 ?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.
?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.Abstract
Background
The success of gene transfection is largely dependent on the development of a vehicle or vector that can efficiently deliver a gene to cells with minimal toxicity.
Methods
A liver cancer-targeted specific peptide (FQHPSF sequence) was successfully synthesized and linked with chitosan-linked polyethylenimine (CP) to form a new targeted gene delivery vector called CPT (CP/peptide). The structure of CPT was confirmed by 1H nuclear magnetic resonance spectroscopy and ultraviolet spectrophotometry. The particle size of CPT/ DNA complexes was measured using laser diffraction spectrometry and the cytotoxicity of the copolymer was evaluated by methylthiazol tetrazolium method. The transfection efficiency evaluation of the CP copolymer was performed using luciferase activity assay. Cellular internalization of the CP/DNA complex was observed under confocal laser scanning microscopy. The targeting specificity of the polymer coupled to peptide was measured by competitive inhibition transfection study. The liver targeting specificity of the CPT copolymer in vivo was demonstrated by combining the copolymer with a therapeutic gene, interleukin-12, and assessed by its abilities in suppressing the growth of ascites tumor in mouse model.
Results
The results showed that the liver cancer-targeted specific peptide was successfully synthesized and linked with CP to form a new targeted gene delivery vector called CPT. The composition of CPT was confirmed and the vector showed low cytotoxicity and strong targeting specificity to liver tumors in vitro. The in vivo study results showed that interleukin-12 delivered by the new gene vector CPT/DNA significantly enhanced the antitumor effect on ascites tumor-bearing imprinting control region mice as compared with polyethylenimine (25 kDa), CP, and other controls, which further demonstrate the targeting specificity of the new synthesized polymer.
Conclusion
The synthesized CPT copolymer was proven to be an effective liver cancer-targeted vector for therapeutic gene delivery, which could be a potential candidate for targeted cancer gene therapy.
Introduction
The basic concept underlying gene therapy is that human disease may be treated by the transfer of a therapeutic gene into the targeted cells or organs with the consequent expression of the transgene or inhibition production of a target protein.Citation1 Gene therapy has gained significant attention over the past two decades as a potential method for treating genetic disorders,Citation2,Citation3 as well as an alternative method to traditional chemotherapy used in treating cancer.Citation4 To a great extent the success of gene transfection is dependent on the development of vectors that can efficiently deliver a gene to cells with minimum toxicity. Initial research concentrated on using viral carriers, including both retroviruses and adenoviruses, as these vectors exhibited high efficiency at delivering both DNA and ribonucleic acid to numerous cell lines.Citation5 Nonetheless, fundamental problems associated with viral vector systems, including toxicity, immunogenicity, potential for oncogenesis due to chromosomal integration, and limitations with respect to scale-up procedures, encouraged the researchers to transfer interest to nonviral vectors for its lower immunogenicity, relatively ease of production, and fewer safety concerns.Citation6,Citation7
Chitosans, a family of linear binary polysaccharides, have been proposed as biocompatible alternative cationic polymers that are suitable for nonviral gene delivery.Citation8 However, this gene delivery system has a significant limitation owing to its low transfection efficiency. One of the primary causes of poor gene delivery efficiency is the insufficient release of chitosans from endosomes into the cytoplasm.Citation9 Another nonviral vector, polyethylenimine (PEI), is regarded to be the most effective cationic polymer for gene delivery.Citation10 Its high proton-buffering capacity results in rapid osmolysis of the endosomes, and the PEI/DNA complexes escape into the cytosol and are subsequently transported into the nucleus.Citation11 Nonetheless, PEI is also associated with dose-dependent toxicity, especially at high molecular weight, and its long-term safety has been a main concern since it is nonbiodegradable. It has, therefore, not yet been used in human studies.Citation12,Citation13 In previous studies, a chitosan/DNA/PEI complex was obtained through incubation and chemical synthesis.Citation14,Citation15 In the initial study, chitosan, DNA, and PEI were incubated by adjusting the nitrogen/phosphorus (N/P) ratio, which is defined as the molar ratio of the amine groups in the copolymer to the phosphate groups in the DNA. The complex showed high transfection efficiency and low toxicity, but the slight PEI/ DNA complex in the solution can’t be removed. The PEI used in the incubation was 25 kDa, which, at this molecular weight, is not biodegradable and may cause safety issues. In order to resolve these problems, a new complex, chitosan-linked PEI (CP), was synthesized in the second step by using the low molecular weight of PEI (1200 Da). The CP/DNA vectors not only had the advantage of both high transfection efficiency and low toxicity, but also significantly extended the transfection time. The tumor suppressive activity was also confirmed by CP-carried CCL22 gene using an ex vivo experiment. However, as such carriers and plasmid DNA complexes are taken up by cells in a nonspecific manner, the in vivo use is restricted by the occurrence of nonspecific side effects. Therefore, there is a need to develop targeted carriers, which can control tissue disposition and reduce the cytotoxicity of aggregation in normal cells when the carriers enter bodies. Moreover, the delivery to the target cell could help in reducing the plasmid DNA dose. Synthetic nonviral vectors provide flexibility in formulation design and can be tailored to interact efficiently with DNA cargo and the specific route of vector administration, and can enhance delivery to specific tissues or cells through the incorporation of a targeting ligand.
