Abstract
Background
Our previous study found that deletion of Sorting nexin 10 (SNX10) can protect against colonic inflammation and pathological damage induced by dextran sulfate sodium (DSS). This inspired us that modulation of SNX10 expression in colonic epithelial cells might represent a promising therapeutic strategy for inflammatory bowel disease (IBD).
Methods
Effective delivery of siRNA/shRNA to silence genes is a highly sought-after means in the treatment of multiple diseases. Here, we encapsulated SNX10-shRNA plasmids (SRP) with polylactide-polyglycolide (PLGA) to make oral nanoparticles (NPs), and then applied them to acute and chronic IBD mice model, respectively. The characteristics of the nanoparticles were assayed and the effects of SRP-NPs on mouse IBD were evaluated.
Results
High-efficiency SNX10-shRNA plasmids were successfully constructed and coated with PLGA to obtain nanoparticles, with a particle size of 275.2 ± 11.4mm, uniform PDI distribution, entrapment efficiency of 87.6 ± 2.5%, and drug loading of 13.11 ± 1.38%, displayed dominant efficiency of SNX10 RNA interference in the colon. In both acute and chronic IBD models, SRP-NPs could effectively reduce the loss of mice body weight, relieve the intestinal mucosal damage and inflammatory infiltration, inhibit the expression of inflammatory cytokines IL-1β, IL-23, TNF-α, and down-regulate the expression of toll-like receptors (TLRs) 2 and 4.
Conclusion
Oral nanoparticles of SNX10-shRNA plasmid displayed dominant efficiency of SNX10 RNA interference in the colon and ameliorate mouse colitis via TLR signaling pathway. SNX10 is a new target for IBD treatment and nanoparticles of SNX10-shRNA plasmid might be a promising treatment option for IBD.
Introduction
Inflammatory bowel disease (IBD), including ulcerative colitis (UC) and Crohn’s disease (CD), is a chronic disease characterized by non-specific inflammation of the gastrointestinal tract, which is closely related to colon cancer which incidence is substantially increasing in recent years with the improvement of living standards.Citation1,Citation2 However, the pathogenesis of IBD remains elusive. Numerous studies indicate that genetic susceptibility plays a very important role in IBD and colon cancer. Carrying genetic variants such as Nod2, IRGM, IL-23R, and XBP1Citation3 results in intestinal mucosal barrier imbalance, intestinal epithelial cell injury, and triggering IBD. Despite the growing number and variety of drugs currently under development, surgical resection of the colon is the only cure for UC.Citation4–Citation6 It is necessary to identify new potential targets for IBD therapy.
Sorting nexins (SNXs) are phosphoinositide-binding proteins containing a phoxhomology-domain, which play important roles in some physiological processes such as protein sorting,Citation7 intracellular sorting transduction,Citation8 the transmembrane transport of macromolecules (extracellular and endocytosis),Citation9 membrane remodeling, and oncogenesis.Citation10 Silencing SNX10 inhibits the differentiation of macrophages into osteoclasts.Citation11 Mutations in the human SNX10 gene can lead to autosomal recessive hereditary osteopetrosis.Citation12 Overexpression of SNX10 induced cells to form giant vacuoles, whereas vesicle trafficking inhibitor Brefeldin A suppresses the formation of the vacuoles, suggesting that SNX10 is involved in homeostasis and membrane trafficking.Citation13 SNX10/V-ATPase participates in the formation of visceral organs and cilia, suggesting that SNX10 may also have important functions in mammalian embryo development.Citation14 A series of studies reported that SNX10 links to autoimmune diseases including non-alcoholic fatty liver and rheumatoid arthritis.Citation15–Citation17 You et al,Citation18 found the progression of ulcerative colitis was inhibited in SNX10−/- mice by regulating the differentiation of macrophages to M2 type, and relieving intestinal inflammation and mucosal damage. As the starting point of IBD, the mucosal barrier system plays a crucial role in maintaining intestinal stabilityCitation19 which consists of the mucus layer, epithelial cell junctional structures, and leukocyte activity. In the study of SNX10 and colon tumors, the correlation between SNX10 deficiency and changes in epithelial cellsCitation20,Citation21 suggests that SNX10 deletion might be the potential mechanism for IBD treatment.
Specifically, knockdown corresponding proteins at the mucosal barrier by interfering RNAs (siRNAs) or short hairpin RNAs (shRNAs) is an attractive therapeutic strategy.Citation22 But the challenges are the low stability of the nucleic acids which could result in its inactivation during delivery to target cells, and side effects or toxicity of oral drug systemic distribution. In addition, there is another challenge for oral formulations for IBD is how to avoid the side effects and toxicity of drug systemic distribution while delivering the drug to the colon site. The main strategies currently adopted are mainly based on the gastrointestinal tract, especially colon physiological conditions.Citation23 Such as Sulfasalazine or Olsalazine, prodrugs of 5-aminosalicylic acid (5-ASA), rely on the activity of coliform bacteria to cleave pro-drugs into active moieties.Citation24 Similarly, non-starch polysaccharide coatings, such as COLAL-PRED® (sodium prednisolone sodium sulfonate) systems, rely on enzyme-specific enzymatic degradation of colon bacteria.Citation25 Another approach involves the use of a specific pH-soluble coating that relies on the pH gradient of the gastrointestinal tract to activate the release, whereas the time-dependent delivery system uses GI transit time as a guide to activate drug release. Recent drug advances have applied nanotechnology to oral dosage form designs to overcome the limitations of conventional formulationsCitation26–Citation29. Polylactide-polyglycolide (PLGA) nanoparticles, as a biodegradable material, can be useful tools to overcome these problems. siRNAs or shRNA-plasmids can be encapsulated into PLGA nanoparticles which could protect loaded nucleic acids from enzymatic degradation by a three-phase degradation process.Citation30 On the other hand, drugs can be effectively delivered to the inflamed region due to the adhesion of nanoparticles to exert a better therapeutic effect.Citation31–Citation33 For example, the high efficiency of PLGA-5-ASA nanoparticles on targeted releasing after oral administration has been confirmed.Citation34 Here, we applied oral PLGA-nanoparticles to the delivery of shRNA plasmids of the novel IBD target SNX10 to achieve therapeutic effects with new mechanisms.
