Abstract
Background
The success of gene therapy asks for the development of multifunctional vectors that could overcome various gene delivery barriers, such as the cell membrane, endosomal membrane, and nuclear membrane. Layer-by-layer technique is an efficient method with easy operation which can be used for the assembly of multifunctional gene carriers. This work describes a pH-sensitive multifunctional gene vector that offered long circulation property but avoided the inhibition of tumor cellular uptake of gene carriers associated with the use of polyethylene glycol.
Methods
Deoxyribonucleic acid (DNA) was firstly condensed with protamine into a cationic core which was used as assembly template. Then, additional layers of anionic DNA, cationic liposomes, and o-carboxymethyl-chitosan (CMCS) were alternately adsorbed onto the template via layer-by-layer technique and finally the multifunctional vector called CMCS-cationic liposome-coated DNA/protamine/DNA complexes (CLDPD) was constructed. For in vitro test, the cytotoxicity and transfection investigation was carried out on HepG2 cell line. For in vivo evaluation, CMCS-CLDPD was intratumorally injected into tumor-bearing mice and the tumor cells were isolated for fluorescence determination of transfection efficiency.
Results
CMCS-CLDPD had ellipsoidal shapes and showed “core-shell” structure which showed stabilization property in serum and effective protection of DNA from nuclease degradation. In vitro and in vivo transfection results demonstrated that CMCS-CLDPD had pH-sensitivity and the outermost layer of CMCS fell off in the tumor tissue, which could not only protect CMCS- CLDPD from serum interaction but also enhance gene transfection efficiency.
Conclusion
These results demonstrated that multifunctional CMCS-CLDPD had pH- sensitivity, which may provide a new approach for the antitumor gene delivery.
Introduction
Gene therapy has offered highly possible promises for treatment of cancers, as many potential therapeutic genes involved in regulation of molecular processes may be introduced by gene transfer, which can arrest angiogenesis, tumor growth, invasion, metastasis, and/or stimulate the immune response against tumors.Citation1 However, the initial enthusiasm for cancer gene therapy has been dampened by poor efficiency of the delivery systems.Citation2 To date, viral vectors are still the most efficient systems for gene transfer but the limitations associated with their use, in terms of risks of mutagenesis, safety, immunogenicity, and high production costs, have encouraged researchers to focus on alternative nonviral systems.Citation3,Citation4
In order to achieve efficient gene delivery into the nucleus of target cells, nonviral systems must overcome several barriers, such as circulation barriers, target tissue distribution, cellular endocytosis, endosomal escape, and nucleus entry. Therefore, various functional elements, such as ligands for specific targeting, fusogenic lipids or pH-sensitive polymers for endosomal escape, and nuclear localization signals for enhanced nuclear entry, are needed to be equipped into the nonviral gene delivery system to form a multifunctional gene vector.Citation5 However, it is difficult to integrate all these functional devices into a single system by simple mixing and to have each function exerted at the appropriate time and correct place.Citation6 To date, two approaches have been widely used to construct multifunctional vectors: chemical modification of block copolymers subsequently assembled with deoxyribonucleic acid (DNA) to form micelles and incorporation of functional devices into lipid-based gene vectors. In this study, the intention was to propose a new approach called layer-by-layer (LbL) self-assembly to construct multifunctional vectors.
LbL technique, first introduced at the beginning of the 1990s by Gero Decher, was used to assemble multilayered films with alternate deposition of polycations and polyanions. Citation7 Based on electrostatic or other intermolecular forces, it presents a new approach to the formation of supramolecular architectures. As DNA is anionic charged and can be self-assembled with cationic polymers, it can be incorporated into multilayered films by LbL technique and these DNA-loaded multilayered films have been used as nonviral gene delivery system.Citation8,Citation9 LbL assembly offers numerous advantages. Firstly, it is entirely aqueous and scarcely involves organic solvents, which is beneficial to maintain the activity of DNA.Citation10 Secondly, it can control DNA loading by controlling film thickness or the number of layers of DNA deposited during fabrication.Citation11 Thirdly, application of functional polyelectrolytes can assign multilayered films with different functions including cell targeting, controlled release of DNA in physiological condition, enhanced membrane penetration, and intracellular targeting.Citation12,Citation13 With the rapid development of LbL technique, gene delivery systems constructed via LbL technique have been extended from planar films to three-dimensional “core-shell” nanoparticles. For example, Trubetskoy et al demonstrated that the recharging of poly-ethylenimine (PEI)/DNA polyplexes using poly(acrylic acid) could be used to increase levels of cell transfection in vitro in the presence of serum and increase levels of gene expression in the lung in vivo.Citation14 Saul et al constructed PEI-DNA- poly(acrylic acid)-PEI quaternary complexes and the increased association of PEI achieved by the multilayer approach lead to substantial increases in expression of transgene for reporter plasmids without the need for excess free polymer typically required for nonviral gene delivery.Citation15 With programmed deposition of required functional polyanions and polycations, LbL assembly has the potential to achieve highly sophisticated programmable control of gene delivery. Therefore, it can be concluded that a multifunctional nanovector could be designed with programmed assembly of functional polyanions and polycations to help overcome biological barriers for gene transfer. Compared with the widely used chemical modification method to fabricate multifunctional gene vectors, LbL is an easy method without complicated chemical modification which avoids losing original functionality of polymers or lipids.Citation16 As multilayers built by the LbL method afford a more stable coating than those prepared by lipid incorporation because of the strong electrostatic attractions between layer to layer and layer to substrate, LbL also solves the problem of randomness and poor reproducibility associated with lipidic incorporation of functional ligands and LbL offers a degree of stable modification to functional ligands.Citation17
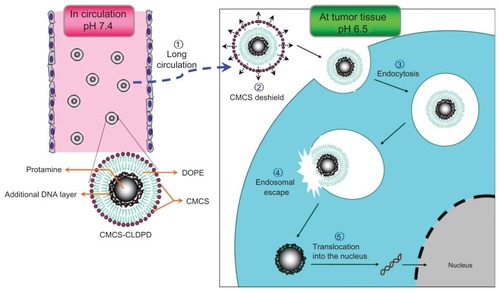
In this study, after choosing the required functional components to overcome the barriers of gene delivery, such as pH-sensitive polymer for long circulation time, fusogenic lipids for endosomal escape, and a nuclear localization signal for enhanced nuclear delivery, a new multifunctional nanovector was constructed via LbL technique. In order to have each function exerted at the appropriate time and correct place, programmed assembly of these functional materials was indispensable. Firstly, protamine (Pro), which showed improvement in both nuclear targeting and intranuclear transcription,Citation18 was used to condense DNA into a compact core as the template for the following assembly. Then, an additional layer of DNA was deposited onto the template to form DNA/Pro/DNA complexes (DPD). This additional DNA layer could increase DNA loading and also reverse the cationic surface charge of Pro/DNA for further assembly. After that, a cationic lipid layer of cationic liposomes for endosomal escape, which were prepared with cationic lipid 6-lauroxyhexyl lysinate (LHLN) previously synthesized by the authors’ group and 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine (DOPE), was assembled onto the surface of anionic DPD, and thus cationic liposome-coated DPD (CLDPD) were obtained. As polysaccharide coating is the most common biomimic strategy, a derivate of chitosan – anionic o-carboxymethyl-chitosan (CMCS) – was chosen as the outermost layer which would be assembled onto CLDPD to form multifunctional CMCS-CLDPD. The CMCS coating offered long circulation property but avoided the inhibition of tumor cellular uptake of gene carriers associated with the use of hydrophilic polymers (ie, polyethylene glycol). It was interesting that the outermost CMCS layer had pH-sensitivity, which possessed cationic charges under pH 6.5 and was anionic charged above pH 7.0. Therefore, in blood circulation, CMCS was anionic charged and adsorbed on the surface of CLDPD, which could protect the cationic CLDPD against serum interaction and offered long circulation capability. When arriving at the acidic environment of tumor tissue, CMCS was cationic charged, repelled with CLDPD, and fell off. The exposed cationic CLDPD combined with negatively-charged cell membrane was internalized via endocytosis. With the help of the fusogenic lipid DOPE, CLDPD could escape from endosomes and release the DPD in cytoserum. The loaded DNA was then translocated into nucleus under the guidance of the nuclear targeting polypeptide Pro. Therefore, a high efficient multifunctional gene vector which offered long circulation, efficient cellular membrane transport, endosomal escape, high DNA loading, and nuclear targeting was fabricated for tumor gene delivery. This delivery process of CMCS-CLDPD is illustrated in . The physicochemical property and in vitro and in vivo transfection efficiency of CMCS-CLDPD were tested in this study. In vitro and in vivo studies showed that the outermost CMCS layer could fall off in the acidic environment of tumor tissue and expose CLDPD, which enhances gene transfection efficiency. Results indicated that the multifunctional CMCS-CLDPD might provide a new approach for antitumor gene delivery.