During the past decades several targeting complexes have been explored by researchers. Park et al prepared modified galactosylated chitosan/DNA complexes, and these systems could efficiently transfect liver cells and express asialogly-coprotein receptors, which specifically recognize the galactose ligands on chitosan.Citation16,Citation17 Zhang et al synthesized folate/ poly(ethylene glycol)/PEI. The folate receptor is known to be overexpressed in a variety of human tumors, nevertheless it is only minimally distributed in normal tissues.Citation18 Their in vivo optical imaging and luciferase assay results demonstrated that the folate-labeled polyplexes resulted in preferential accumulation of transgenes in folate receptor-positive tumors, and a minicircle DNA-mediated approach achieved 2.3-fold higher gene expressions in tumors than conventional plasmid. Bonoiu et al utilized gold nanorod/ small interfering ribonucleic acid complexes (nanoplexes) that targeted the dopaminergic signaling pathway in the brain. Their nanoplexes were a new approach to treat drug addiction and other addictive behaviors. Their study showed that the gold nanorod could cross an in vitro model of the blood–brain barrier, which suggests success in vivo.Citation19
In the present study, a novel CP/peptide (CPT) copolymer was designed and developed by linking a specific peptide (FQHPSF sequence) to CP.Citation14 This peptide was selected from a phage display peptide library, which was specific to liver tumors.Citation20 The specificity and the efficiency of this polymeric gene vector were examined in HepG2 (human hepatoblastoma cells), HeLa (human cervical adenocarcinoma cell line), and A549 (human lung carcinoma cells) cells in vitro. To further evaluate the potential of the new vector in cancer gene therapy, CPT was used to deliver interleukin-12 (IL-12) plasmid and its antitumor effect in vivo was tested. The results show that the therapeutic effect of the new gene vector on ascites tumor-bearing mice was significantly enhanced as compared with the control groups.
Materials and methods
Materials
Chitosan (molecular weight 100 kDa) was purchased from Yuhuan Marine Biochemistry Co, Ltd (Yuhuan, China) and was dissolved in 0.5% acetic acid to give a final concentration of 2%. PEI (molecular weight 25 kDa, 1200 Da) was obtained from Sigma-Aldrich Corporation (St Louis, MO). 1,1′-Carbonyldiimidazole and N-succinimidyl 3-(2-pyridyldithio)propionate was purchased from Thermo Scientific Pierce Protein Research Products (Rockford, IL). The luciferase activity assay kit and protein assay kit were obtained from Biyuntian Biologic Co, Ltd (Nanjing, China). Gibco® Dulbecco’s modified Eagle medium (DMEM) and trypsin were obtained from Invitrogen Life Technologies (Carlsbad, CA). Fetal bovine serum was purchased from Sijiqing Biological Engineering Materials Co, Ltd (Hangzhou, China). Fluorescein isothiocyanate-labeled DNA and FQHSPFC peptide was synthesized by Shanghai Sangon Biological and Engineering Technology Services Co, Ltd (Shanghai, China). All other chemicals were analytical grade.
The pGL3 and pEGFP-N1 plasmid DNA were kindly provided by the Institute of Infectious Diseases, Zhejiang University (Hangzhou, China). The CCL22 and IL-12 plasmid DNA were a generous gift from Dr Shinsaku Nakagawa (Osaka University, Osaka, Japan). The plasmid DNA were amplified using Escherichia coli DH5α and purified using AxyPrep™ Plasmid Maxiprep Kit (Axygen Biosciences, Union City, CA). The purity of the plasmids consisting of supercoiled and open circular forms was checked by electrophoresis (EC250-90; EC Apparatus, Holbrook, NY) on a 1.0% agarose gel and the concentration of DNA was determined by measuring ultraviolet absorbance (DU70; Beckman Coulter, Brea, CA) at 260 nm and 280 nm. DNA from protamine was utilized for the measurement of particle sizes.