Toll-like receptors (TLRs) play an important role in innate immunity, and when TLRs recognize ligands, they dimerize to recruit downstream signaling molecules. The TIR domain of myeloid-differentiation factor 88 (MyD88) binds to the TIR of TLRs and its N-terminus can transmit signals downstream through the death domain (DD), eventually activating NF-κB and affecting the production of related inflammatory cytokines.Citation35 And TLR mRNA levels are positively correlated with inflammatory activity.Citation36 Under normal conditions, the gut flora is recognized by TLR2 and TLR4 and controls IEC homeostasis.Citation37 But in IBD, they tend to worsen the situation. Previous studies have also shown that IBD induced by IL-10 deletion is dependent on the TLR/MyD88 signaling pathway and that the loss of TLR2 and TLR4 is beneficial in reducing the DSS-induced colitis. Some clinical studies have shown that elevated mRNA and protein levels of TLR2 were detected in the cytoplasm of UC patients;Citation38,Citation39 TLR4 expression was highly up-regulated in the ileum and rectum of UC patients and in the terminal ileum of CD patients.Citation40 Accumulating evidence suggests that TLR signaling is closely related to IBD.Citation41
Based on this background, we successfully prepared oral SNX10-shRNA plasmid (SRP) PLGA nanoparticles (NPs) and observed its curative effect on mouse colitis. We found that SRP-NPs could effectively inhibit the inflammatory reaction and tissue damage in mice with acute and chronic IBD. In intestinal epithelial cells, SRP-NPs could inhibit the phosphorylation of p38, down-regulate the expression of TLR2 and TLR4, and affect downstream signaling pathways, which might be the mechanisms that SRP-NPs could reduce excessive inflammation.
Materials and Methods
Cells and Mice
RAW264.7 and HCT-116 cells were purchased from the Cell Culture Collection of Chinese Academy of Sciences. RAW264.7 was cultured in DMEM containing 10% FBS and HCT-116 cells were cultured in 1640 medium with 10% FBS, 37°C, 5% CO2.C57BL/6 male mice (8 weeks old, 20 ± 2g of body weight) were purchased from Shanghai Biomodel Organism Science and Technology Development Co., Ltd., IL10−/- male mice (8 weeks old, 20 ± 2g body weight) were purchased from Jackson Laboratory, USA. Mice were maintained in the Animal Experiment Center of the School of Pharmacy, Fudan University. All animal procedures were performed following the Guide for the Care and Use of Laboratory Animals published by the National Institutes of Health (NIH) and were approved by the Ethics Committee of the Experimental Research, Shanghai Medical College, Fudan University. All efforts were made to reduce the number of animals used and to minimize animals’ suffering.
Construction of SNX10-shRNA Plasmid (SRP)
The interference sequence of SNX10 is S3. 5ʹ-CAGGGCTTGGAAGATTTCCTCAGAA. The vector information is GV102, hU6-MCS-CMV-GFP-SV40-Neomycin.
Evaluation of SRP Interference Efficiency
HCT-116 cells in the logarithmic growth phase were digested with 0.25% trypsin and passaged at a ratio of 1: 3 (RAW264.7 cells were blown down and passaged at the same ratio). The SRP was coated with Lipofectamine 3000 Reagent (Invitrogen) Transfection at 1: 1ratio, added to the attached cells, and incubated for 48 hours, cell RNA was extracted and used for qRT-PCR analysis.
Preparation of SRP-NPs
200 mg of PLGA was dissolved in 1 mL of dichloromethane, then mixed with 200 mL (500 ng/µL) of the plasmid-containing solution and dissolved it by sonication. Followed by adding 2 mL solution containing 1% PVA, 10% sucrose, and sonicated, then added to a solution containing 0.5% PVA, 10% sucrose, dissolved 5–10 min, and rotary evaporation to remove the organic solvent for 4 h. The final solution was centrifuged at 14,500 rpm for 45 min at 4°C, then frozen at −80°C for 4 h, and lyophilized for 48 h.
SRP-NPs Characterization Measurement
The PLGA-SNX10-shRNA particle size and distribution were detected by a Malvern Zetasizer 3000E laser particle size analyzer. SRP-NPs suspension was slurried on a glass slide, air dried, sprayed with gold, and then the nanoparticles were carefully observed with an electron microscope.
Determination of Encapsulation Efficiency and Drug Loading
The supernatant was collected when the nanoparticles were prepared, the volume of the solution was accurately determined, and the absorbance of the solution was measured by an ultraviolet spectrophotometer to obtain the content of the SRP-NPs free from the supernatant. Encapsulation efficiency was calculated according to the formula:
EN, entrapment efficiency; Cf, Free drug concentration; Ct, Total concentration.
The drug loading was calculated according to the formula:
LE, percentage of drug loaded in the liposomes; We, dose encapsulated in the liposomes; Wm, the total weight of the drug loaded liposomes.
Stability of Nanoparticles
The optimized SRP-NPs suspension prepared by centrifugation was centrifuged and the NPs pellets were dispersed in 50 mL phosphate buffer (PBS, pH 6.8, 7.0, 7.4, 7.6, 7.8) at different pHs respectively, then sit at room temperature for 0.5, 1, 1.5, 2, 3, 4, 5, 6, 7, 8, 9, 10, 12, 14, 16 days respectively. UV-Vis was used to detect the nanosuspension at different wavelengths of light transmittance. T/Ti% is the ratio of the transmittance of SRP nanoparticles measured at their respective pH to the transmittance of nanoparticles in deionized water.
Detection of SRP-NPs’ Efficiency in vivo and in vitro
Experiments were performed on human HCT-116 cells and murine RAW264.7 cells, respectively. The drugs were given according to the control group (1 mg/mL MP-NPs), low dose group (0.2 mg/mL SRP-NPs) and high dose group (1 mg/mL SRP-NPs). After 10 hours, the medium containing drug was replaced with normal medium for 48 h, and RNA was extracted for qRT-PCR to detect the expression of SNX10 at the mRNA level.
Twenty-four C57BL/6 mice were randomly divided into three groups: control group (intragastric administration of 3 mg/20g of MP-NPs), low dose group (1 mg/20g of SRP-NPs), and high dose group (3 mg/20g of SRP-NPs). After three consecutive days of administration, mice were sacrificed and the heart, spleens, lungs, liver, kidneys, small intestine, and colon were collected, and total RNA was extracted for qRT-PCR. The Ri/R ratio was calculated to evaluate the effects of oral SRP-NPs on different organs.
Establishment of Acute IBD Mouse Model
Forty C57BL/6 mice were randomly divided into 5 groups: normal group, disease model group, blank group, low dose group, and high dose group. 3 days prior to modeling, mice were treated by intragastric administration of 3 mg/20g/day of MP-NPs for mice in the blank group, 1 mg/20g/day of SRP-NPs for mice in the low dose group, and 3 mg/20g/day of SRP-NPs for mice in high dose group. All except the normal group were given a free drink of 2.5% (w/v) DSS solution for seven days. Mice were sacrificed by cervical dislocation, mice colon were collected.