Figure 1 Schematic representation of multifunctional o-carboxymethyl-chitosan cationic liposome-coated deoxyribonucleic acid/protamine/deoxyribonucleic acid complexes delivery process.
Abbreviations: CMCS, o-carboxymethyl-chitosan; CLDPD, cationic liposome-coated deoxyribonucleic acid/protamine/deoxyribonucleic acid complexes; DNA, deoxyribonucleic acid; DOPE, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine.
Materials and methods
Materials
The pEGFP-N1 vector was provided by Zhejiang University (Hangzhou, China). PicoGreen® reagent was purchased from Invitrogen Life Technologies (Carlsbad, CA). Pro was obtained from Sigma-Aldrich Corporation (St Louis, MO). DOPE was obtained from Avanti Polar Lipids, Inc, (Alabaster, AL). CMCS (Mr 50000, carboxymethyl degree of substitution 60%) was purchased from Jinan Haidebei Marine Bioengineering Co, Ltd, (Jinan, China). Agarose was purchased from Biowest (Nuaille, France). Goldview was obtained from Beijing Saibaisheng Biological Engineering Co, Ltd, (Beijing, China). MTT (3-[4,5-dimethyl-2- thiazolyl]-2,5-diphenyl-2H- tetrazolium bromide) was purchased from Sigma-Aldrich. Lipofectamine™ 2000 was obtained from Invitrogen. Human liver carcinoma (HepG2) and mouse melanoma (B16) cells were obtained from the American Type Culture Collection (Manassas, VA). DNase I enzyme and buffer were obtained from Beijing Yinfeng Century Scientific Develop Co, Ltd, (Beijing, China). Heparin was delivered by the Institute of Biochemical and Biotechnological Drug of Shandong University (Jinan, China). Deionized water was used throughout the experiment. All other chemicals and reagents used were of analytical purity grade or higher, obtained commercially.
Animals
Kunming mice weighing 18–22 g were supplied by the Medical Animal Test Center of the New Drugs Evaluation Center, Shandong University (Jinan, China). All animal experiments complied with the requirements of the National Act on the Use of Experimental Animals (China).
Preparation of CMCS-CLDPD
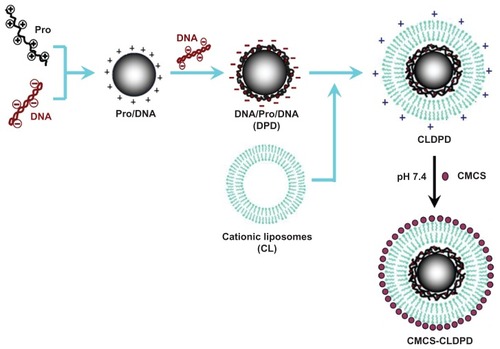
Initially, 100 μL of 50 μg/mL aqueous solution of plasmid DNA was added dropwise to 100 μL of 100 μg/mL Pro solutions under gentle vortexing for 20 seconds. The sample was incubated at room temperature for 30 minutes to facilitate the formation of Pro/DNA complexes.Citation19 Then, an isovolumic solution of DNA was added dropwise to Pro/DNA and the sample was incubated at room temperature for 30 minutes to facilitate the formation of DPD. After that, DPD were stored at 4°C until use. LHLN was synthesized using the method described previously.Citation20 LHLN was dissolved in dehydrated alcohol and mixed with a helper lipid DOPE (1:1 molar ratio). The mixture was evaporated to dryness in a round-bottomed flask using a rotary evaporator at room temperature. The resulting lipid film was dried by nitrogen for an additional 10 minutes to evaporate any dehydrated alcohol. After that, DPD was added to the round-bottomed flask. The flask was then gently vibrated to facilitate hydration of the lipid film and sonicated using a Sonic Dismembrator (Model 500; Thermo Fisher Scientific, Pittsburgh, PA) for 5 minutes at room temperature to form homogenized CLDPD. The obtained CLDPD suspension was put into an Eppendorf tube (Eppendorf AG, Hamburg, Germany). Under weak vortex conditions, CMCS was added dropwise and incubated for 30 minutes at room temperature. Finally, CMCS-CLDPD was obtained. The preparation process of CMCS-CLDPD is illustrated in .
Figure 2 Schematic representation of the assembly process of multifunctional o-carboxymethyl-chitosan cationic liposome-coated deoxyribonucleic acid/protamine/deoxyribonucleic acid complexes.
Abbreviations: CMCS, o-carboxymethyl-chitosan; CLDPD, cationic liposome-coated deoxyribonucleic acid/protamine/deoxyribonucleic acid complexes; DNA, deoxyribonucleic acid; Pro, protamine.
Characterization of DPD, CLDPD, and CMCS-CLDPD
The surface morphology of DPD, CLDPD, and CMCS-CLDPD was examined by transmission electronic microscopy (JEM-1200EX; JEOL Ltd, Tokyo, Japan). Samples were prepared respectively by placing a drop of three kinds of nanoparticle suspensions onto a copper grid and air drying, followed by negative staining with one drop of 3% aqueous solution of sodium phosphotungstate (Sigma-Aldrich) for contrast enhancement. The air-dried samples were then directly examined under the transmission electron microscope. Mean particle size and zeta potential of the three kinds of nanoparticle suspensions were analyzed by photon correlation spectroscopy with a Zetasizer® 3000 (Malvern Instruments, Malvern, UK). All measurements were carried out in triplicate. The average particle size was expressed as volume mean diameter and the reported value was represented as mean ± standard deviation (n = 3).
