Abstract
Nanoparticles have an enormous potential for development in biomedical applications, such as gene or drug delivery. We developed and characterized aminopropyltriethoxysilane-functionalized silicon dioxide nanoparticles (APTES-SiNPs) for gene therapy. Lipofectamine® 2000, a commonly used agent, served as a contrast. We showed that APTES-SiNPs had a gene transfection efficiency almost equal to that of Lipofectamine 2000, but with lower cytotoxicity. Thus, these novel APTES-SiNPs can achieve highly efficient transfection of plasmid DNA, and to some extent reduce cytotoxicity, which might overcome the critical drawbacks in vivo of conventional carriers, such as viral vectors, organic polymers, and liposomes, and seem to be a promising nonviral gene therapy vector.
Introduction
Nanomedicine is an emerging new field created by fusion of nanotechnology and medicine in a straightforward manner by adding, correcting, or replacing genes. A key problem in gene therapy is the efficient delivery of genetic material to target cells in a patient without significant toxicity and side effects. A number of novel delivery strategies have been proposed to enhance the efficiency of plasmid DNA (pDNA) transfection. Nevertheless, the reliability of the carrier is still a critical concern for any gene therapy. An ideal gene delivery system should be biodegradable, nontoxic, nonimmunogenic, stable, and target-specific, should assist to improve gene expression, and be able to be used to treat genetic diseases which require systemic administration of therapy.
Nanoscale gene carriers traditionally include viral vectors, organic polymers, and liposomes.Citation1,Citation2 Viral vectors have been used widely for effective DNA delivery, but their potential immune response, difficult recombination, and high cost have hampered their applications.Citation3 Nonviral polymer carriers have more potential for gene and drug delivery because of such advantages as easy preparation and reduced risk of immune response. Cationic polymers and liposomes have recently shown promise,Citation4,Citation5 and solid lipid nanoparticles have also been shown to be a useful vehicle for gene therapy.Citation6 However, any benefits due to their cationic nature are offset by their instability in serum and high cytotoxicity profile. A number of approaches have been attempted to overcome the cytotoxicity limitation and to make nonviral cationic systems safer.Citation7 An ideal method with both high transfection efficiency and relative safety in vitro and in vivo has not been found, so more attention needs to be devoted to finding new materials.
Fortunately, inorganic nanoparticles such as those made from hydroxyapatite and silicon, offer promise as gene and drug transfer carriers because of their biocompatibility, lack of toxicity, and stable physical and chemical properties.Citation8 Silicon nanoparticles have been used recently as gene carriers in the transfection of pDNA.Citation9
Furthermore, enormous effort has been devoted to surface modification of various polymer drug vectors in order to improve their biocompatibility, resist protein adsorption, and increase their circulation time and internalization efficiency.Citation10 Common functionalization procedures for preparing surfaces for protein binding utilize silane compounds with terminal functional groups that interact electrostatically or covalently with protein surface groups to increase protein adsorption. One common chemical used for surface functionalization is aminopropyltriethoxysilane (APTES), which has the chemical formula NH2 (CH2)3–Si(OC2H5)3.Citation11–Citation14 The silane end of the APTES molecule binds covalently to surface silicon atoms, and the amino end of the molecule increases protein adsorption on the surface by electrostatic interactions. The structural transition of a single dsDNA molecule immobilized on an APTES-treated substrate has been demonstrated. DNA binding to the APTES linker is much stronger than that on an alkylthiol/substrate.Citation15 Also, APTES can dissolve in both polar and nonpolar solvents, and also has high solubility in cell membranes.Citation16,Citation17
Here we report a pDNA carrier based on APTES-modified silica nanoparticles (APTES-SiNPs), the surface of which was functionalized with TX-100 in order to enhance its monodispersity. It was produced by a guided self-assembly approach to the manufacture of DNA nanostructures on a silicon substrate. Because of the unique profiles of APTES and the silica nanoparticles, the novel APTES-SiNP vector has a large surface area to volume ratio, tends to agglomerate, and absorbs more pDNA.
In order to evaluate the transfection efficiency and safety of APTES-SiNPs, we investigated the ability of these novel nanoparticles to condense the plasmid encoding for enhanced green fluorescent protein (pEGFP) and transfer it into human vascular smooth muscle cells (HVSMCs). Using this approach, we compared the stability, plasmid immobilization, transfection efficiency, and cytotoxicity of this formulation with widely used conventional Lipofectamine® 2000 in order to evaluate its potential as a gene transfection vector.