Establishment of Chronic IBD Mouse Model
After gene identification, IL10−/- mice were divided into normal KO group, disease model group, blank control group, low dose group, and high dose group. From three days before the DSS modeling, mice were intragastrically treated by SRP-NPs at the above-mentioned concentrations and then fed by the diet containing Piroxicam (200 ppm) (except the control WT and the KO group) for 4 days, followed by normal diets with re-administration of SRP-NPs every 14 days after the first administration for 2 months before sacrificed.
Western Blot
An equal amount of proteins (30 μg) were separated on 10% of SDS-PAGE, then were transferred onto nitrocellulose membranes (Millipore). Membranes were blocked in 2% BSA-TBS buffer for 1 h, RT, then incubated with the desired primary antibody overnight at 4°C, followed by incubation with appropriate secondary antibodies for 1 hour at room temperature. Blots were developed using enhanced chemiluminescence (Yesen, Shanghai) and visualized with the GeneGnome XRQ (Syngene). The specific primary antibodies used in Western blotting analysis are as follows: TLR4 (1:1000, Abcam), TLR6 (1:1000, Abcam), TLR9 (1:1000, Abcam), SNX10 (1:500, Santa Cruz), MyD88 (1:1000, Abcam), IκB (1:1000, Abcam), p-p38 (1:1000, CST), β-actin (1:1000, Beyotime, China).
RT-qPCR
For RT-qPCR analysis, the cells or tissues were lysed in Trizol reagent to extract total RNA, followed by reverse transcription using HiScript II 1st Strand cDNA Synthesis Kit (Vazyme Biotech, Shanghai). Real-time PCR was performed by using ChamQ SYBR qPCR Master Mix (Vazyme Biotech, Shanghai). The profile of thermal cycling consisted of initial denaturation at 95°C for 2 min, and 40 cycles at 95°C for 15 s and 60°C for 30s. Primers were synthesized from Invitrogen. The specificity of each primer pair was confirmed by melting curve analysis and agarose-gel electrophoresis. β-actin was used as an internal control.
ELISA
The blood of mice was let stand for 2h, centrifuged for 15 min at 3000 rpm to collect serum. The samples above were analyzed of protein activity with ELISA kit as the manufacturer’s (DAKAWE) protocol.
Statistics
Statistical analyses were performed using SPSS 13 (Statistical Program for Social Sciences). The Independent-Samples t-test or nonparametric Mann–Whitney U-test were used to compare the two groups, and a one-way ANOVA followed by Bonferroni post hoc test for multiple comparisons was used. P < 0.05 was the significance level. All the numerical data presented are representative of at least 3 repeat experiments and are expressed as mean ± SEM.
Results
Preparation and Characterization of SRP-NPs
We designed an interference sequence (3.5ʹ-AGGGCTTGGAAGATTTCCTCAGAA) of human SNX10 mRNA and expressed it into the GV102 vector (hU6-MCS-CMV-GFP-SV40-Neomycin) (). The interference efficiency of SRP in human colon cancer epithelial cell line HCT116 and mouse mononuclear macrophage leukemia cell lines RAW264.7 were tested. The expression efficiency of SNX10 was 92.36 ± 6.88% in the Mock group and 21.39 ± 5.26% in the SRP interference group compared with that in the control group of HCT-116 cells, respectively. Compared with that in the control group in RAW264.7 cells, the efficiency of SNX10 expression was 94.42 ± 12.75% in the mock group, and 25.69 ± 4.87%in theSNX10-shRNA group (). In order to ensure that the SRP could be successfully delivered to the colon, we utilized a PLGA material that had adhesion to the intestinal mucosa to encapsulate the SRP into nanoparticles by using a double emulsion-solvent evaporation method, which is a well-established technique for fabricating drug-loaded nanoparticles. The inner aqueous phase (containing SRP) was added to the organic phase (containing PLGA) to form a first water/oil emulsion under sonication. We then added this first emulsion to the aqueous phase (containing PVA) and performed sonication to form a water/oil/water emulsion based on the Gibbs-Marangoni effect and a capillary breakup mechanism. Next, organic solvent (dichloromethane) was removed by evaporation to finally obtain PLGA micro/nanoparticles. The method could change the particle size by ultrasonic time, power, emulsifier concentration, and stirring speed. When the stirring speed reached 10,000 r · min−1 and the PVA concentration was 9%, the particle size could be controlled below 300 nm. The seal rate could reach more than 90%.Citation42
Figure 1 Experiments of preparation, characterization, and stability of SNX10-shRNA plasmids nanoparticles. (A) Schematic illustration of SRP-NPs. (B) After 48 h of transfection, the expression of SNX10 in RAW264.7 and HTC-116 cells was determined by qRT-PCR. Determination of (C) particle size, (D) PDI values, (E) ζ-potential, (F) entrapment efficiency, (G) drug loading of SRP-NPs at different weight ratios. (H) SEM photo of SRP-NPs at the weight ratio of 1 showed that the nanoparticles are spherical and uniform particle size of about 275 nm, consistent with the measured data by particle size analyzer. Bars=1000 nm. (I) The turbidity of the nanoparticles was measured by the turbid metric method to investigate the stability under different pH storage conditions. *P < 0.05, **P < 0.01, ***P < 0.001 vs control group, ns- not significant.
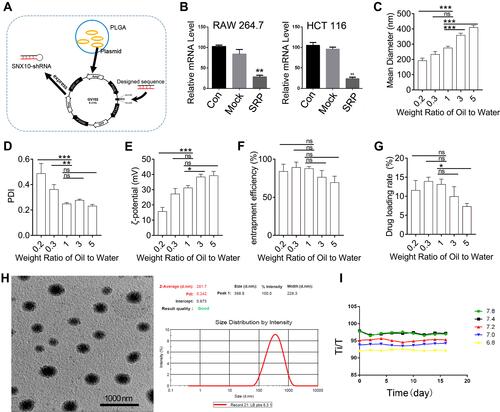
To optimize the ratio of oil to water, parameters of the nanoparticles in different ratios were measured (–). With the increase of the mass ratio of PLGA/water phase, the particle size, zeta potential, and drug loading increase, while the entrapment efficiency of drugs decreases. However, the loading of nanoparticles has a certain threshold. Once this threshold is exceeded, the nanoparticles will not completely encapsulate the drug, leading to little increase or even reduction in encapsulation efficiency and drug loading. Considering the particle size of nanoparticles, drug loading, encapsulation efficiency, and PDI, the NPs at weight ratio 1 was the most suitable choice. The prepared nanoparticles had a particle size of 275.2 ± 11.4 nm and a uniform particle size distribution. The diameters of the nanoparticles are concentrated at 265–285 nm and the zeta potential is 31.2 ± 1.5 mv. The entrapment efficiency of SRP was 87.6 ± 2.5% and drug loading was 13.1 ± 1.4%. SEM photo showed that SRP-NPs were spherical and had a uniform size at about 275 nm (), which was consistent with the previous test data.