pH sensitivity analysis of CMCS-CLDPD
In previous studies focused on drug delivery, the determination of drugs released from carriers at different pH values was often used to evaluate the pH-sensitivity of carriers.Citation21 It was expected that the outermost layer of CMCS would fall off from the surface of CMCS-CLDPD in the acidic environment of tumors. Therefore, in order to evaluate the pH sensitivity of CMCS-CLDPD, the in vitro release of DNA from CMCS-CLDPD in 50 mM phosphate buffer solution (PBS) with different pH values (pH 5.5, 6.0, 6.5, 7.0, 7.4) was determined. Typically, an aliquot of CMCS-CLDPD (equivalent to 2.5 μg DNA) was suspended in 1 mL PBS at pH 5.5 in Eppendorf tubes in a 37°C shaking water bath at 100 rpm. Separate tubes were used for each time point. At predetermined time intervals, the CMCS-CLDPD suspensions were centrifuged (15,000 rpm for 30 minutes) and the amount of DNA released in the supernatant was analyzed using fluorospectrophotometry with PicoGreen™ double-stranded DNA quantitative reagent.Citation22 Background readings were obtained using the supernatants from PBS at pH 5.5. Fluorescence was measured by fluorescence spectrophotometer (model 850; Hitachi High-Technologies, Tokyo, Japan) at excitation and emission wavelengths of 480 nm and 520 nm, respectively. The amount of DNA was calculated according to the linear calibration curve of DNA (F = 0.0850C + 0.1132, R = 0.9996). The in vitro release of DNA at other values (pH 6.0, 6.5, 7.0, 7.4) was respectively performed with the same procedure. Also, zeta potential of CMCS-CLDPD after incubation in PBS with different pH values (pH 5.5, 6.0, 6.5, 7.0, 7.4) was analyzed by photon correlation spectroscopy.
DNase I protection assay
To test whether CMCS-CLDPD could protect loaded DNA from nuclease digestion, DNase I-mediated digestion was evaluated using agarose gel electrophoresis. In brief, 20 μL of CMCS-CLDPD containing 1 μg of DNA was incubated with DNase I (0.2 U/μg DNA) in DNase I buffer. The suspensions were incubated in a shaking water bath at 37°C and 100 rpm for 1, 2, and 4 hours, respectively. Naked DNA (1 μg) treated with DNase I at 0.2 U/μg DNA in DNase I buffer were incubated in a shaking water bath at 37°C and 100 rpm for 30 minutes. The enzymatic digestion reaction was terminated with ethylenediaminetetraacetic acid solution (0.5 M, pH 8.0). To separate DNA from CMCS-CLDPD, TE buffer (10 mM tris(hydroxymethyl)aminomethane-hydrochloride, 1 mM ethylenediaminetetraacetic acid, pH 7.4) containing 1% heparin and 1% Triton X-100 (Sigma-Aldrich) was added to the suspensions. The suspensions were then placed in a 37°C shaking water bath at 100 rpm for 2 hours. After that, the suspension was analyzed by gel electrophoresis as described above. Untreated DNA was applied to the gel as a control.
Stability study in the presence of serum
To estimate the stability of CMCS-CLDPD, the turbidity change of CMCS-CLDPD suspension in the serum was evaluated in a previous report.Citation23 A freshly prepared CMCS-CLDPD solution (250 μL) was added to 1 mL of serum at pH 7.4 and the mixed solution was incubated at 37°C with mild stirring. The turbidity change of the mixed solution was measured using an ultraviolet-visible spectrophotometer (UV-2102PC; UNICO, Dayton, NJ) at 450 nm.
Cells culture
HepG2 and B16 cells were cultured in Roswell Park Memorial Institute (RPMI) 1640 medium supplemented with 10% fetal bovine serum, streptomycin at 100 μg/mL, and penicillin at 100 U/mL. All cells were cultured in a 37°C incubator with 5% carbon dioxide.
In vitro cell viability
Cell viability studies were investigated by MTT assay using HepG2 cells as model cells.Citation24 HepG2 cells were seeded in Corning® Costar® 96-well plates (Sigma-Aldrich) at a density of 8 × 103 viable cells per well in 0.2 mL RPMI 1640 supplemented with 10% fetal bovine serum and antibiotics. After incubation in a 5% carbon dioxide incubator at 37°C for 24 hours, the culture medium was replaced by 200 μL fresh RPMI 1640 containing different concentrations of CMCS-CLDPD (0.1–0.5 mg/mL) and the corresponding CLDPD. The cells were then incubated for another 24 hours. With the same DNA dose, PEI/DNA polyplexes (N/P ratio 10.0) – widely applied in gene delivery – were used as positive control.Citation25 Cell viability was then assessed by MTT assay: 5 mg/mL of MTT in PBS was added to each well and the plate was incubated for an additional 4 hours at 37°C in a 5% carbon dioxide incubator. Then the MTT-containing medium was removed and the crystals formed by living cells were dissolved in 100 μL dimethyl sulfoxide. Absorbance at 570 nm was determined by a microplate reader (Model 680; Bio-Rad Laboratories, Hercules, CA). Untreated cells were taken as a control with 100% viability and cells without addition of MTT were used as a blank to calibrate the spectrophotometer to zero absorbance. Relative cell viability (%) compared to control cells was calculated by (Abssample/Abscontrol) × 100. All experiments were repeated in triplicate.
In vitro transfection studies
The transfection activity of CMCS-CLDPD was evaluated using plasmid DNA, encoding enhanced green fluorescence protein (EGFP) as reporter gene in the transfection studies. HepG2 cells were seeded in six-well plates at a density of 3 × 105 cells per well and incubated overnight at 37°C in 2 mL of complete RPMI 1640 culture medium. At a confluence level of 70%–80%, the growth medium was removed and the cells were washed twice with PBS buffer. The medium was replaced with serum free medium (pH 7.4), serum free medium (pH 6.5), and 20% serum medium (pH 7.4), respectively. CMCS-CLDPD containing 5 μg of DNA was then added. All transfections were carried out in three wells each. Naked DNA and PEI/DNA (N/P 10.0) were used as negative control and positive control, respectively. The cells were incubated with the test complexes for 4 hours at 37°C before replacing with fresh medium for 48 hours. Fluorescence in cells was observed using an inverted fluorescence microscope (excitation: 540 nm and emission: 580 nm; BX40; Olympus Corporation, Tokyo, Japan). The fluorescent images were obtained using a DP70 digital imaging system (Olympus) analyzed by Olysia BioReport imaging software (v 3.2; Olympus). After that, all cells were harvested for trypsinization and washed in PBS three times, and cell-associated fluorescence was quantitatively determined by a FACSCalibur™ flow cytometer (BD Biosciences, San Jose, CA) by counting 10,000 events. Only the viable cells were gated for fluorescence analysis.
In vivo transfection studies
Each Kunming mouse was inoculated subcutaneously in the subaxillary region with 8 × 106 B16 cells. The tumor growth was monitored by measuring tumor length (L) and width (W) with a caliper every 2 days. The tumor volume (V) was calculated as V = 1/2 × L2 × W. When the tumor reached 300–400 mm3, mice were randomly assigned to four groups (six mice in each group). CMCS-CLDPD and CLDPD (100 μL, containing 10 μg pEGFP) were intratumorally injected four times with a 12-hour interval. Naked DNA was used as a negative control and PEI/DNA (N/P 10.0) was used as a positive control, both of which were intratumorally injected with the same dosage of pEGFP. The mice were sacrificed 72 hours or 96 hours after the first injection and the tumor tissue samples were taken out. The tumor tissues were homogenized by pressing the samples through a 30 μm cell mesh with the plunger of a 10 mL syringe; erythrocyte lysis buffer was added during homogenization to lyse the red blood cells. The homogenates were washed three times with PBS containing 0.5% bovine serum albumin and then filtered (30 μm CellTrics®; Partec GmbH, Munster, Germany). The cells were finally obtained after centrifugation (4°C, 1000 rpm, 5 minutes) and were seeded into 24-well plates in 1 mL of Dulbecco’s Modified Eagle’s Medium with 10% fetal bovine serum. The fluorescent cells were observed using an inversion fluorescence microscope and pictures taken. The pictures were analyzed with Image-Pro Plus software (v 7.0; Media Cybernatics Inc, Silver Spring, MD) to calculate total fluorescence intensity of tumor cells.
