. Phosphate-buffered solution and sodium dodecyl sulfate were considered as negative and positive controls in the experiment, respectively. (B) The extent of hemolysis was recorded by measuring optical density at λmax 570 nm using an ultraviolet-visible spectrophotometer. The graph was normalized with respect to the optical density of sodium dodecyl sulfate (positive control, 100% hemolysis).
Note: Data are presented as the mean of three repeats for each incubation (mean ± standard error of the mean, *P < 0.05, compared with the PLGA curcumin nanoformulation).
Abbreviations: CD, β-cyclodextrin; CUR, curcumin; PLGA, polylactic-co-glycolic acid; HSA, human serum albumin; PBS, phosphate-buffered solution; SDS, sodium dodecyl sulfate.
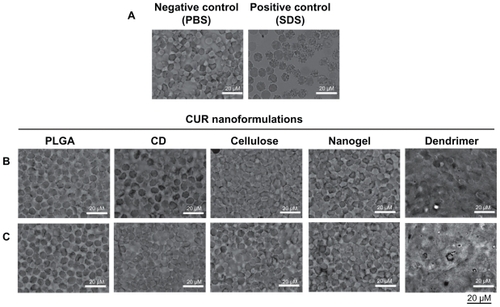
Red blood cells exposed to curcumin nanoformulations as well as negative and positive hemolysis-inducing agents were analyzed for morphological variations providing visual evidence of hemolysis (). Untreated red blood cells (negative control) have a normal biconcave shape with no visible surface abnormalities, whereas a positive hemolytic agent (sodium dodecyl sulfate) exposed to red blood cells clearly exhibits ghost cell morphology with numerous surface projections (). At low concentrations of all five curcumin nanoformulations (50 μM), exposure did not show aberrant morphological variations (), implying that hemolytic activity was not present. There were no morphological alterations observed, even in red blood cells treated with 100 μM of PLGA, β-cyclodextrin, cellulose, and nanogel curcumin nanoformulation, and no toxic effects were apparent (). However, a significant change in morphology and loss of biconcavity was observed when red blood cells were treated with the dendrimer curcumin nanoformulation, suggesting an increase in ghost cells and heavy hemolysis. This may be due to a higher rate of interaction or penetration of the dendrimer nanoparticles through the membranes of the red blood cells.Citation68,Citation69 This behavior is attributable to either direct interaction with enzymes or a change in ATPase activity by the dendrimer formulation.Citation70 This activity may also depend on the type of dendrimer generated.Citation71
Figure 5 Morphological variation of red blood cells incubated with curcumin nanoformulations for 2 hours. Phase contrast images of red blood cells incubated with (A) controls (phosphate-buffered solution and sodium dodecyl sulfate), (B) curcumin nanoformulations (50 μM), and (C) curcumin nanoformulations (100 μM). Bar equals 20 microns.
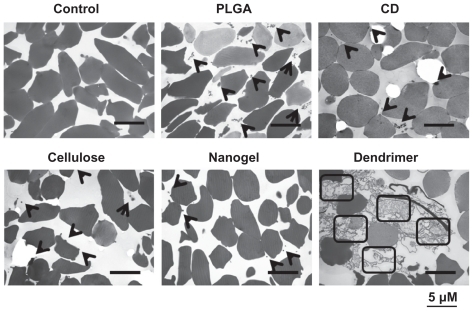
Nanoparticle interactions with erythrocytes cause abnormal membrane proteins and lipids that lead to the destruction of erythrocyte integrity and hemolysis. To investigate why some curcumin formulations are toxic to erythrocytes at 100 μM, binding of nanoparticles and red blood cells was investigated using transmission electron microscopy. Nanoparticles can usually be internalized within cancer cells as well as by many other primary cells. However, in this study, all the curcumin nanoformulations failed to internalize in erythrocytes but instead attached to the membranes of the red blood cells (). There were few PLGA, β-cyclodextrin, cellulose, or nanogel curcumin molecules on the membranes of erythrocytes (, black arrows) compared with controls (). A change in the morphology of the red blood cells from concave to spherical was observed, and secretion of some membrane proteins due to a higher rate of interaction with dendrimer curcumin nanoformulations (represented by squares) implies increased toxicity. This mainly occurs as a result of interaction between the dendrimer and the protein components of the plasma membrane, causing a change in the properties of the red blood cell membrane.Citation71
Figure 6 Attachment of curcumin nanoparticles to red blood cells. Transmission electron microscopic images of cross-sections of red blood cells after incubation with phosphate-buffered solution and curcumin nanoformulations (100 μM). Bar equals 5 microns.