Materials and methods
Materials
All chemicals, ie, APTES (Alfa Aesar, Ward Hill, MA), methyltrimethoxysilane (Alfa Aesar), and Triton X-100 (Sigma-Aldrich, St Louis, MO) were used without further purification. All the glassware (glass bottle and small pieces of glass substrate) was cleaned and sonicated in ethanol for 5 minutes, rinsed with double distilled water, soaked in a H2O/HNO3 (65%)/H2O2 (1:1:1, v/v/v) solution, rinsed again with doubly distilled water, and finally dried in air.
Synthesis of ATPES-SiNPs
APTES 0.1 mL and TX-100 0.3 mL were dissolved in 20 mL of double distilled water and stirred for 10 minutes. Methyl-trimethoxysilane 2 mL was added, followed by stirring for 10 minutes, with subsequent addition of 28% NH3H2O to reach a pH of 11. The solution was stirred for 5 hours, then left overnight at room temperature. Finally, the product was collected by centrifugation, rinsed with double distilled water and ethanol, and allowed to dry at room temperature.
Agarose gel electrophoresis
Complex formation between the nanoparticles and pDNA was analyzed by 1% agarose gel electrophoresis. The pDNA-APTES complex was prepared according to the following process. Briefly, 1 μL of APTES-SiNP in solution (5 mg/500 μL in water) was added to 1 μL of pDNA aqueous solution (0.1 μg/μL) and 8 μL water (pH 7.4) in a microcentrifuge tube. The resulting pDNA/APTES-SiNP complexes were left at room temperature for 30 minutes to facilitate complexation. The samples were centrifuged at 5000 rpm for 5 minutes, after which the suspension was obtained and analyzed by loading onto 1% agarose gel with ethidium bromide (0.1 μg/mL) and running with Tris-acetate buffer at 100 V for 30 minutes. DNA retardation was observed by irradiation with ultraviolet light.
Transfection of cultured cells
HVSMCs were cultured in 75 cm2 flasks in Dulbecco’s modified Eagle’s medium (Gibco BRL, Carlsbad, CA) supplemented with 10% fetal bovine serum, 50 Ag/mL penicillin, 50 Ag/mL streptomycin, and 100 Ag/mL neomycin at 37°C in a humidified 5% CO2-containing atmosphere. For the transfection experiments, the cells were trypsinized and seeded into a 6-well plate at a density of 2.5 × 105 cells/well overnight before the transfection procedure was performed. The cells were washed with phosphate-buffered solution twice prior to the addition of Optimen® 2 mL. The transfection procedure consisted of 12 μL pEGFP (185 μg/mL) with 12 μL APTES-SiNP solution (APTES-SiNPs 5 mg/Tris-EDTA 500 μL), and 18 μL water (pH 7.4) in one microcentrifuge tube. The contents were then mixed together gently and incubated at room temperature for 30 minutes. The cells were then incubated at 37°C in 5% CO2. After 5 hours, the complexes were aspirated and replaced with culture medium. After 48 hours, the cells were assayed for green fluorescent protein expression, both qualitatively by fluorescence microscopy and quantitatively by fluorescence-activated cell sorting. The cells were washed twice with phosphate-buffered solution and collected by trypsinization, centrifuged at 2000 g for 2 minutes, and washed twice with phosphate-buffered solution. The cells were then resuspended in phosphate-buffered solution and analyzed by flow cytometry (Beckman Coulter, Brea, CA).
Statistical analysis
All experimental measurements were collected in triplicate. The values are expressed as the mean ± standard deviation. The statistical significance of differences in cell viability was examined using one-way analysis of variance followed by Fisher’s least significant difference test. The significance level was set at P < 0.05.
Results and discussion
Nanoparticle size and zeta potential measurement
The nanoparticles were characterized by scanning electron microscopy and elemental analysis (x-ray photoelectron spectrometry, ). The mean size of the APTES-SiNPs was found to be 174.5 nm by dynamic light scattering.
Figure 1 Scanning electron microscopic images of amino-modified silicon nanoparticles.
Note: Bars, 500 nm.