Statistical analysis
All studies were repeated a minimum of three times and measured at least in triplicate. Results were reported as mean ± standard deviation. Statistical significance was analyzed using Student’s t-test. Differences between experimental groups were considered significant when P < 0.05.
Results
Optimization of CMCS-CLDPD
The influence of DNA on the formation of DPD
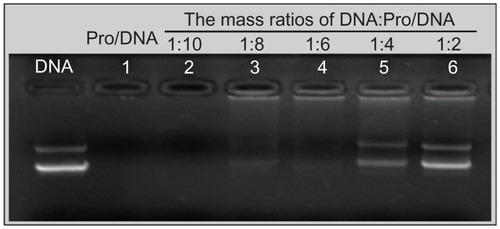
Pro/DNA complexes were prepared at N/P mass ratio of 2.5. Gel retarding analysis was used to optimize the appropriate mass ratio (DNA: Pro/DNA ternary complexes) required for the formulation of DPD. Once DNA was deposited onto the surface of Pro/DNA complexes, the mobility of DNA was retarded and it remained in the hole of the agarose gel. Fluorescence could be seen in the holes from Lane 3 to Lane 6 (). The part of DNA which did not bind onto the surface of Pro/DNA complexes migrated to the positive electrode in the same manner with the control DNA. As illustrated in , an additional DNA layer was successfully deposited onto the surface of Pro/DNA when the mass ratio was above 1:8. However, too high mass ratio might cause too much free DNA in the suspension, which was unfavorable to the deposition of additional cationic LHLN-liposomes layer.
Mass ratios of LHLN-liposomes to DPD for preparation of CLDPD
As the mass ratios of LHLN-liposomes to DPD increased, the particle size of CLDPD increased (). Zeta potential of DPD was −19.3 ± 2.8 mV (). However, when the mass ratio increased to 10:1, zeta potential reversed to positive charges, demonstrating that the cationic LHLN-liposomes were successfully adsorbed onto the surface of DPD. When the mass ratio was higher than 50:1, zeta potential of CLDPD remained unchanged, demonstrating that the surface of DPD had been saturated with cationic LHLN-liposomes. The transmission electron microscopic images of CLDPD prepared with different mass ratios are illustrated in . It was found that CLDPD prepared at mass ratio 25:1 were particles without spherical morphology, which showed that they were partially encapsulated with LHLN-liposomes. When the mass ratio was 50:1, CLDPD showed spherical shapes with a fairly narrow size distribution. However, when the mass ratio increased to 75:1, CLDPD severely aggregated and the particle size was 469.3 ± 6.3 nm, demonstrating that this mass ratio was too high for the assembly of CLDPD. Therefore, the mass ratio of 50:1, which offered high surface charge for CLDPD, was finally chosen for further assembly.
Table 1 Particle size and zeta potential of cationic liposome-coated deoxyribonucleic acid/protamine/deoxyribonucleic acid complexes prepared with different mass ratios
Mass ratios of CMCS to CLDPD for preparation of CMCS-CLDPD
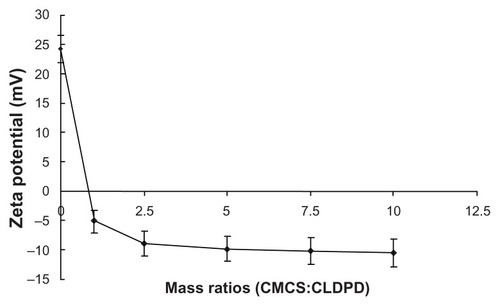
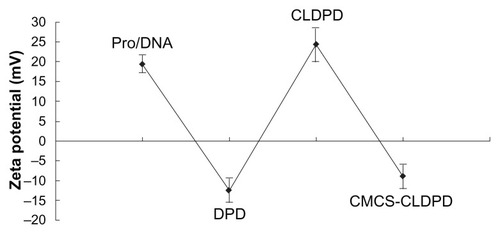
Zeta potential reversed when the mass ratio of CMCS to CLDPD was 1:1, demonstrating the successful deposition of CMCS on the surface of CLDPD (). When the mass ratio was higher than 2.5:1, zeta potential of CMCS-CLDPD remained almost the same and it could be concluded that CMCS had already saturated the surface of CLDPD. In order to avoid the existence of excess CMCS which might influence the biological properties of gene vectors, the mass ratio of 2.5:1 was chosen as the optimization of CMCS-CLDPD.
Figure 5 Zeta potential of o-carboxymethyl-chitosan cationic liposome-coated deoxyribonucleic acid/protamine/deoxyribonucleic acid complexes with different mass ratios.
Abbreviations: CMCS, o-carboxymethyl-chitosan; CLDPD, cationic liposome-coated deoxyribonucleic acid/protamine/deoxyribonucleic acid complexes.
Physicochemical properties of CMCS-CLDPD
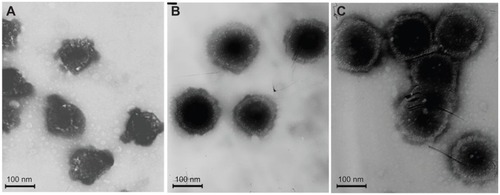
CMCS-CLDPD were successfully prepared via LbL technique and characterized in terms of morphology, particle size, and surface charge. As illustrated in , Pro/DNA complex, DPD, CLDPD, and CMCS-CLDPD all showed spherical or ellipsoidal shapes. With the sequential deposition of DNA, LHLN-liposomes, and CMCS layer, a layered structure was found on the surface of the corresponding DPD, CLDPD, and CMCS-CLDPD, which demonstrated the formation of CMCS-CLDPD with multilayered structure. The physicochemical characteristics of different nanoparticles are summarized in . As the assembly process proceeded, the particle size increased from 100.2 ± 3.4 nm of Pro/DNA to 210.9 ± 6.8 nm of CMCS-CLDPD. All the particles exhibited narrow size distributions (polydispersity index <0.3). As shown in , when Pro/DNA complex was coated with a layer of DNA, zeta potential reversed to −12.5 ± 3.2 mV and switched back to positive when further coated by another layer of LHLN-liposomes. After the deposition of the last layer of CMCS, the zeta potential reversed from positive to negative values again and CMCS-CLDPD achieved a zeta potential of −8.9 ± 2.7 mV.
Table 2 The particle size and zeta potential of different nanoparticles
Figure 6 Transmission electron microscopic imaging of different nanoparticles: (A) Protamine (Pro)/deoxyribonucleic acid (DNA) complex, (B) DNA/Pro/DNA complexes, (C) Cationic liposome-coated DNA/Pro/DNA complexes, and (D) o-carboxymethyl-chitosan cationic liposome-coated DNA/Pro/DNA complexes.
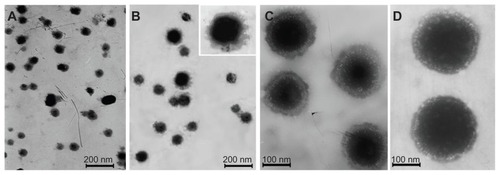
Figure 7 The changes of zeta potential during assembly process of o-carboxymethyl-chitosan cationic liposome-coated deoxyribonucleic acid/protamine/deoxyribonucleic acid complexes.