The turbid metric method was used to investigate the stability of nanoparticles under different pH conditions by measuring the turbidity of the nanoparticles. Graphs were drawn by measuring the relative light transmittance (T/Ti%) of the nanoparticles at different pH conditions within 16 days. In the pH 6.8–7.8, the T/Ti value of SRP did not change much in 16 days (), indicating that SRP can be stored in the above pH conditions.
SRP-NPs Reduce Expressions of SNX10 in vitro and in vivo
SRP-NPs were applied to HCT-116 and RAW264.7 cells and their interference efficiency was tested. The results showed that compared with that in the control group, the expression of SNX10 mRNA in HCT-116 cells was decreased by relatively 48% and 95% in the low-dose and high-dose groups, respectively. In RAW264.7 cells, SNX10 mRNA expression was decreased by 52% in the low-dose group and by 97% in the high-dose group. These results indicated that SRP-NPs could suppress the expression of SNX10 in both epithelioid cells and macrophages in a dose-dependent manner ().
Figure 2 Interference efficiency of SRP-NPs in vivo and in vitro. (A) Experiments were performed on human HCT-116 cells and murine RAW264.7 cells, respectively. Con means the control group (1 mg/mL MP-NPs), L means the low dose group (0.2 mg/mL SRP-NPs) and H means the high dose group (1 mg/mL SRP-NPs). The level of SNX10 mRNA was detected by qRT-PCR after 48 h of treatment. (B) HCT116 cells were incubated with PBS, naked SRP (containing an equivalent amount of SRP-NPs in the high dose group), and SRP-NPs for 1, 8 h, and then cellular uptake of TOTO-3-labeled nanoparticles was determined by a confocal microscope. (C) After 3 days of oral administration, the expression of SNX10 of colon mucosa was determined by qRT-PCR. Con (control groups, 3 mg/20 g, MP-NPs); L (low dose group, 0.2 mg/20 g, SRP-NPs); H (high dose group,3 mg/20g, SRP-NPs). *P < 0.05, **P < 0.01, ***P < 0.001 vs the control group or L group. (D) Mice (n=16) were randomly divided into two groups. The control group received 3 mg/20 g of MP-NPs, and the drug group received 3 mg/20 g of SRP-NPs. After 3 days of oral administration, the expression of SNX10 of several organs was determined by qRT-PCR. Ri/R: Value of SNX10 mRNA relative to the control group.**P < 0.01 vs control group.
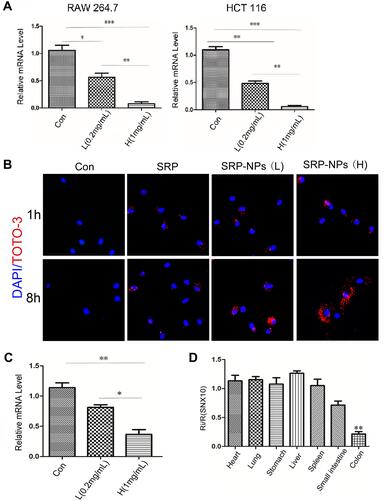
To monitor cellular uptake of SRP-NPs in HCT116, we employed TOTO-3 red fluorescence dyed plasmid for visualization of intracellular distribution and localization of plasmid and gene carriers. The results showed that, in comparison with untreated control groups, cells now with SRP only displayed minimal TOTO-3 fluorescence signal. However, cells that were subjected to high-dose SRP-NPs exhibited much more red fluorescence internalization, indicating efficient cellular uptake of nanoparticles ().
In vivo, SNX10 mRNA expression of mice colonic mucosa in the high-dose group decreased by about 78%, indicating that high-dose SRP-NPs significantly inhibited SNX10 expression in the colon (), but not in the lungs, stomach, liver, and spleen. There was no significant change of Ri/R in the small intestine which was about 0.7 ().
SRP-NPs is Effective in Acute IBD Models
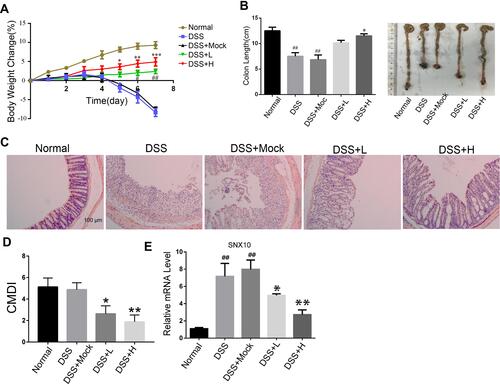
To observe the effects of SRP-NPs on IBD, we treated mice with DSS and administered blank, low-dose, and high-dose drugs respectively. As shown in , the mice body weight changes in the drug group were significantly higher than that in the model group and the blank group, and the high-dose group was higher than the low-dose group. This protective effect was also displayed in the colon length of mice (). The proximal colon was harvested for histopathology (). According to previous studies,Citation43 colon mucosa damage index (CMDI) scoring was assessed in a blinded fashion. Compared with the normal group, the histopathological characteristics, such as damage of epithelial cells and recesses, infiltration of a large number of inflammatory cells, were obviously observed in the model group and the blank group, but in the drug groups, those damages were significantly relieved (). The efficiency of SRP-NPs on SNX10 mRNA expression in colon mucosa was confirmed by RT-qPCR. As shown in , SNX10 mRNA was obviously reduced in SRP-NP treated groups in a does dependent manner.