Altogether, these results provide an interesting insight into the correlation between various curcumin nanoformulations and protein interactions that may impact in vivo efficacy. From the analysis of protein binding and hemolysis experiments, it can be concluded that all the curcumin nanoformulations behave differently than the dendrimer curcumin formulation. Our study clearly suggests that PLGA-based and nanogel-based curcumin formulations have lower protein binding, with no signs of hemolysis, demonstrating their suitability for therapeutic application. In vivo studies of PLGA and polyNIPAM nanogel curcumin formulations have shown improved bioavailability, and therefore are more effective systems for delivering curcumin into tumors.Citation12,Citation14,Citation16,Citation72,Citation73 These data clearly support the overall conclusion of this study. Our future research efforts will include evaluation of the in vivo pharmacokinetics of these curcumin nanoformulations to clarify the underlying mechanisms for potential cancer therapeutics.
Conclusion
This work underlines the importance of selection of efficient curcumin nanoformulations for biomedical applications, in particular for cancer therapeutics. The selection process is based on assessing the interaction or binding of nanoformulations with common human blood proteins and erythrocytes. Overall, the study results indicate that PLGA and nanogel curcumin nanoformulations have lower interaction(s) with proteins and red blood cells and may predict better efficiency for in vivo therapeutic targets compared with other curcumin nanoformulations. Further, we believe that antibody-guided therapies using these curcumin formulations may be very useful for targeted tumor therapy and could replace conventional chemotherapeutics in the future.
Acknowledgments
The authors thank Cathy Christopherson and Jenna Hultgren for editorial assistance, Amber Kruse and Hillary Newby for maintenance of the cell cultures, and Robert Japs for his help with transmission electron microscopy characterization. The authors are also grateful to Susan Puumala and Ashley Miller, Methodology and Data Analysis Center, for statistical analyses. This work was partially supported by grants from Department of Defense (PC073887, PC073643), Governor’s Cancer 2010, the National Institutes of Health Research Project Grant Program (RO1) (CA142736), and the Centers of Biomedical Research Excellence (P20 RR024219).
Disclosure
The authors report no conflicts of interest in this work.
References
- ClinicalTrialsgov [homepage on the Internet]Bethesda, MDNational Library of Medicine (US) Available from: http://clinicaltrials.gov/ct2/homeAccessed Nov 2, 2011
- HatcherHPlanalpRChoJTortiFMTortiSVCurcumin: from ancient medicine to current clinical trialsCell Mol Life Sci200865111631165218324353
- Bar-SelaGEpelbaumRSchafferMCurcumin as an anti-cancer agent: review of the gap between basic and clinical applicationsCurr Med Chem201017319019720214562
- MaheshwariRKSinghAKGaddipatiJSrimalRCMultiple biological activities of curcumin: a short reviewLife Sci200678182081208716413584
- AbuzeidWMDavisSTangALSensitization of head and neck cancer to cisplatin through the use of a novel curcumin analogArch Otolaryngol Head Neck Surg2011137549950721576562
- BavaSVPuliappadambaVTDeeptiANairAKarunagaranDAntoRJSensitization of taxol-induced apoptosis by curcumin involves down-regulation of nuclear factor-kappaB and the serine/threonine kinase Akt and is independent of tubulin polymerizationJ Biol Chem200528086301630815590651