Abbreviations: CMCS, o-carboxymethyl-chitosan; CLDPD, cationic liposome-coated deoxyribonucleic acid/protamine/deoxyribonucleic acid complexes; DNA, deoxyribonucleic acid; Pro, protamine.
pH sensitivity analysis of CMCS-CLDPD
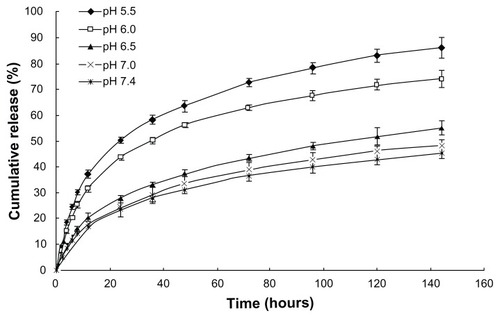
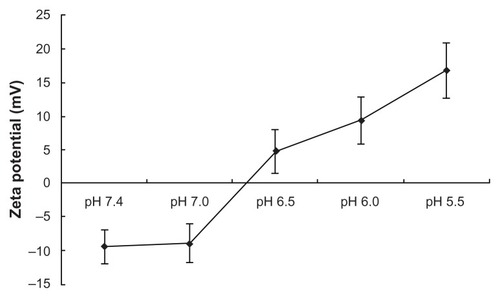
At the same time point (t > 12 hours), the cumulative release of DNA in PBS with different pH values followed the inequality: pH 5.5 > pH 6.0 > pH 6.5 > (pH 7.0, pH 7.4) (P < 0.05; ). It could be concluded that the loaded DNA was released from CMCS-CLDPD more quickly in acid medium than in neutral medium. In order to demonstrate whether this pH sensitivity release was caused by the disassociation of the outermost layer of CMCS, zeta potential of CMCS-CLDPD after incubation in PBS with different pH values was determined. As illustrated in , zeta potential of CMCS-CLDPD reversed from negative to positive values when the pH of medium was below 6.5, indicating that CMCS could fall off from the surface of CMCS-CLDPD at pH below 6.5. Therefore, CMCS-CLDPD showed pH sensitivity and the outermost layer of CMCS could fall off in response to acid medium.
Stability of CMCS-CLDPD against DNase I
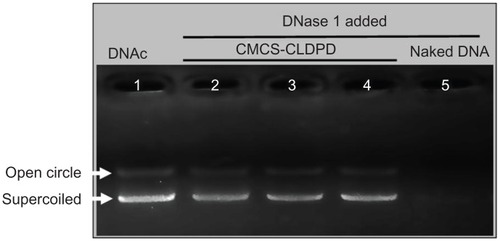
For effective gene expression, DNA in the gene vehicle should be protected from degradation by enzymes.Citation26 To test whether CMCS-CLDPD could protect the loaded plasmid DNA from digestion by nucleases, they were exposed to DNase I at various time intervals. As illustrated in , naked plasmid DNA (lane 5) was completely digested within 30 minutes of incubation with the DNase I (0.2 U/μg DNA). However, after 2 hours of degradation, DNA in CMCS-CLDPD still showed a relatively complete structure as indicated by the presence of supercoiled DNA. Results showed that CMCS-CLDPD could protect DNA against degradation by DNase I, which is one of the crucial factors for efficient gene delivery in vitro as well as in vivo.
Figure 10 Agarose gel electrophoresis analysis of o-carboxymethyl-chitosan cationic liposome-coated deoxyribonucleic acid/protamine/deoxyribonucleic acid complexes after incubation with DNase I. Lane 1: deoxyribonucleic acid control; Lane 2–4: o-carboxymethyl-chitosan cationic liposome-coated deoxyribonucleic acid/protamine/deoxyribonucleic acid complexes incubated with DNase I for 0.5, 1, and 2 hours, respectively; Lane 5: naked deoxyribonucleic acid incubated with DNase I for 0.5 hours.
Abbreviations: CMCS, o-carboxymethyl-chitosan; CLDPD, cationic liposome-coated deoxyribonucleic acid/protamine/deoxyribonucleic acid complexes; DNA, deoxyribonucleic acid; DNAc, deoxyribonucleic acid control.
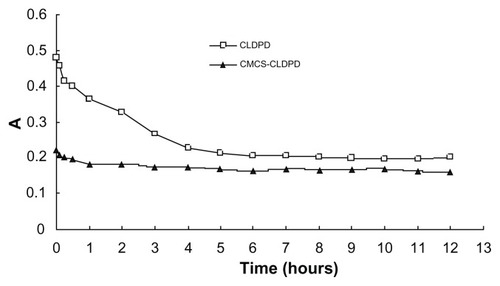
Serum stability of CMCS-CLDPD
Serum stability of CMCS-CLDPD was evaluated by determining the turbidity changes of CMCS-CLDPD after incubation with serum. CLDPD was used as control to estimate the effect of CMCS on preventing aggregation of CLDPD induced by serum. shows the turbidity changes of CLDPD and CMCS-CLDPD after incubation with serum. CLDPD with high positive charges started to aggregate once mixed with serum. As time increased, large aggregates began to precipitate, resulting in turbidity decrease. However, particle aggregation induced by serum protein was completely prevented after the coating of CMCS layer, and the turbidity of CMCS-CLDPD remained almost the same as time went by. A possible explanation could be that high anionic surface charge of CMCS prevented serum protein adsorption onto CMCS-CLDPD. Therefore, CMCS-CLDPD had high serum stability and might be suitable for in vivo gene delivery.
Cytotoxicity of CMCS-CLDPD
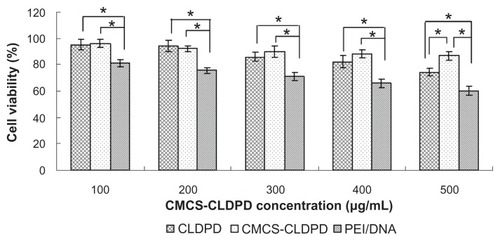
The in vitro cytotoxicity of CMCS-CLDPD at various concentrations (100, 200, 300, 400, and 500 μg/mL) was evaluated in HepG2 cells by MTT assay. As shown in , the cell viabilities of HepG2 cells in the presence of CMCS-CLDPD over the studied concentration range (100–500 μg/mL) and their corresponding CLDPD were all significantly higher than that of PEI/DNA (P < 0.05) with the same DNA dose, demonstrating that they exhibit lower cytotoxicity than PEI/DNA. CMCS-CLDPD with 400 μg/mL concentration and the corresponding CLDPD were used for in vitro transfection test. At an even higher concentration of 500 μg/mL, CMCS-CLDPD showed higher cell viability than the corresponding CLDPD (P < 0.05).
Figure 12 Cell viabilities of o-carboxymethyl-chitosan cationic liposome-coated deoxyribonucleic acid/protamine/deoxyribonucleic acid complexes with different concentrations and the corresponding cationic liposome-coated deoxyribonucleic acid/protamine/deoxyribonucleic acid complexes in HepG2 cells (mean ± standard deviation, n = 3). Polyethylenimine/deoxyribonucleic acid (N/P ratio 10.0) prepared with the same deoxyribonucleic acid dose was used as positive control.
Note: *P < 0.05.