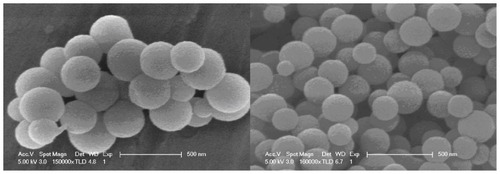
Several approaches have been proposed to increase the surface area of the solid-phase support to enhance the capacity for DNA. Examples include the use of smaller-sized particles. Decreasing the particle size has proved to be an effective method of increasing the surface area for DNA absorption.Citation18 However, too small a particle size can cause extremely high back pressure during the extraction procedure. The monotonic decrease of the size of small particles to increase the surface area of the solid phase has consequently been limited in practical application. Nanomaterials with unique properties such as a large surface area, pore structure, embedded effect, and size effect have been used effectively in bioanalysis and drug delivery.Citation19,Citation20
After amino functionalization, zeta potential measurements carried out using a Beckman Delsa Nano C at an APTES-SiNP concentration of 1 g/L at 20°C demonstrated that the APTES-SiNP particles had a positive surface potential of +41.8 mV. We assessed pDNA condensation and integrity using gel electrophoresis (). The positive potential might have resulted from protonation of amino groups on the surface of APTES-SiNPs. Therefore, the positively charged silica nanoparticles could be obtained directly without further modification and could combine with the negatively charged pDNA as a carrier for DNA delivery. The positively charged silica APTES-SiNPs could combine with negatively charged pDNA by electrostatic interactions to form APTES-SiNP-pDNA complexes.
Figure 2 Agarose gel electrophoresis assay demonstrating APTES-SiNP-DNA complexation. A constant amount of DNA was complexed with nanoparticles at different ratios of 50:1, 30:1, 20:1, 10:1, and 1:1 (10 mM, pH 7.4). No 50:1 or 30:1 APTES-SiNP-DNA band is observed because of fluorescence quenching by nanoparticle complexation.
Abbreviations: APTES, aminopropyltriethoxysilane; SiNP, silicon dioxide nanoparticle.
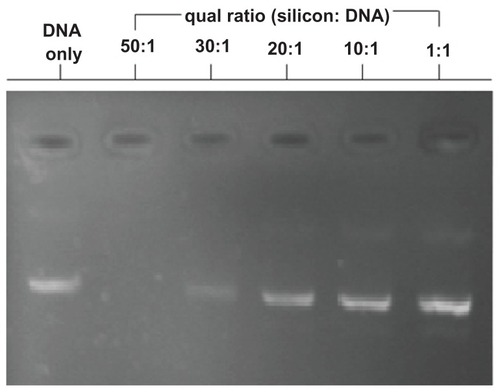
A suitable ratio of silicon to pDNA was about 30:1, indicating that the positive charge density of APTES-SiNPs is not very high, considering their large diameter. Generally speaking, a lower positive charge density would reduce cytotoxicity.
Gel electrophoresis analysis
Binding of APTES-SiNPs to polyanionic pDNA was studied using the electrophoretic mobility of pDNA in agarose gel. The complexes were formed in deionized water by mixing a fixed amount of pDNA with increasing amounts of APTES-SiNPs in such a way as to obtain APTES-SiNPs:pDNA weight ratios ranging from 50:1 to 1:1, corresponding to the w/w ratio indicated in . The efficiency of pDNA complexation with APTES-SiNPs after 30 minutes of incubation was evaluated by the amount of APTES-SiNPs required to retard migration of pDNA toward the cathode during agarose gel electrophoresis. Efficient complexation of pEGFP by APTES-SiNPs led to pDNA immobilization. As can be seen in , at the higher two ratios of APTES-SiNPs:pDNA assayed, stable complexes were formed. The absence of DNA in lines 2 to 3 confirms that DNA was completely immobilized inside the complexes.
In summary, APTES-SiNPs have size and zeta potential values similar to those of other carriers and the same capacity to complex pDNA. Two bands () were characterized for their capacity to condense pDNA and to transfect HVSMCs.
Cytotoxicity
Gene vectors should not induce cytotoxic effects in gene therapy,Citation21 so the low cytotoxicity of APTES-SiNPs:pDNA is very important. Nonviral delivery systems have certain drawbacks, such as cytotoxic reactions, especially when cationic lipids are involved.Citation22 Cationic formulations have been reported to affect cell proliferation, differentiation, and proapoptotic genes in human epithelial cells. The polycationic nature of synthetic nonviral gene transfer systems induces cytotoxicity by necrosis and apoptosis.