Figure 3 The effects of SRP-NPs on an acute mouse model of IBD. (A) 40 mice were randomly divided into five groups with eight mice in each group: Normal, DSS, DSS + Mock (3 mg/20 g MP-NPs), DSS + L (1 mg/20 g SRP-NPs), and DSS + H group (3 mg/20 g SRP-NPs). After 3 days of continuous administration as described above, 2.5% DSS was administered. Weight loss was measured daily and expressed as the average percentage of initial body weight ± SEM. (B) The length of colons from mice was measured on day 7 after euthanasia. (C) Representative histological images of mice on the 7th day were compared. (D) Colon mucosa damage index (CMDI) scoring was assessed in a blinded fashion. (E) The mRNA expression of colon mucosa SNX10 was measured by qRT-PCR. *P<0.05, **P < 0.01, ***P < 0.001 vs DSS+Mock group; #P < 0.05, ##P < 0.01vs Normal group; n = 8 in each group.
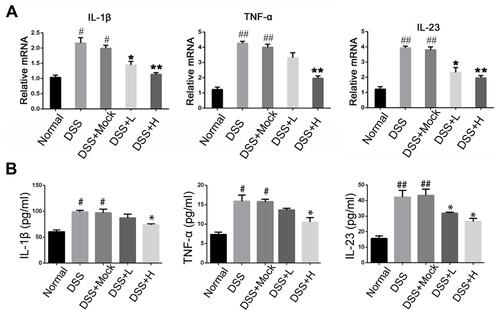
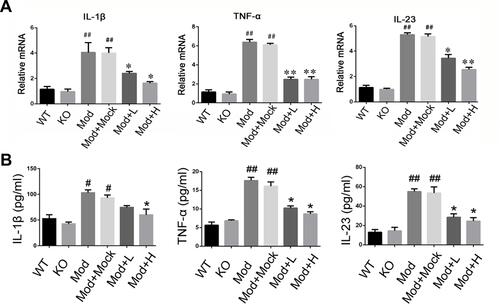
We examined mRNA levels of inflammatory cytokines and found it significantly increased in the IBD model, and IL-1β, TNF-α, and IL-23 expression were decreased in the drug-treated group (). ELISA tests on peripheral blood serum yielded similar results (). These data suggest that SRP-NPs can effectively suppress the systemic inflammatory response caused by acute colitis with dose-dependence.
Figure 4 Levels of inflammatory cytokines in acute IBD models. (A) The mRNA expression of IL-1β, TNF-α, and IL-23 of colon mucosa were measured by qRT-PCR. (B) The concentration of IL-1β, TNF-α, and IL-23in the serum of mice were measured by ELISA.*P<0.05, **P < 0.01vs DSS+Mock group; #P < 0.05, ##P < 0.01vs Normal group; n = 8 in each group.
SRP-NPs is Effective in Chronic IBD Mice
Mice deficient of IL-10 develop chronic colitis under conventional conditions. However, the incidence of spontaneous colitis in IL-10−/-mice on a colitis resistant C57BL/6J inbred background is low and prolonged. Here, we use piroxicam to accelerate the course of IL-10-deficient mice, a method that has been reported to resemble human IBD with regard to its heterogeneous pathogenesis and the importance of IL-10 deficiency and decreased epithelial integrity in disease development.
We administered SRP-NPs orally to mice on the basis of this model and conducted measurements similar to the previous model. The results showed that, compared with the model group, the expression of SNX10 mRNA in the drug group mice was effectively inhibited (). In addition, we found significant increases of SXN10 mRNA levels in the model group compared with the control group (), indicating that colonic inflammatory environment can up-regulate the expression of SNX10. In body weight change, colon length, pathological analysis, tissue and peripheral blood inflammatory cytokines, all the administration groups showed relief of the conditions (– and and ). In the chronic IBD model, SRP-NPs also played a therapeutic role in a dose-dependent manner.
Figure 5 The effects of SRP-NPs on a chronic mouse model of IBD. (A) 40 IL10−/-mice were randomly divided into five groups with eight mice in each group: KO, Mod, Mod + Mock (3 mg/20 g MP-NPs), Mod + L (1 mg/20 g SRP-NPs), and Mod + H group (3 mg/20 g SRP-NPs).After 3 days of continuous drug administration as described above, piroxicam (200 ppm) was added to the diet for 4 days. Weight loss was measured for 60 days and expressed as the average percentage of initial body weight ± SEM. (B) The length of colons from mice was measured at day 60 after euthanasia. (C) Representative histological images of mice on the 60th day were compared. (D) Colon mucosa damage index scoring (CMDI) was assessed in a blinded fashion. (E) The mRNA expression of SNX10 of colon mucosa was measured by qRT-PCR. *P<0.05, **P < 0.01, ***P < 0.001 vs Mod+Mock group; ##P < 0.01, ###P < 0.001 vs KO group; n = 8 in each group.
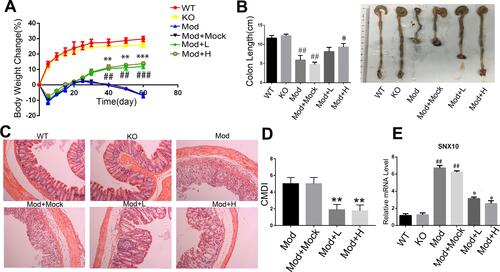
Figure 6 Levels of inflammatory cytokines in chronic IBD models. (A) The mRNA expression of IL-1β, TNF-α, and IL-23 of colon mucosa were measured by qRT-PCR. (B) The concentration of IL-1β, TNF-α, and IL-23 in the serum of mice were measured by ELISA. *P<0.05, **P < 0.01, vs DSS+Mock group; #P < 0.05, ##P < 0.01vs Normal group; n = 8 in each group.
SRP-NPs Down-Regulate the TLR Signaling Pathway
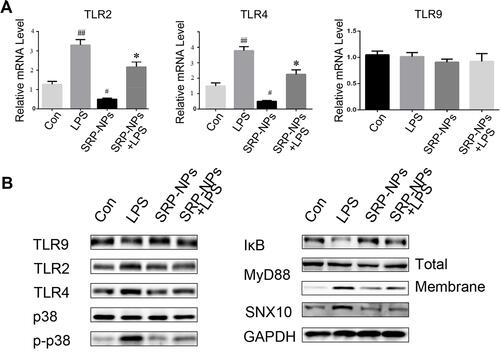
We conducted a preliminary investigation of the mechanism of SRP-NPs’ therapeutic effect on IBD. As innate immune receptors, TLRs, when activated by pathogen-associated molecular patterns (PAMPs), evoke a potent pro-inflammatory response involving rapid induction of MyD88 and NF-κB-dependent signaling pathways, and the resultant expression of a broad array of pro-inflammatory adhesion molecules, chemokines, and cytokines.Citation44 Previous studies have also shown that IBD induced by IL10 deletion is dependent on the TLR/MyD88 signaling pathwayCitation45 and that the loss of TLR2 and TLR4 is beneficial in reducing DSS-induced colitis.Citation46 Therefore, we speculated that SRP-NPs might improve IBD by downregulating the TLR signaling pathway. LPS (1 μg/mL) was added into HCT-116 cells medium to mimic the inflammatory environment. p38 kinase pathways were involved in TLR2 mRNA up-regulation.Citation47 The expression of TLR2 and TLR4 was significantly reduced, which led to a decrease in recruitment of Myd88 and an increase in intracellular IκB ( and ). A previous studyCitation47 has shown that increased TLR2 mRNA expression in stimulated T cells can be antagonized by MAP kinase inhibitors.Citation47 And in our study, we also found that phosphorylation of p38 was reduced, which might contribute to the decreased expression of TLR2 and TLR4.