Abbreviations: CMCS, o-carboxymethyl-chitosan; CLDPD, cationic liposome-coated deoxyribonucleic acid/protamine/deoxyribonucleic acid complexes; DNA, deoxyribonucleic acid; PEI, polyethylenimine.
In vitro transfection results of CMCS-CLDPD
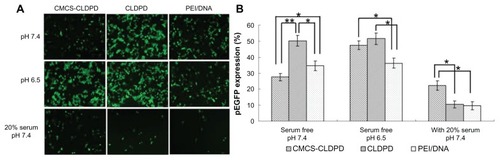
Pictures from inversion fluorescence microscope presenting the green fluorescence in transfected HepG2 cells are shown in . It was found that CMCS-CLDPD, CLDPD, and PEI/DNA could successfully transfer HepG2 cells in 48 hours. The gene transfection efficiency was further quantitatively determined by flow cytometry (). When the pH value of the transfection medium without serum was 7.4, the transfection efficiency of different vectors followed the relationship: CLDPD > PEI/DNA > CMCS-CLDPD (P < 0.05). However, when the pH value of the transfection medium without serum was 6.5, both CMCS-CLDPD and CLDPD showed higher transfection efficiency than PEI/DNA (P < 0.05) and CMCS-CLDPD obtained almost the same value as CLDPD (P > 0.05). It could be concluded that the outermost anionic layer of CMCS might fall off at pH 6.5 and then expose the cationic CLDPD, which enhances transfection efficiency. In the transfection medium with 20% serum (pH 7.4), CMCS-CLDPD displayed the highest transfection efficiency, which showed that CMCS-CLDPD had the best stability in serum and could highly transfect HepG2 cells even in the presence of serum.
Figure 13 (A) Fluorescent micrographs and (B) Flow cytometry results of HepG2 cells transfected by pEGFP with o-carboxymethyl-chitosan cationic liposome-coated deoxyribonucleic acid/protamine/deoxyribonucleic acid complexes and corresponding cationic liposome-coated deoxyribonucleic acid/protamine/deoxyribonucleic acid complexes (containing 5 μg deoxyribonucleic acid) in the absence of serum with pH 6.5 and pH 7.4, respectively, and also in the presence of 20% serum (pH 7.4). Polyethylenimine/deoxyribonucleic acid (N/P ratio 10.0) prepared with 5 μg deoxyribonucleic acid was used as positive control. Gene expression was examined after 48 hours posttransfection.
Notes: *P < 0.05; **P < 0.01.
Abbreviations: CMCS, o-carboxymethyl-chitosan; CLDPD, cationic liposome-coated deoxyribonucleic acid/protamine/deoxyribonucleic acid complexes; DNA, deoxyribonucleic acid; PEI, polyethylenimine.
In vivo transfection results of CMCS-CLDPD
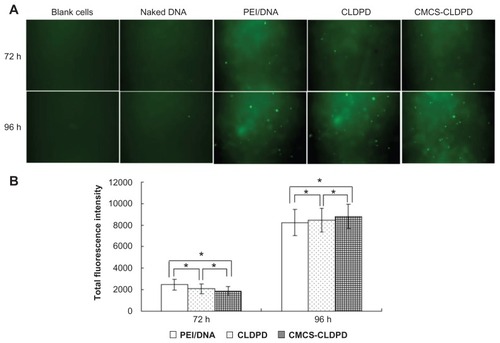
After intratumoral injection of naked DNA, PEI/DNA, CLDPD, and CMCS-CLDPD into tumor-bearing mice, mice were sacrificed and tumor cells were isolated at predetermined time intervals. The fluorescent cells transfected with pEGFP were observed using an inverted fluorescence microscope. As illustrated in , blank tumor cells isolated from mice without any treatment had some slight green autofluorescence and were used as a negative control. Naked DNA could barely transfect tumor cells and caused almost no green fluorescence. After administration, PEI/DNA, CLDPD, and CMCS-CLDPD showed higher transfection efficiencies at 96 hours than at 72 hours (P < 0.05). Anionic- charged CMCS-CLDPD displayed similar transfection efficiency to cationic-charged CLDPD (P > 0.05).
Figure 14 (A) Fluorescent micrographs and (B) Total fluorescence intensity of tumor cells isolated from tumor-bearing mice transfected by pEGFP after intratumoral injection of o-carboxymethyl-chitosan cationic liposome-coated deoxyribonucleic acid/protamine/deoxyribonucleic acid complexes and corresponding cationic liposome-coated deoxyribonucleic acid/protamine/deoxyribonucleic acid complexes (containing 40 μg deoxyribonucleic acid). Naked deoxyribonucleic acid and polyethylenimine/deoxyribonucleic acid (N/P ratio 10.0) with 40 μg deoxyribonucleic acid administered via intratumoral injection were used as negative control and positive control, respectively.
Note: *P > 0.05.
Abbreviations: CMCS, o-carboxymethyl-chitosan; CLDPD, cationic liposome-coated deoxyribonucleic acid/protamine/deoxyribonucleic acid complexes; DNA, deoxyribonucleic acid; h, hours; PEI, polyethylenimine.
Discussion
The purpose of this study was attempting to construct a multifunctional vector for efficient gene delivery via LbL technique, using alternate layers of polyanions and polycations with required functionalities. With a deep understanding of gene delivery process, it is acknowledged that efficient gene delivery asks for multifunctional gene vectors that possess long circulation time, cellular or tissue targeting, endosomal escape, and nuclear targeting.Citation27 Thus, in this study, Pro with nuclear targeting capability was used to condense DNA to form a cationic core, which was used as assembly template. Then, additional layers of DNA for higher loading, LHLN-liposomes for endosomal escape, and pH-sensitive CMCS for long circulation time were sequentially assembled onto the template. DNA was subtly used as an additional layer which could not only reverse the surface charge from positive to negative values for further deposition of cationic LHLN-liposomes but also increase the DNA loading of the vectors. In a previous study, LHLN was respectively used as a cationic modifier to prepare solid lipid nanoparticles and PLA-polyethylene glycol nanoparticles, which improved transfection efficiency and lowered cytotoxicity.Citation20,Citation28 Recently, as a cationic lipid it was mixed with fusogenic lipid DOPE, which always had endosomal escape capability, to prepare cationic LHLN-liposomes.Citation29 It was demonstrated that they showed comparable transfection efficiency to commercial Lipofectamine™ 2000 but with lower cytotoxicity. Based on these results, LHLN-liposomes were served as a cationic functional material to assemble the multifunctional gene vector.
Traditionally, attachment of polyethylene glycol had previously been found to be very useful for reducing undesired serum interactions and improving circulation time of gene carriers.Citation30,Citation31 However, polyethylene glycol shielding lowered transfection efficiency due to reduced cell-surface interactions.Citation32,Citation33 This hurdle has been partly overcome by inclusion of targeting ligands, but these targeted and pegylated carriers do not reach the activity of the uncoated positively- charged vectors.Citation34,Citation35 It is obvious that polyethylene glycol chains should be detached after reaching the target cell. For tumor gene delivery, intratumoral pH value in solid tumors may drop to 6.5, one pH unit lower than in normal blood (7.4), because of hypoxia and massive cell death inside the tumor.Citation36 Therefore, pH-sensitive linkages that were quite stable at pH around 7.4 but degrade at pH around 6.5 were developed to detach polyethylene glycol chains from gene carriers in the acidic conditions of tumor tissues based on the diorto esters,Citation37 vinyl esters,Citation38 and hydrazones.Citation39 But the chemical approaches are complicated and laborious.