Therefore, evaluation of the cytotoxicity of cationic gene delivery systems is essential. In the present study, the toxicity of APTES-SiNPs was compared with that of Lipofectamine 2000 in HVSMCs by flow cytometry in vitro, as shown in . We observed that Lipofectamine 2000 was much more toxic than APTES-SiNP to cells.
Figure 3 Cell viability assay. Cytotoxicity of Lipofectamine® 2000 and APTES-SiNPs on human vascular smooth muscle cells. After 48 hours of incubation, cell viability was measured by flow cytometry. Silicon nanoparticles had negligible toxicity to human vascular smooth muscle cells compared with Lipofectamine 2000.
Notes: *P < 0.05 for APTES-SiNPs versus control; §P < 0.05 for Lipofectamine 2000 versus APTES-SiNPs.
Abbreviations: APTES, aminopropyltriethoxysilane; SiNPs, silicon dioxide nanoparticles.
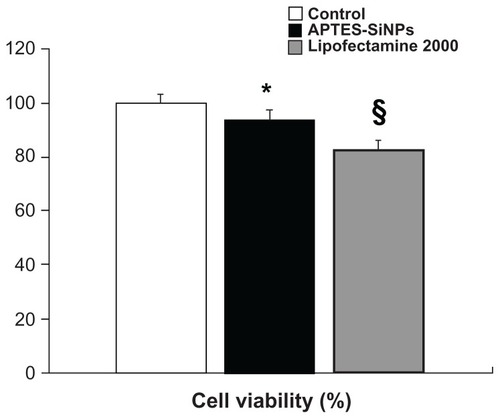
Previous studies have shown that mesoporous silica nanoparticles are nontoxic and biocompatible,Citation23,Citation24 sufficiently excreted via the renal route,Citation25 and promising candidates for drug delivery applications.Citation25,Citation26 Our study results are in accordance with those of a previous study showing that APTES-SiNPs have almost no adverse effects on human cells. However, there is an increasing body of evidence suggesting that the surface properties of nanoparticles can have considerable toxicity.Citation27 The toxicity of cationic polymers has been reported to be a function of the interactions of the polymers with cell membranes and/or the efficiency of cellular uptake.Citation28,Citation29 As show in , APTES-SiNPs actually do have some adverse effects on human cells, mainly ascribable to APTES. However, quaternized APTES derivatives show less cytotoxicity than regular APTES because they maintain a cationic charge inside the polymeric backbone shielded by surface hydroxyl groups. This could explain why the cytotoxicity of APTES is low after it is fixed onto silicon nanoparticles.
The higher cell viability observed for APTES-SiNPs is probably due to the better cell compatibility of the silicon particles. As a consequence, APTES-SiNPs maintain the same low level of cytotoxicity even at high particle concentrations and a long cell contact time.
Transfection
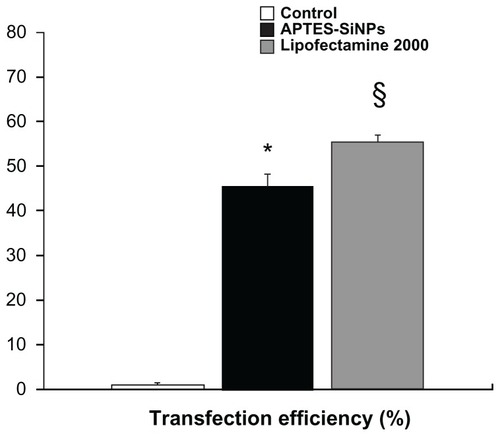
In the present study, we found that the nature of the chemical groups on the surface of APTES-SiNPs is a critical factor affecting transfection efficiency. We modified the surface of the silicon nanoparticles and evaluated the ability of the mica to deliver the pEGFP gene into HVSMCs. Our results show that the APTES-SiNPs were able to deliver pEGFP into cells ( and ). They also suggest that the positive charge introduced by an amino group might allow the APTES-SiNPs to bind to anionic pDNA and the cell membrane via electrostatic interactions, which could be the initial step for gene transfection.
Figure 4 Human vascular smooth muscle cells transfected with pEGFP delivered with APTES-SiNPs. A combined transmission and fluorescence image is shown. Fluorescence spectra of pEGFP taken in human vascular smooth muscle cells (magnification 400×).