Figure 7 SRP-NPs down-regulate the TLR signaling pathway. (A) HCT116 cells were treated with LPS at 1 ug/mL for 0.5 h and SRP at 500 μg/mL. After 6 hours, the mRNA expression of TLR2, TLR4, and TLR9 of cells was measured by qRT-PCR. #P<0.05, ##P<0.01 vs control group; *P<0.05 vs LPS. (B) The above-treated cells were treated in radio-immunoprecipitation assay buffer (RIPA), and total or membrane proteins were extracted for Western blot analysis.
Discussion
IBD is a chronic recurrent disease of unknown etiology, with an increasing worldwide incidence year by year. But so far the treatment of IBD still depends on surgical resection with a high recurrence rate and lacks completely curable drugs. Clinical drug treatment usually uses salicylic acid inhibitors, immunosuppressive agents, and biological agents. 5-ASA lacks treatment for CD and causes side effects such as rash and neutropenia; Corticosteroids are only useful in the treatment of acute attacks but ineffective in maintaining remission on UC; Immunosuppressive agents such as Azathioprine and 6-mercaptopurine effect slowly, and Cyclosporin A shows quick effects but is prone to cause serious adverse reactions. Since the introduction of biologics, therapeutic standards have shifted from non-specific immune modulators to signal-based anti-inflammatory therapies.Citation48,Citation49 TNF inhibitors and anti-integrins have initiated a new therapeutic era in the treatment of flares and maintenance of clinical remission, but they are effective only in some certain subgroups of patients.Citation50 Indeed, a significant number of patients relapse after cessation of treatment.Citation51 Therefore, looking for an alternative therapeutic target beyond TNF has become a central issue in the current research. At present, there are some drugs that target IL-6, IL-23, JAK1/JAK3, SMAD7, α4/β7, and S1P1/S1P5 in clinical research.
We have been concerned about the SNX10, a PX domain-only phosphoinositide-binding protein, which has been proved to be associated with a variety of autoimmune diseases. A study has shown that deletion of SNX10 can effectively prevent DSS-induced inflammation and pathological damage in the colon of mice.Citation18 We tried to find a direct and effective means of interference to prove the potential of SNX10 as an IBD target.
RNAi technology has been widely used in gene therapy at present, which mainly achieves the effect of treating diseases by inhibiting the over-expression of pathogenic genes while avoiding the adverse effects of using antibodies directed against signaling pathways. Compared with the previous antisense nucleotide technology, RNAi displayed the advantages of high accuracy, efficiency, and small side effects. RNAi often only requires a small amount of RNA, which can effectively and specifically interfere with the expression of a certain gene in an organism. There are already some drugs that use RNAi technology under clinical trials, for example, siRNA-027 can specifically inhibit the expression of sFlt-1 mRNA to inhibit the development of new blood vessels in the vessels.Citation52
RNAi can be achieved by constructing the SNX10-shRNA plasmid (SRP), but it requires a carrier that can deliver the SRP precisely to the colon to avoid the fate of degradation elsewhere in the GI tract and the potential threat of systemic effects. Here, we chose PLGA, an orally administered bioadhesive material, to prepare the SRP-NPs by double emulsion volatilization. PLGA nanoparticles have a certain dependence on the particle size, so we select the best conditions through a series of measurements. The final nanoparticles with a diameter of about 275 nm, displayed a good PDI, encapsulation efficiency, and drug loading. In both human and murine cells, SRP effectively interferes with SNX10 mRNA expression. In normal mice, SRP-NPs efficiently interfered with SNX10 mRNA expression only in the colon rather than other organs, demonstrating that SRP was effectively delivered to the colon by the nanoparticles.
In this study, we mainly evaluated the effects of SRP on two colitis models, the acute IBD induced by DSS and the chronic IBD induced by IL-10 knockout. Our results showed that SRP-NPs could alleviate the mice body weight loss and colon pathological damage in both models, and down-regulate colonic mucosa mRNA and peripheral blood concentration of IL-1β, IL-23, and TNF-α.
The therapeutic mechanism of SRP-NPs is worthy of discussion. The aforementioned mention that SXN10 can affect IBD by affecting the polarization of macrophages may not be suitable here. We hypothesize that SRP-NPs exert anti-inflammatory effects through signaling pathways that exist in epithelial cells. Toll-like receptors in intestinal epithelial cells play an important role in IBD as PAMPs receptors, and TLR-MyD88-NF-κB is a classic signaling pathway for inflammation. Through the stimulation of LPS, we found that SRP-NPs can directly affect the expression of TLR2 and TLR4 in the HCT-116 cells, consistent with a study that pointed out that MAP kinase inhibitors can reduce the expression of TLR2 mRNA.Citation47
Conclusion
Oral nanoparticles of SNX10-shRNA plasmid display dominant efficiency of SNX10 RNA interference in the colon and ameliorate mouse colitis via TLR signaling pathway. SNX10 is a new target for IBD treatment and nanoparticles of SNX10-shRNA plasmid might be a promising treatment option for IBD.
Acknowledgments
This study was supported by the following grants: National Natural Science Foundation of China General Program (No. 81973523, 81773744, and 81971488); Natural Science Research Project of Jiangsu Province Major Program (17KJA360002); The Science and Technology Development Fund, Macau SAR (file No.: 130/2017/A3, 0099/2018/A3).
Disclosure
The authors report no conflicts of interest for this work.