In this study, an attempt was made to propose a new pH-sensitive deshielding of hydrophilic polymer based on CMCS. CMCS, a water-soluble derivative of chitosan obtained by introducing –CH2–COOH groups onto –OH along the chitosan molecular chain, is an amphoteric polyelectrolyte containing both carboxyl and amino groups.Citation40 It can change the charged states of the carboxyl and amino groups by varying pH. Therefore, CMCS has good pH and ion sensitivity, and is used as a material that can help provide pH sensitivity in aqueous solution.Citation41 In the present study, a kind of CMCS with carboxymethyl degree of substitution 60% was used. It was found that when the pH value was below or equal to 6.5, the amino groups protonated and CMCS fell off from the surface of CMCS-CLDPD (; reversed surface charge at pH 6.5) because of the electrostatic repulsion between the positively-charged CLDPD and CMCS. However, when the pH value was above 7.0, carboxyl groups ionized and CMCS was negatively charged which strongly bound with cationic CLDPD. Therefore, in blood circulation with pH 7.4, CMCS would strongly bind with cationic CLDPD and protect the gene carrier from serum interaction (). When in the acidic environment of tumor tissue, CMCS was deshielded and CLDPD exposed, which then enhanced gene delivery efficiency. This new responsive approach is an easy method without use of organic solvents and complicated chemical modification, which helps to maintain biological activity of DNA. It would be of great value for tumor gene therapy.
For LbL technique, electrostatic interactions are widely reported to be the main driving force for the multilayer formation process between two oppositely charged polyelectrolytes. Citation42 Morphology, particle size, and zeta potential of vectors are often determined in order to assess whether an additional layer could be successfully deposited on its previous template. As illustrated in , the layered and “core-shell” structure visually exhibited the successful assembly of multilayered CMCS-CLDPD. Moreover, the increased particle size and reversed zeta potential of particles shown in and strongly demonstrate the construction of CMCS-CLDPD. During the assembly process, charge overcompensation by adsorbing polyelectrolytes, which leads to a reversal of the net surface charge, is considered to be a prerequisite for polyelectrolyte multilayer formation as it allows the subsequent adsorption of oppositely-charged polyelectrolytes through electrostatic association.Citation43 The amount of diffusing polyelectrolyte added during a deposition step should not only exceed the minimum amount needed to neutralize the surface charges of its adsorbing multilayer but also create sufficient new charges (ie, net surface charge) for further buildup.Citation44 Usually, the surface of multilayer for further buildup was saturated by adsorbed polyelectrolytes and its zeta potential no longer changed with addition of excess polyelectrolytes.
There is an increasing body of evidence demonstrating that the surface properties of nanoparticles can lead to considerable toxicity.Citation45 Cationic agents with high charge density have been shown to induce membrane toxicity due to the electrostatic interactions with the negatively-charged glycocalyx of the cellular surface.Citation46,Citation47 Therefore, lower positive charge density or even negative surface charge could reduce cytotoxicity.Citation48 This explanation is in accordance with the results illustrated in . The deposition of anionic layer of CMCS on the surface of cationic CLDPD resulted in CMCS-CLDPD being negatively charged, so at the highest concentration in the test, cell viability of CMCS-CLDPD was higher than CLDPD (P < 0.05). Both CMCS-CLDPD and CLDPD suspensions showed lower cytotoxicity than PEI/DNA (P < 0.05), most probably due to good biodegradability and low toxicity of CMCSCitation49 and cationic lipid LHLNCitation20 used in the preparation.
It was demonstrated that intratumoral pH value in solid tumors was around 6.5.Citation50 Therefore, in the in vitro transfection test, the pH value of cell culture medium without serum was adjusted to 6.5 to mimic the acidic microenvironment of tumor cells. It was found that CMCS-CLDPD achieved similar transfection efficiency to CLDPD at pH 6.5 but with lower transfection efficiency at pH 7.4 (). In order to further evaluate the pH sensitivity of CMCS-CLDPD in vivo, in vivo transfection efficiency was examined in tumor-bearing mice. Mice bearing B16 cells were subjected to an intratumoral injection of CMCS-CLDPD and CLDPD, and the gene expression efficiency in the isolated tumor cells was assayed. Anionic-charged CMCS-CLDPD displayed similar transfection efficiency to cationic-charged CLDPD (P > 0.05), which indicated that CMCS-CLDPD had pH sensitivity and the outermost layer of CMCS could fall off in the tumor tissue. Also, CMCS-CLDPD showed similar transfection efficiency to PEI/DNA (P > 0.05), which demonstrated that CMCS-CLDPD had a great advantage over PEI/DNA in tumor tissue as CMCS-CLDPD displayed comparable transfection activity to PEI/DNA but with lower cytotoxicity ().
Conclusion
In this work, a multifunctional nanovector with high transfection efficiency was successfully constructed via LbL technique. LbL technique is an easy method without use of organic solvents and complicated chemical modification which helps to maintain biological activity of DNA. When the appropriate functional polyelectrolytes, such as nuclear localization peptide for enhanced nuclear delivery, fusogenic lipids or pH-sensitive polymers for endosomal escape, and ligands for specific targeting, were designed to construct multilayered vectors, the assembly vectors might contain nuclear targeting, endosomal escape, and cell uptake simultaneously to express high gene transfection efficiency. Therefore, LbL technique may furthermore be of great application value for the preparation of high efficient and low cytotoxic multifunctional gene vectors. CMCS was demonstrated to be a pH-sensitive polymer which would be of great importance for the construction of antitumor gene vectors. Moreover, CMCS has –COOH and –NH2 groups which can be easily modified with a wide range of biomolecules and chemical ligands to achieve active targeting to tissues and cells. Further research will be directed towards the surface-modification of CMCS-CLDPD with various kinds of ligands to achieve active targeting to tumor tissues for efficient gene delivery.
Acknowledgments
The work was supported by Program for New Century Excellent Talents in University of China (NCET-08-0334) and Shandong Province Natural Science Foundation (ZR2009CM011).
Disclosure
The authors report no conflicts of interest in this work.