Abbreviations: APTES, aminopropyltriethoxysilane; SiNPs, silicon dioxide nanoparticles; pEGFP, plasmid encoding for enhanced green fluorescent protein.
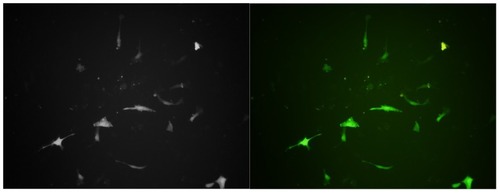
Figure 5 Cell transfection efficiency. The percentage of pEGFP expression was determined by flow cytometry after APTES-SiNP transfection in human vascular smooth muscle cells.
Notes: *P < 0.05 for APTES-SiNPs versus control; §P < 0.05 for Lipofectamine 2000 versus APTES-SiNPs.
Abbreviations: APTES, aminopropyltriethoxysilane; SiNPs, silicon dioxide nanoparticles; pEGFP, plasmid encoding for enhanced green fluorescent protein.
We have developed an optimal method to protect DNA from cleavage by using bioconjugated amino-modified silicon nanoparticles. Our results clearly show that DNA could be easily enriched onto the positively charged surface of the nanoparticles for further manipulation. The DNA strands are protected from enzymatic cleavage and have the same properties as free DNA strands when released from the nanoparticles. The plasmids released are biologically active. This bionanotechnology is simple and efficient for protecting DNA strands. It will be highly useful in DNA separation, purification, manipulation, and detection, and possibly in genetic engineering and gene therapy.
The DNA delivery pathway involves the following steps. First, nanoparticles are adsorbed onto the cell membrane. The nanoparticles are then taken up by cells via endocytosis.Citation30,Citation31 Certain intracellular processes can avoid transport of DNA across the cell membrane into the nucleus. Endosomal degradation of DNA can occur during endocytosis inside an endosome if DNA does not escape from the endosome before fusion with lysosomes (in which pH is <5).Citation32
In a previous study it was reported that hydrophilic APTES nanoparticles are actively taken up by tumor cells in vitro, as seen by fluorescence staining of the cytoplasm.Citation33 This cytoplasmic staining has been observed for a few types of tumor cells that we have investigated, an example of which is HVSMCs. Although the mechanism for this type of cellular uptake is not yet fully understood, the net negative charge on the nanoparticles is thought to play a crucial role. A similar observation was made in a recent study that demonstrated high specific uptake of anionic magnetic nanoparticles by cells in vitro.Citation34 In gene delivery, the genetic material should migrate to the nucleus as soon as it is released inside the cytoplasm of a cell.Citation35 Again, optical imaging can play a significant role in studying this nuclear migration of DNA. In the present study, we irreversibly tagged DNA with fluorescent APTES-SiNPs and unlabeled silicon nanoparticle complexes, as described in the experimental program. The post-release nuclear trafficking of DNA is demonstrated in the confocal image of HVSMCs. After comparison of this image with the image of HVSMCs stained with amino-encapsulated nanoparticles, it can be seen that there is considerable staining inside the nucleus in addition to cytoplasmic staining (). However, these amino-modified nanoparticles are not able to penetrate the cell nucleus. Thus, we believe that this nuclear staining is a result of migration of the labeled DNA released inside the nucleus. A recent study report on the mobility of DNA molecules inside cells indicates that free DNA molecules can move in the cytoplasm, and that a small portion of them can reach the intranuclear space.Citation36
This nuclear accumulation of foreign DNA can be increased by tagging with nucleus-targeting peptides.Citation37 Finally, to confirm that the DNA delivered is functional, pEGFP expression was checked by transfection of cultured cells. We observed that pEGFP was transfected successfully using APTES-SiNPs as the delivery vector. The observed green cellular fluorescence was from the expressed pEGFP, which was confirmed by localized spectroscopy (). Our investigation shows that the nanomedicine approach using APTES-SiNPs is a promising direction for nonviral gene delivery.