References
- Molodecky NA, Soon IS, Rabi DM, et al. Increasing incidence and prevalence of the inflammatory bowel diseases with time, based on systematic review. Gastroenterology. 2012;142(1):46–54 e42; quiz e30. doi:10.1053/j.gastro.2011.10.001
- Gong W, Lv N, Wang B, et al. Risk of ulcerative colitis-associated colorectal cancer in China: a multi-center retrospective study. Dig Dis Sci. 2012;57(2):503–507. doi:10.1007/s10620-011-1890-9
- Burton PR, Clayton DG, Cardon LR; Wellcome Trust Case Control, C.; Australo-Anglo-American Spondylitis, C. Association scan of 14,500 nonsynonymous SNPs in four diseases identifies autoimmunity variants. Nat Genet. 2007;39(11):1329–1337.
- Hanauer SB. Medical therapy for ulcerative colitis 2004. Gastroenterology. 2004;126(6):1582–1592. doi:10.1053/j.gastro.2004.02.071
- Nakamura K, Honda K, Mizutani T, Akiho H, Harada N. Novel strategies for the treatment of inflammatory bowel disease: selective inhibition of cytokines and adhesion molecules. World J Gastroenterol. 2006;12(29):4628–4635. doi:10.3748/wjg.v12.i29.4628
- Pescovitz MD. Daclizumab: humanized monoclonal antibody to the interleukin-2 receptor. Expert Rev Clin Immunol. 2005;1(3):337–344. doi:10.1586/1744666X.1.3.337
- Hara S, Kiyokawa E, Iemura S, et al. The DHR1 domain of DOCK180 binds to SNX5 and regulates cation-independent mannose 6-phosphate receptor transport. Mol Biol Cell. 2008;19(9):3823–3835. doi:10.1091/mbc.e08-03-0314
- Lunn ML, Nassirpour R, Arrabit C, et al. A unique sorting nexin regulates trafficking of potassium channels via a PDZ domain interaction. Nat Neurosci. 2007;10(10):1249–1259. doi:10.1038/nn1953
- Kurten RC, Cadena DL, Gill GN. Enhanced degradation of EGF receptors by a sorting nexin, SNX1. Science. 1996;272(5264):1008–1010. doi:10.1126/science.272.5264.1008
- Bergant M, Banks L. SNX17 facilitates infection with diverse papillomavirus types. J Virol. 2013;87(2):1270–1273. doi:10.1128/JVI.01991-12
- Aker M, Rouvinski A, Hashavia S, et al. An SNX10 mutation causes malignant osteopetrosis of infancy. J Med Genet. 2012;49(4):221–226. doi:10.1136/jmedgenet-2011-100520
- Strzepa A, Szczepanik M. IL-17-expressing cells as a potential therapeutic target for treatment of immunological disorders. Pharmacol Rep. 2011;63(1):30–44. doi:10.1016/S1734-1140(11)70396-6
- Zhu C, Morse LR, Battaglino RA. SNX10 is required for osteoclast formation and resorption activity. J Cell Biochem. 2012;113(5):1608–1615. doi:10.1002/jcb.24029
- Chen Y, Wu B, Xu L, et al. A SNX10/V-ATPase pathway regulates ciliogenesis in vitro and in vivo. Cell Res. 2012;22(2):333–345. doi:10.1038/cr.2011.134
- You Y, Li WZ, Zhang S, et al. SNX10 mediates alcohol-induced liver injury and steatosis by regulating the activation of chaperone-mediated autophagy. J Hepatol. 2018;69(1):129–141. doi:10.1016/j.jhep.2018.01.038
- Zhou C, Wang Y, Peng J, Li C, Liu P, Shen X. SNX10 plays a critical role in MMP9 secretion via JNK-p38-ERK signaling pathway. J Cell Biochem. 2017;118(12):4664–4671. doi:10.1002/jcb.26132
- Zhou C, You Y, Shen W, et al. Deficiency of sorting nexin 10 prevents bone erosion in collagen-induced mouse arthritis through promoting NFATc1 degradation. Ann Rheum Dis. 2016;75(6):1211–1218. doi:10.1136/annrheumdis-2014-207134
- You Y, Zhou C, Li D, et al. Sorting nexin 10 acting as a novel regulator of macrophage polarization mediates inflammatory response in experimental mouse colitis. Sci Rep-Uk. 2016;6.
- Neurath MF, Travis SPL. Mucosal healing in inflammatory bowel diseases: a systematic review. Gut. 2012;61(11):1619–1635. doi:10.1136/gutjnl-2012-302830
- Zhang S, Yang Z, Bao W, et al. SNX10 (sorting nexin 10) inhibits colorectal cancer initiation and progression by controlling autophagic degradation of SRC. Autophagy. 2020;16(4):735–749. doi:10.1080/15548627.2019.1632122
- Le Y, Zhang S, Ni J, et al. Sorting nexin 10 controls mTOR activation through regulating amino-acid metabolism in colorectal cancer. Cell Death Dis. 2018;9(6):666. doi:10.1038/s41419-018-0719-2
- Lieberman J, Song E, Lee SK, Shankar P. Interfering with disease: opportunities and roadblocks to harnessing RNA interference. Trends Mol Med. 2003;9(9):397–403. doi:10.1016/S1471-4914(03)00143-6
- Yang LB, Chu JS, Fix JA. Colon-specific drug delivery: new approaches and in vitro/in vivo evaluation. Int J Pharm. 2002;235(1–2):1–15. doi:10.1016/S0378-5173(02)00004-2
- Sandborn MJ. Rational selection of oral 5-aminosalicylate formulations and prodrugs for the treatment of ulcerative colitis. Am J Gastroenterol. 2002;97(12):2939–2941. doi:10.1111/j.1572-0241.2002.07092.x
- Hanauer SB, Sparrow M. COLAL-PRED Alizyme. Curr Opin Investig Drugs. 2004;5(11):1192–1197.