- ChanMMFongDSopranoKJHolmesWFHeverlingHInhibition of growth and sensitization to cisplatin-mediated killing of ovarian cancer cells by polyphenolic chemopreventive agentsJ Cell Physiol20031941637012447990
- ChatterjeeSJPandeySChemo-resistant melanoma sensitized by tamoxifen to low dose curcumin treatment through induction of apoptosis and autophagyCancer Biol Ther201111221622821088500
- DeebDDJiangHGaoXDivineGDulchavskySAGautamSCChemosensitization of hormone-refractory prostate cancer cells by curcumin to TRAIL-induced apoptosisJ Exp Ther Oncol200552819116471035
- YallapuMMMaherDMSundramVBellMCJaggiMChauhanSCCurcumin induces chemo/radio-sensitization in ovarian cancer cells and curcumin nanoparticles inhibit ovarian cancer cell growthJ Ovarian Res201031120429876
- AnandPThomasSGKunnumakkaraABBiological activities of curcumin and its analogues (Congeners) made by man and Mother NatureBiochem Pharmacol200876111590161118775680
- BansalSSGoelMAqilFVadhanamMVGuptaRCAdvanced drug delivery systems of curcumin for cancer chemopreventionCancer Prev Res (Phila)2011481158117121546540
- BishtSMaitraASystemic delivery of curcumin: 21st century solutions for an ancient conundrumCurr Drug Discov Technol20096319219919496751
- AnandPKunnumakkaraABNewmanRAAggarwalBBBioavailability of curcumin: problems and promisesMol Pharm20074680781817999464
- KurienBTScofieldRHIncreasing aqueous solubility of curcumin for improving bioavailabilityTrends Pharmacol Sci200930733433519523694
- AnandPNairHBSungBDesign of curcumin-loaded PLGA nanoparticles formulation with enhanced cellular uptake, and increased bioactivity in vitro and superior bioavailability in vivoBiochem Pharmacol201079333033819735646
- AntonyBMerinaBIyerVSJudyNLennertzKJoyalSA Pilot Cross-Over Study to Evaluate Human Oral Bioavailability of BCM- 95CG (Biocurcumax), A Novel Bioenhanced Preparation of CurcuminIndian J Pharm Sci200870444544920046768
- BegumANJonesMRLimGPCurcumin structure-function, bioavailability, and efficacy in models of neuroinflammation and Alzheimer’s diseaseJ Pharmacol Exp Ther2008326119620818417733
- ShaikhJAnkolaDDBeniwalVSinghDKumarMNNanoparticle encapsulation improves oral bioavailability of curcumin by at least 9-fold when compared to curcumin administered with piperine as absorption enhancerEur J Pharm Sci2009373–422323019491009
- YanYDKimJAKwakMKYooBKYongCSChoiHGEnhanced oral bioavailability of curcumin via a solid lipid-based self-emulsifying drug delivery system using a spray-drying techniqueBiol Pharm Bull20113481179118621804203
- YallapuMMGuptaBKJaggiMChauhanSCFabrication of curcumin encapsulated PLGA nanoparticles for improved therapeutic effects in metastatic cancer cellsJ Colloid Interface Sci20103511192920627257
- YallapuMMJaggiMChauhanSCbeta-Cyclodextrin-curcumin self- assembly enhances curcumin delivery in prostate cancer cellsColloids Surf B Biointerfaces201079111312520456930
- GaoYLiZSunMPreparation and characterization of intravenously injectable curcumin nanosuspensionDrug Deliv201118213114220939679
- KakkarVSinghSSinglaDKaurIPExploring solid lipid nanoparticles to enhance the oral bioavailability of curcuminMol Nutr Food Res201155349550320938993
- XieXTaoQZouYPLGA Nanoparticles Improve the Oral Bioavailability of Curcumin in Rats: Characterizations and MechanismsJ Agric Food Chem201159179280928921797282
- YallapuMMDobberpuhlMRMaherDMJaggiMChauhanSCDesign of Curcumin loaded Cellulose Nanoparticles for Prostate CancerCurr drug metab2011 [Epub ahead of print]
- BriggerIDubernetCCouvreurPNanoparticles in cancer therapy and diagnosisAdv Drug Deliv Rev200254563165112204596