Conclusion
In recent years, the transfection effect of nonviral carriers has been emphasized whilst overlooking the critical issue of cytotoxicity. Consequently, even though we have found many highly promising efficient carriers in vitro, their potential immune response, difficult recombination, and high cost has hampered their application in vivo.Citation3 Undoubtedly, the basic requirement of a new carrier is transfection efficiency, but its safety is likely to play a pivotal role in its clinical application. We need to devote more attention to modification of the structural aspects of these carriers to minimize their toxicity and also find new materials, such as silicon nanoparticles, because transfection efficiency can be easily improved by increasing the weight ratio, whereas toxicity cannot be tackled so easily. Most new carriers found in recent years have been discarded upon practical application, for similar reasons.
In the present study, positively charged APTES-SiNPs with an average diameter of 174.5 nm were synthesized as pDNA carriers using a water-in-oil microemulsion method. The positively charged APTES-SiNPs were able to combine with the negatively charged pDNA by electrostatic interactions to form a bioconjugate favorable for cellular uptake. The APTES-SiNPs were able to protect the pDNA in the complex from nuclease degradation. In vitro investigation suggested that the APTES-SiNP carriers could efficiently transfect pEGFP into HVSMCs, although the gene transfection efficiency was a little lower than that achieved by the more widely used Lipofectamine 2000 (). It is worthwhile to emphasize that the toxicity of the APTES-SiNP carriers was lower than that of Lipofectamine 2000, and that they showed good biocompatibility and almost no cytotoxicity to HVSMCs. Further, the surface of APTES-SiNPs can also be chemically modified with various functional groups, such as cancer-specific antibodies or ligands, for targeting purposes.Citation38–Citation42 Considering all of the data, APTES-SiNPs may be superior to Lipofectamine 2000 as carriers of pDNA in HVSMCs, and may be a promising nonviral gene therapy vector.
Disclosure
The authors report no conflicts of interest in this work.
References
- BertlingWMGareisMPaspaleevaVUse of liposomes, viral capsids, and nanoparticles as DNA carriersBiotechnol Appl Biochem19911333904051883530
- LuoDSaltzmanWMSynthetic DNA delivery systemsNat Biotechnol2000181333710625387
- AndersonWFHuman gene therapyNature1998392Suppl 667925309579858
- LiWSzokaFCJrLipid-based nanoparticles for nucleic acid deliveryPharm Res200724343844917252188
- LiSDHuangLNon-viral is superior to viral gene deliveryJ Control Release2007123318118317935817
- del Pozo-RodriguezAPujalsSDelgadoDA proline-rich peptide improves cell transfection of solid lipid nanoparticle-based non-viral vectorsJ Control Release20091331525918854203
- PackDWHoffmanASPunSStaytonPSDesign and development of polymers for gene deliveryNat Rev Drug Discov20054758159316052241
- AllenTMCullisPRDrug delivery systems: entering the mainstreamScience200430356651818182215031496
- RoyIOhulchanskyyTYBharaliDJOptical tracking of organically modified silica nanoparticles as DNA carriers: a nonviral, nanomedicine approach for gene deliveryProc Natl Acad Sci U S A2005102227928415630089
- StormGten KateMTWorkingPKBakker-WoudenbergIADoxorubicin entrapped in sterically stabilized liposomes: effects on bacterial blood clearance capacity of the mononuclear phagocyte systemClin Cancer Res1998411111159516959
- GanSYangPYangWPhotoactivation of alkyl C-H and silanization: a simple and general route to prepare high-density primary amines on inert polymer surfaces for protein immobilizationBiomacromolecules20091051238124319317482
- LiuTWangSChenGImmobilization of trypsin on silica-coated fiberglass core in microchip for highly efficient proteolysisTalanta20097751767177319159796
- JangLSLiuHJFabrication of protein chips based on 3-aminopropyltriethoxysilane as a monolayerBiomed Microdevices200911233133818821014
- LibertinoSGiannazzoFAielloVXPS and AFM characterization of the enzyme glucose oxidase immobilized on SiO(2) surfacesLangmuir20082451965197218205419