Animals
Female imprinting control region mice (4–5 weeks old) were purchased from the Laboratory Animal Center of Zhejiang University. All of the experimental procedures were in accordance with the Zhejiang University guidelines for the welfare of experimental animals.
Cell line
HeLa, A549, HepG2, and H22 (peripheral hepatoma cells) cells were obtained from the Institute of Biochemistry and Cell Biology, Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences (Shanghai, China). The cells were cultured in DMEM containing 10% fetal bovine serum at 37°C in a humidified 5% carbon dioxide-containing atmosphere.
Synthesis of CPT
CP was synthesized in a similar way to previously reported.Citation15 The average ratio of chitosan to PEI was 2.5, as determined by 1H nuclear magnetic resonance (Avance™ 600, Bruker BioSpin GmbH, Rheinstetten, Germany).Citation15 CP (100 mg) was dissolved in mixed solvents of 3 mL phosphate buffered saline and 3 mL dimethyl sulfoxide. N-succinimidyl 3-(2-pyridyldithio)propionate (1 mg) was dissolved in 1 mL dimethyl sulfoxide. The two solutions were mixed and 0.2 mL of triethylamine was added. The mixture was stirred in the dark for 1 hour. The FQHPSF peptide (6 mg) was dissolved in 5 mL of dimethyl sulfoxide and added dropwise into the solution for 2 hours under stirring in the dark. Three hours later the product was purified by dialysis in water for 24 hours. The light white powder was yielded by lyophilizing. All reactions were performed at room temperature, in the dark, and were nitrogen protected.
Characterization of the copolymer
The composition of the prepared CPT copolymer was estimated by 1H nuclear magnetic resonance spectroscopy (Avance 600) and ultraviolet spectrophotometry (Beckman Coulter).
Preparation of the CPT/DNA complex
All CPT/DNA complexes were freshly prepared prior to use. The charge ratio (N/P) is defined as the molar relation of amine groups in the cationic molecule, which represent the positive charges, to phosphate groups in the DNA, which represent the negative charges. For calculation of N/P ratios, 330 Da was utilized as an average mass per charge for DNA. Complexes were prepared by adding copolymer solution to equal volumes of calf thymus DNA (Sigma-Aldrich) (for size and zeta potential measurements), to pGL3 control solution (for luciferase assay), or to CCL22 and IL-12 therapy gene (for gene therapy), with gentle vortexing and incubation at room temperature for 30 minutes.
Gel retardation assay
Different amounts of CPT (charge ratio 2–10) were combined with DNA (0.5 g of pGL3) and the effect of CPT on condensation of DNA was investigated by electrophoresis on a 1% agarose gel with tris(hydroxymethyl)aminomethane running buffer at 100 V for 30 minutes. DNA was visualized with ethidium bromide (0.2 g/mL).
Measurement of particle sizes and zeta potential
The particle sizes and surface charge (represented by the surface zeta potential) of the synthesized CPT/DNA complexes (weight/weight [w/w] = 20) was measured using laser diffraction spectrometry (Zetasizer® 3000HS; Malvern Instruments Ltd, Malvern, United Kingdom). The volume of the samples was 1 mL, containing a final DNA concentration of 50 μg/mL.
Cell viability assays
The CP, CPT, and PEI solutions were filtered through 0.22- μm aseptic filter membranes (Millex-GV; Millipore Corp., Bedford, MA) before they were incubated with cells. For cell viability assay, HeLa, HepG2, and A549 cells were seeded into 96-well plates at a density of 5 × 103 cells/well. After 24 hours, CP/DNA (w/w = 10), CPT/DNA (w/w = 20), and PEI/DNA (N/P = 10) were added with 100 μL fresh serum-free DMEM to replace the culture medium. The concentration of DNA was 0.2 μg per well for the entire complex. Cells were incubated at 37°C in a humidified 5% carbon dioxide-containing atmosphere for 6 hours. The culture medium was then replaced by fresh DMEM with 10% fetal bovine serum. After being incubated for 42 hours, 20 μL of methylthiazol tetrazolium solution (5 mg/mL) was added to each well and the absorbance was measured according to the manufacturer’s instruction. The cell viability was calculated as following:
OD595 (sample) represents a measurement from the group treated with vectors and OD595 (control) represents the group treated with phosphate buffered saline only. All the experiments were carried out in triplicate to ascertain the reproducibility.