- LiangXJChenCZhaoYJiaLWangPCBiopharmaceutics and therapeutic potential of engineered nanomaterialsCurr Drug Metab20089869770918855608
- ZhangJWuLChanHKWatanabeWFormation, characterization, and fate of inhaled drug nanoparticlesAdv Drug Deliv Rev201063644145521118707
- BalaIHariharanSKumarMNPLGA nanoparticles in drug delivery: the state of the artCrit Rev Ther Drug Carrier Syst200421538742215719481
- DebbagePTargeted drugs and nanomedicine: present and futureCurr Pharm Des200915215317219149610
- PaudelKRRauniarGPBhattacharyaSKDasBPRecent advancement in drug delivery systemKathmandu Univ Med J (KUMJ)20086226226718769102
- ChoKWangXNieSChenZGShinDMTherapeutic nanoparticles for drug delivery in cancerClin Cancer Res20081451310131618316549
- ThierryBDrug nanocarriers and functional nanoparticles: applications in cancer therapyCurr Drug Deliv20096439140319534706
- XieYBagbyTRCohenMSForrestMLDrug delivery to the lymphatic system: importance in future cancer diagnosis and therapiesExpert Opin Drug Deliv20096878579219563270
- YallapuMMVasirJKJainTKVijayaraghavaluSLabhasetwarVSynthesis, Characterization and Antiproliferative Activity of Rapamycin- Loaded Poly (N-Isopropylacrylamide)-Based Nanogels in Vascular Smooth Muscle CellsJ Biomed Nanotechnol200841624
- PanyamJLabhasetwarVBiodegradable nanoparticles for drug and gene delivery to cells and tissueAdv Drug Deliv Rev200355332934712628320
- VasirJKLabhasetwarVBiodegradable nanoparticles for cytosolic delivery of therapeuticsAdv Drug Deliv Rev200759871872817683826
- ZhangJWuLChanHKWatanabeWFormation, characterization, and fate of inhaled drug nanoparticlesAdv Drug Deliv Rev201163644145521118707
- EmerichDFThanosCGTargeted nanoparticle-based drug delivery and diagnosisJ Drug Target200715316318317454354
- ShekunovBNanoparticle technology for drug delivery: from nanoparticles to cutting-edge delivery strategies – part IIIDrugs20058540240315883922
- SinghRLillardJWJrNanoparticle-based targeted drug deliveryExp Mol Pathol200986321522319186176
- MukerjeeAVishwanathaJKFormulation, characterization and evaluation of curcumin-loaded PLGA nanospheres for cancer therapyAnticancer Res200929103867387519846921
- PrajaktaDRatneshJChandanKCurcumin loaded pH-sensitive nanoparticles for the treatment of colon cancerJ Biomed Nanotechnol20095544545520201417
- SouKOyajobiBGoinsBPhillipsWTTsuchidaECharacterization and cytotoxicity of self-organized assemblies of curcumin and amphiphatic poly(ethylene glycol)J Biomed Nanotechnol20095220220820055098
- LynchICedervallTLundqvistMCabaleiro-LagoCLinseSDawsonKAThe nanoparticle-protein complex as a biological entity; a complex fluids and surface science challenge for the 21st centuryAdv Colloid Interface Sci2007134–135167174
- EhrenbergMSFriedmanAEFinkelsteinJNOberdorsterGMcGrathJLThe influence of protein adsorption on nanoparticle association with cultured endothelial cellsBiomaterials200930460361019012960
- HorieMNishioKFujitaKProtein adsorption of ultrafine metal oxide and its influence on cytotoxicity toward cultured cellsChem Res Toxicol200922354355319216582
- KastantinMMissirlisDBlackMAnanthanarayananBPetersDTirrellMThermodynamic and kinetic stability of DSPE-PEG(2000) micelles in the presence of bovine serum albuminJ Phys Chem B201011439126321264020828210
- LacerdaSHParkJJMeuseCInteraction of gold nanoparticles with common human blood proteinsACS Nano20104136537920020753
- MonopoliMPWalczykDCampbellAPhysical-chemical aspects of protein corona: relevance to in vitro and in vivo biological impacts of nanoparticlesJ Am Chem Soc201013382525253421288025