- NguyenTHKimYUKimKJChoiSSInvestigation of structural transition of dsDNA on various substrates studied by atomic force microscopyJ Nanosci Nanotechnol2009932162216819435096
- YamazakiMItoTDeformation and instability in membrane structure of phospholipid vesicles caused by osmophobic association: mechanical stress model for the mechanism of poly(ethylene glycol)-induced membrane fusionBiochemistry1990295130913142322565
- BoniLTHahJSHuiSWMukherjeePHoJTJungCYAggregation and fusion of unilamellar vesicles by poly(ethylene glycol)Biochim Biophys Acta198477534094186466682
- LevyMSO’KennedyRDAyazi-ShamlouPDunnillPBiochemical engineering approaches to the challenges of producing pure plasmid DNATrends Biotechnol200018729630510856925
- SantraSWangKTapecRTanWDevelopment of novel dyedoped silica nanoparticles for biomarker applicationJ Biomed Opt20016216016611375725
- CuiZMumperRJPlasmid DNA-entrapped nanoparticles engineered from microemulsion precursors: in vitro and in vivo evaluationBioconjug Chem20021361319132712440869
- ConwellCCHuangLRecent advances in non-viral gene deliveryAdv Genet20055331816240988
- TorchilinVPMultifunctional nanocarriersAdv Drug Deliv Rev200658141532155517092599
- BarnesCAElsaesserAArkuszJReproducible comet assay of amorphous silica nanoparticles detects no genotoxicityNano Lett2008893069307418698730
- SlowingIIWuCWVivero-EscotoJLLinVSMesoporous silica nanoparticles for reducing hemolytic activity towards mammalian red blood cellsSmall200951576219051185
- HeXNieHWangKTanWWuXZhangPIn vivo study of biodistribution and urinary excretion of surface-modified silica nanoparticlesAnal Chem200880249597960319007246
- BurnsAAViderJOwHFluorescent silica nanoparticles with efficient urinary excretion for nanomedicineNano Lett20099144244819099455
- KabanovAVPolymer genomics: an insight into pharmacology and toxicology of nanomedicinesAdv Drug Deliv Rev200658151597162117126450
- FischerDLiYAhlemeyerBKrieglsteinJKisselTIn vitro cytotoxicity testing of polycations: influence of polymer structure on cell viability and hemolysisBiomaterials20032471121113112527253
- JevprasesphantRPennyJJalalRAttwoodDMcKeownNBD’EmanueleAThe influence of surface modification on the cytotoxicity of PAMAM dendrimersInt J Pharm20032521–226326612550802
- LoyterAScangosGJuricekDKeeneDRuddleFHMechanisms of DNA entry into mammalian cells. II. Phagocytosis of calcium phosphate DNA co-precipitate visualized by electron microscopyExp Cell Res198213912232347084316
- CoonrodALiFQHorwitzMOn the mechanism of DNA transfection: efficient gene transfer without virusesGene Ther1997412131313219472555
- SchmidSLFuchsRMalePMellmanITwo distinct subpopulations of endosomes involved in membrane recycling and transport to lysosomesCell198852173833345561
- RoyIOhulchanskyyTYPudavarHECeramic-based nanoparticles entrapping water-insoluble photosensitizing anticancer drugs: a novel drug-carrier system for photodynamic therapyJ Am Chem Soc2003125267860786512823004
- WilhelmCBilloteyCRogerJPonsJNBacriJCGazeauFIntracellular uptake of anionic superparamagnetic nanoparticles as a function of their surface coatingBiomaterials20032461001101112504522
- DavisSSBiomedical applications of nanotechnology – implications for drug targeting and gene therapyTrends Biotechnol19971562172249183864
- RoyIOhulchanskyyTYBharaliDJOptical tracking of organically modified silica nanoparticles as DNA carriers: A nonviral, nanomedicine approach for gene deliveryProc Natl Acad Sci USA2005102227928415630089
- ZantaMABelguise-ValladierPBehrJPGene delivery: a single nuclear localization signal peptide is sufficient to carry DNA to the cell nucleusProc Natl Acad Sci U S A199996191969874777
- KlichkoYLiongMChoiEMesostructured silica for optical functionality, nanomachines, and drug deliveryJ Am Ceram Soc200992S1S2S1019834571
- CotiKKBelowichMELiongMMechanised nanoparticles for drug deliveryNanoscale200911163920644858
- LiongMLuJKovochichMMultifunctional inorganic nanoparticles for imaging, targeting, and drug deliveryACS Nano20082588989619206485
- Nguyen-HuuTLettCAugerPPoggialeJCSpatial synchrony in host-parasitoid models using aggregation of variablesMath Biosci2006203220422116723140
- LuJChoiETamanoiFZinkJILight-activated nanoimpeller-controlled drug release in cancer cellsSmall20084442142618383576