- Mandal H, Katiyar SS, Swami R, et al. epsilon-Poly-L-Lysine/plasmid DNA nanoplexes for efficient gene delivery in vivo. Int J Pharmaceut. 2018;542(1–2):142–152. doi:10.1016/j.ijpharm.2018.03.021
- Martins JP, Das Neves J, de la Fuente M. The solid progress of nanomedicine. Drug Deliv Transl Res. 2020;10(3):726–729. doi:10.1007/s13346-020-00743-2
- Abeer MM, Rewatkar P, Qu Z, et al. Silica nanoparticles: A promising platform for enhanced oral delivery of macromolecules. J Control Release. 2020;326:544–555. doi:10.1016/j.jconrel.2020.07.021
- Steed KP, Hooper G, Monti N, Benedetti MS, Fornasini G, Wilding IR. The use of pharmacoscintigraphy to focus the development strategy for a novel 5-ASA colon targeting system (“TIME CLOCK (R)” system). J Control Release. 1997;49(2–3):115–122. doi:10.1016/S0168-3659(97)00062-X
- Danhier F, Ansorena E, Silva JM, Coco R, Le Breton A, Preat V. PLGA-based nanoparticles: an overview of biomedical applications. J Control Release. 2012;161(2):505–522. doi:10.1016/j.jconrel.2012.01.043
- Elzatahry AA, Eldin MSM, Soliman EA, Hassan EA. Evaluation of alginate-chitosan bioadhesive beads as a drug delivery system for the controlled release of theophylline. J Appl Polym Sci. 2009;111(5):2452–2459. doi:10.1002/app.29221
- Pujara N, Wong KY, Qu Z, et al. Oral delivery of beta-lactoglobulin-nanosphere-encapsulated resveratrol alleviates inflammation in winnie mice with spontaneous ulcerative colitis. Mol Pharm. 2020. doi:10.1021/acs.molpharmaceut.0c00048
- Qu Z, Wong KY, Moniruzzaman M, et al. One-pot synthesis of ph-responsive eudragit-mesoporous silica nanocomposites enable colonic delivery of glucocorticoids for the treatment of inflammatory bowel disease. Adv Ther-Germany. 2020;2000165. doi:10.1002/adtp.202000165
- Mahajan N, Sakarkar D, Manmode A, Pathak V, Ingole R, Dewade D. Biodegradable nanoparticles for targeted delivery in treatment of ulcerative colitis. Adv Sci Lett. 2011;4(2):349–356. doi:10.1166/asl.2011.1247
- Hacker H, Redecke V, Blagoev B, et al. Specificity in Toll-like receptor signalling through distinct effector functions of TRAF3 and TRAF6. Nature. 2006;439(7073):204–207. doi:10.1038/nature04369
- Sanchez-Munoz F, Fonseca-Camarillo G, Villeda-Ramirez MA, et al. Transcript levels of toll-like receptors 5, 8 and 9 correlate with inflammatory activity in ulcerative colitis. BMC Gastroenterol. 2011;11:138. doi:10.1186/1471-230X-11-138
- De Jager PL, Franchimont D, Waliszewska A, et al. The role of the Toll receptor pathway in susceptibility to inflammatory bowel diseases. Genes Immun. 2007;8(5):387–397. doi:10.1038/sj.gene.6364398
- Tan Y, Zou KF, Qian W, Chen S, Hou XH. Expression and implication of toll-like receptors TLR2, TLR4 and TLR9 in colonic mucosa of patients with ulcerative colitis. J Huazhong Univ Sci Technolog Med Sci. 2014;34(5):785–790. doi:10.1007/s11596-014-1353-6
- Fan Y, Liu B. Expression of Toll-like receptors in the mucosa of patients with ulcerative colitis. Exp Ther Med. 2015;9(4):1455–1459. doi:10.3892/etm.2015.2258
- Frolova L, Drastich P, Rossmann P, Klimesova K, Tlaskalova-Hogenova H. Expression of Toll-like receptor 2 (TLR2), TLR4, and CD14 in biopsy samples of patients with inflammatory bowel diseases: upregulated expression of TLR2 in terminal ileum of patients with ulcerative colitis. J Histochem Cytochem. 2008;56(3):267–274. doi:10.1369/jhc.7A7303.2007
- Erridge C, Duncan SH, Bereswill S, Heimesaat MM. The induction of colitis and ileitis in mice is associated with marked increases in intestinal concentrations of stimulants of TLRs 2, 4, and 5. PLoS One. 2010;5:2. doi:10.1371/journal.pone.0009125
- Zhang XQ, Dahle CE, Baman NK, Rich N, Weiner GJ, Salem AK. Potent antigen-specific immune responses stimulated by codelivery of CpG ODN and antigens in degradable microparticles. J Immunother. 2007;30(5):469–478. doi:10.1097/CJI.0b013e31802fd8c6
- Siegmund B, Fantuzzi G, Rieder F, et al. Neutralization of interleukin-18 reduces severity in murine colitis and intestinal IFN-gamma and TNF-alpha production. Am J Physiol Regul Integr Comp Physiol. 2001;281(4):R1264–73. doi:10.1152/ajpregu.2001.281.4.R1264
- Levin A, Shibolet O. Toll-like receptors in inflammatory bowel disease-stepping into uncharted territory. World J Gastroentero. 2008;14(33):5149–5153. doi:10.3748/wjg.14.5149
- Rakoff-Nahoum S, Hao L, Medzhitov R. Role of toll-like receptors in spontaneous commensal-dependent colitis. Immunity. 2006;25(2):319–329. doi:10.1016/j.immuni.2006.06.010
- Heimesaat MM, Fischer A, Siegmund B, et al. Shift towards pro-inflammatory intestinal bacteria aggravates acute murine colitis via Toll-like receptors 2 and 4. PLoS One. 2007;2(7):e662. doi:10.1371/journal.pone.0000662
- Matsuguchi T, Takagi K, Musikacharoen T, Yoshikai Y. Gene expressions of lipopolysaccharide receptors, toll-like receptors 2 and 4, are differently regulated in mouse T lymphocytes. Blood. 2000;95(4):1378–1385. doi:10.1182/blood.V95.4.1378.004k08_1378_1385
- Coskun M, Steenholdt C, de Boer NK, Nielsen OH. Pharmacology and optimization of thiopurines and methotrexate in inflammatory bowel disease. Clin Pharmacokinet. 2016;55(3):257–274. doi:10.1007/s40262-015-0316-9
- Nielsen OH, Coskun M, Steenholdt C, Rogler G. The role and advances of immunomodulator therapy for inflammatory bowel disease. Expert Rev Gastroenterol Hepatol. 2015;9(2):177–189. doi:10.1586/17474124.2014.945914
- Olesen CM, Coskun M, Peyrin-Biroulet L, Nielsen OH. Mechanisms behind efficacy of tumor necrosis factor inhibitors in inflammatory bowel diseases. Pharmacol Ther. 2016;159:110–119. doi:10.1016/j.pharmthera.2016.01.001
- Torres J, Boyapati RK, Kennedy NA, Louis E, Colombel JF, Satsangi J. Systematic review of effects of withdrawal of immunomodulators or biologic agents from patients with inflammatory bowel disease. Gastroenterology. 2015;149(7):1716–1730. doi:10.1053/j.gastro.2015.08.055
- Check E. A crucial test. Nat Med. 2005;11(3):243–244. doi:10.1038/nm0305-243