- YallapuMMFoySPJainTKLabhasetwarVPEG-functionalized magnetic nanoparticles for drug delivery and magnetic resonance imaging applicationsPharm Res201027112283229520845067
- BihariPVippolaMSchultesSOptimized dispersion of nanoparticles for biological in vitro and in vivo studiesPart Fibre Toxicol200851418990217
- ZhangXYinLTangMPuYOptimized method for preparation of TiO2 nanoparticles dispersion for biological studyJ Nanosci Nanotechnol20101085213521921125873
- RusoJMTaboadaPVarelaLMAttwoodDMosqueraVAdsorption of an amphiphilic penicillin onto human serum albumin: characterisation of the complexBiophys Chem2001921–214115311527586
- AlmeidaJPChenALFosterADrezekRIn vivo biodistribution of nanoparticlesNanomedicine (Lond)20116581583521793674
- JokerstJVLobovkinaTZareRNGambhirSSNanoparticle PEGylation for imaging and therapyNanomedicine (Lond)20116471572821718180
- SemeteBBooysenLLemmerYIn vivo evaluation of the biodistribution and safety of PLGA nanoparticles as drug delivery systemsNanomedicine20106566267120230912
- ChambersEMitragotriSLong circulating nanoparticles via adhesion on red blood cells: mechanism and extended circulationExp Biol Med (Maywood)2007232795896617609513
- CiochinaADBredeteanODimitriuDCIacobGConsiderations on in vitro and in vivo magnetic nanoparticles hemocompatibility testingRev Med Chir Soc Med Nat Iasi2009113127928521495331
- KoziaraJMOhJJAkersWSFerrarisSPMumperRJBlood compatibility of cetyl alcohol/polysorbate-based nanoparticlesPharm Res200522111821182816132346
- MayerAVadonMRinnerBNovakAWintersteigerRFrohlichEThe role of nanoparticle size in hemocompatibilityToxicology20092582–313914719428933
- ShelmaRSharmaCPDevelopment of lauroyl sulfated chitosan for enhancing hemocompatibility of chitosanColloids Surf B Biointerfaces201184256157021367586
- FischerHCChanWCNanotoxicity: the growing need for in vivo studyCurr Opin Biotechnol200718656557118160274
- GraingerDWNanotoxicity assessment: all small talk?Adv Drug Deliv Rev200961641942119386273
- FischerDLiYAhlemeyerBKrieglsteinJKisselTIn vitro cytotoxicity testing of polycations: influence of polymer structure on cell viability and hemolysisBiomaterials20032471121113112527253
- WangWXiongWZhuYXuHYangXProtective effect of PEGylation against poly(amidoamine) dendrimer-induced hemolysis of human red blood cellsJ Biomed Mater Res B Appl Biomater2010931596420186802
- SutherlandEDixonBSLeffertHLSkallyHZaccaroLSimonFRBiochemical localization of hepatic surface-membrane Na+,K+-ATPase activity depends on membrane lipid fluidityProc Natl Acad Sci U S A19888522867386772847169
- ChenCHZuklieBMRothLGElucidation of biphasic alterations on acetylcholinesterase (AChE) activity and membrane fluidity in the structure-functional effects of tetracaine on AChE-associated membrane vesiclesArch Biochem Biophys199835111351409500847
- KlajnertBSadowskaMBryszewskaMThe effect of polyamidoamine dendrimers on human erythrocyte membrane acetylcholinesterase activityBioelectrochemistry2004651232615522688
- CiolkowskiMRozanekMSzewczykMKlajnertBBryszewskaMThe influence of PAMAM-OH dendrimers on the activity of human erythrocytes ATPasesBiochim Biophys Acta20111808112714272321806962
- BishtSMizumaMFeldmannGSystemic administration of polymeric nanoparticle-encapsulated curcumin (NanoCurc) blocks tumor growth and metastases in preclinical models of pancreatic cancerMol Cancer Ther2010982255226420647339
- LimKJBishtSBarEEMaitraAEberhartCGA polymeric nanoparticle formulation of curcumin inhibits growth, clonogenicity and stem-like fraction in malignant brain tumorsCancer Biol Ther201111546447321193839