References
- ParkKKimWJChoYHCancer gene therapy using adeno-associated virus vectorsFront Biosci2008132653265917981740
- MorilleMPassiraniCDufortSTumor transfection after systemic injection of DNA lipid nanocapsulesBiomaterials20113292327233321185595
- WolffJARozemaDBBreaking the bonds: non-viral vectors become chemically dynamicMol Ther200816181517955026
- BondiMLCraparoEFSolid lipid nanoparticles for applications in gene therapy: a review of the state of the artExpert Opin Drug Deliv20107171820017658
- GaoYGuWWChenLXuZLiYA multifunctional nano device as non-viral vector for gene delivery: in vitro characteristics and transfectionJ Control Release2007118338138817321625
- KogureKAkitaHHarashimaHMultifunctional envelope-type nano device for non-viral gene delivery: concept and application of Programmed PackagingJ Control Release2007122324625117651856
- DecherGFuzzy nanoassemblies: toward layered polymeric multicompositesScience1997277533012321237
- JewellCMLynnDMMultilayered polyelectrolyte assemblies as platforms for the delivery of DNA and other nucleic acid-based therapeuticsAdv Drug Deliv Rev200860997999918395291
- LinQKRenKFJiJHyaluronic acid and chitosan-DNA complex multilayered thin film as surface-mediated nonviral gene delivery systemColloids Surf B Biointerfaces200974129830319740634
- SaurerEMJewellCMKuchenreutherJMLynnDMAssembly of erodible, DNA-containing thin films on the surfaces of polymer microparticles: toward a layer-by-layer approach to the delivery of DNA to antigen-presenting cellsActa Biomater20095391392418838346
- BlacklockJYouYZZhouQHMaoGOupickyDGene delivery in vitro and in vivo from bioreducible multilayered polyelectrolyte films of plasmid DNABiomaterials200930593995019013638
- ZhangJChuaLSLynnDMMultilayered thin films that sustain the release of functional DNA under physiological conditionsLangmuir200420198015802115350066
- LiPZhangNLayer-by-layer self-assembly vectors for gene deliveryCurr Gene Ther2011111587321182463
- TrubetskoyVSWongSCSubbotinVRecharging cationic DNA complexes with highly charged polyanions for in vitro and in vivo gene deliveryGene Ther200310326127112571634
- SaulJMWangCHKNgCPPunSHMultilayer nanocomplexes of polymer and DNA exhibit enhanced gene deliveryAdv Mater20082011925
- KeJHYoungTHMultilayered polyplexes with the endosomal buffering polycation in the core and the cell uptake-favorable polycation in the outer layer for enhanced gene deliveryBiomaterials201031359366937220864166
- CaiKRechtenbachAHaoJBossertJJandtKDPolysaccharide-protein surface modification of titanium via a layer-by-layer technique: characterization and cell behaviour aspectsBiomaterials200526305960597115913761
- MasudaTAkitaHHarashimaHEvaluation of nuclear transfer and transcription of plasmid DNA condensed with protamine by microinjection: the use of a nuclear transfer scoreFEBS Lett2005579102143214815811332
- YeJWangALiuCChenZZhangNAnionic solid lipid nanoparticles supported on protamine/DNA complexesNanotechnology2008192828570821828742
- YuWLiuCYeJZouWZhangNXuWNovel cationic SLN containing a synthesized single-tailed lipid as a modifier for gene deliveryNanotechnology2009202121510219423923
- KimDLeeESOhKTGaoZGBaeYHDoxorubicin-loaded polymeric micelle overcomes multidrug resistance of cancer by double-targeting folate receptor and early endosomal pHSmall20084112043205018949788
- LevyMSLotfianPO’KennedyRLo-YimMYShamlouPAQuantitation of supercoiled circular content in plasmid DNA solutions using a fluorescence- based methodNucleic Acids Res20002812E5710871380
- HanHDShinBCChoiHSDoxorubicin-encapsulated thermosensitive liposomes modified with poly(N-isopropylacrylamide-co-acrylamide): drug release behavior and stability in the presence of serumEur J Pharm Biopharm200662111011616183268
- SrinivasanCBurgessDJOptimization and characterization of anionic lipoplexes for gene deliveryJ Control Release20091361627019331848
- HuYLiKWangLYinSZhangZZhangYPegylated immuno-lipopolyplexes: a novel non-viral gene delivery system for liver cancer therapyJ Control Release20101441758120144670
- ParkMRHanKOHanIKDegradable polyethylenimine-alt-poly(ethylene glycol) copolymers as novel gene carriersJ Control Release2005105336738015936108
- KamiyaHAkitaHHarashimaHPharmacokinetic and pharmacodynamic considerations in gene therapyDrug Discov Today200382199099614643162
- LiuCChenZYuWZhangNNovel cationic 6-lauroxyhexyl lysinate modified poly(lactic acid)-poly(ethylene glycol) nanoparticles enhance gene transfectionJ Colloid Interface Sci2011354252853521094495
- LiPLiuDSunXLiuCLiuYZhangNA novel cationic liposome formulation for efficient gene delivery via a pulmonary routeNanotechnology2011222424510421543837
- KunathKvon HarpeAPetersenHThe structure of PEG-modified poly(ethylene imines) influences biodistribution and pharmacokinetics of their complexes with NF-kappa B decoy in micePharm Res200219681081712134951
- van VlerkenLEVyasTKAmijiMMPoly(ethylene glycol)-modified nanocarriers for tumor-targeted and intracellular deliveryPharm Res20072481405141417393074
- OgrisMWalkerGBlessingTKircheisRWolschekMWagnerETumor-targeted gene therapy: strategies for the preparation of ligand-polyethylene glycol-polyethylenimine/DNA complexesJ Control Release2003911–217318112932649
- OupickyDOgrisMHowardKADashPRUlbrichKSeymourLWImportance of lateral and steric stabilization of polyelectrolyte gene delivery vectors for extended systemic circulationMol Ther20025446347211945074
- GuoWLeeRLReceptor-targeted gene delivery via folate-conjugated polyethylenimineAAPS Pharm Sci199914E19
- RudolphCSchillingerUPlankCNonviral gene delivery to the lung with copolymer-protected and transferrin-modified polyethylenimineBiochim Biophys Acta200215731758312383945
- VaupelPKallinowskiFOkunieffPBlood flow, oxygen and nutrient supply, and metabolic microenvironment of human tumors: a reviewCancer Res19894923644964652684393
- GuoXSzokaFCJrSteric stabilization of fusogenic liposomes by a low-pH sensitive PEG – diortho ester – lipid conjugateBioconjug Chem200112229130011312691
- TorchilinVMultifunctional and stimuli-sensitive pharmaceutical nanocarriersEur J Pharm Biopharm200971343144418977297
- FellaCWalkerGFOgrisMWagnerEAmine-reactive pyridylhydrazone- based PEG reagents for pH-reversible PEI polyplex shieldingEur J Pharm Sci2008344–530932018586470
- LiYDuYTangYWangXA novel pH-sensitive and freeze-thawed carboxymethyl chitosan/poly(vinyl alcohol) blended hydrogel for protein deliveryPolym Int2009581011201125
- ChenLDuYWuHXiaoLRelationship between molecular structure and moisture-retention ability of carboxymethyl chitin and chitosanJ Appl Polym Sci200283612331241
- DubasSTSchlenoffJBFactors controlling the growth of polyelectrolyte multilayersMacromolecules1999322481538160
- SteitzRJaegerWvon KlitzingRInfluence of charge density and ionic strength on the multilayer formation of strong polyelectrolytesLangmuir2001171544714474
- KujawaPMoraillePSanchezJBadiaAWinnikFMEffect of molecular weight on the exponential growth and morphology of hyaluronan/chitosan multilayers: a surface plasmon resonance spectroscopy and atomic force microscopy investigationJ Am Chem Soc2005127259224923415969601
- KabanovAVPolymer genomics: an insight into pharmacology and toxicology of nanomedicinesAdv Drug Deliv Rev200658151597162117126450
- ChoksakulnimitrSMasudaSTokudaHTakakuraYHashidaMIn vitro cytotoxicity of macromolecules in different cell culture systemsJ Control Release1995343233241
- DaumNNeumeyerAWahlBBurMLehrCMIn vitro systems for studying epithelial transport of macromoleculesMethods Mol Biol200948015116419085125
- Vega-VillaKRTakemotoJKYanezJARemsbergCMForrestMLDaviesNMClinical toxicities of nanocarrier systemsAdv Drug Deliv Rev200860892993818313790
- ChenSCWuYCMiFLLinYHYuLCSungHWA novel pH-sensitive hydrogel composed of N,O-carboxymethyl chitosan and alginate cross-linked by genipin for protein drug deliveryJ Control Release200496228530015081219
- MatsumuraYKataokaKPreclinical and clinical studies of anticancer agent-incorporating polymer micellesCancer Sci2009100457257919462526