Tumor specific transfection
To evaluate the specific transfection of CPT, CPT/DNA (1 μg of pGL3 plasmid with mass ratio of 20:1) was prepared to transfect into HeLa cells, A549 cells, and HepG2 cells. PEI/ DNA (N/P = 10, 1 μg DNA) was also prepared using similar methods. The transfection procedure was the same as previously mentioned.Citation14 After 24 hours, the luciferase assay was carried out according to the manufacturer’s instruction. Light units due to luciferase activity were measured with a chemiluminometer (AutoLumat LB 953; Berthold Technologies GmbH, Bad Wildbad, Germany). All the experiments were carried out in triplicate to ascertain the reproducibility.
Competitive inhibition transfection
To study the targeting specificity of the polymer coupled to the peptide, a competitive test with free oligopeptide was performed. The free FQHPSF peptide (0.4 mg/well or 0.8 mg/ well) was added into the plates of HepG2 cells before the transfection of CPT/DNA.
Confocal laser scanning microscopy
The HepG2 cells were plated on glass coverslips in 24-well plates at 5 × 104 cells/well. The cells were incubated for 18 hours. The media were then replaced with serum-free media containing polymer/fluorescein isothiocyanate-labeled DNA plasmid (1 μg) complex (w/w = 10). After incubation for 0.5, 2, 4, and 6 hours, the cells were washed in phosphate buffered saline and fixed with 4% paraffin. The nucleus was stained with propidium iodide for 30 minutes in room temperature and washed three times. The coverslips were mounted on microslides. Cells were observed with a confocal laser scanning microscope (MRC-1024; Bio-Rad Laboratories, Hercules, CA).
Tumor regression for in vivo experiment
The mice were intraperitoneally inoculated with 1.5 × 106 H22 cells. After 2 days, CPT/IL-12 (mass ratio 20:1), CP/IL-12 (mass ratio 10:1), PEI/IL-12 (N/P = 4), and naked DNA IL-12 were prepared for intraperitoneal administration in mice. The dose of IL-12 was 50 μg per mouse. Mice weights were measured every 2 days. Each group included at least six mice. The luciferase activity of H22 cells in ascites extract and normal culture medium were also detected.
Statistical analysis
All the experiments were repeated in triplicate. The data were presented as mean ± standard deviation. Student’s t-test was employed to determine the difference between independent groups. Statistical calculations were computed with a statistics software package (v. 7.0; SPSS Inc, Chicago, IL). All statistical tests were two-tailed tests. Differences were considered statistically significant when P < 0.05.
Results and discussion
Synthesis and characterization of the copolymer
A nonviral polymeric gene vector, CP, was previously developed by crosslinking low molecular branch PEI (1800 Da) with chitosan, which was proven to have low cytotoxicity and long-term high gene transfection efficiency.Citation18 To further improve its targeting ability, a liver tumor specific peptide selected from a phage display library,Citation20 was coupled to CP to get the final gene vector CPT (). The structures of the peptide, CP, and CPT were confirmed by 1H nuclear magnetic resonance spectroscopy. As shown in , two proton peaks corresponding to the benzene ring appeared at 7.0 ppm and 8.5 ppm in both peptide and CPT, indicating that the peptide was successfully grafted to CPT. However, quantitative analysis of the peak at 3.0–2.5 ppm showed that the values decreased in CPT compared with CP, indicating the loss of PEI during the CPT synthesis procedure. It was reported previously that the improved transfection efficiency of CP was mainly attributed to PEI.Citation14 Therefore, the loss of PEI may affect the transfection efficiency of CPT. The amount of peptide grafted to CP was calculated by ultraviolet spectrophotometry for its benzene ring on cysteine. It was found that about 4% of the peptide was linked to CP ().
Figure 1 Reaction scheme for the synthesis of chitosan/polyethylenimine/peptide complex.
Abbreviations: CP, chitosan-linked polyethylenimine; CPT, chitosan/polyethylenimine/peptide complex.
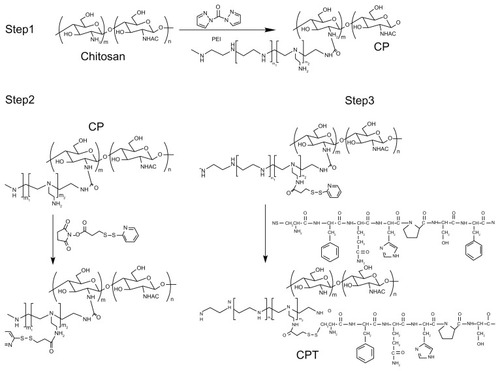
Figure 2 1H nuclear magnetic resonance spectra of chitosan-linked polyethylenimine, peptide, and chitosan/polyethylenimine/peptide complex in heavy water (D2O) at room temperature.
Abbreviations: CP, chitosan-linked polyethylenimine; CPT, chitosan/polyethylenimine/peptide complex.
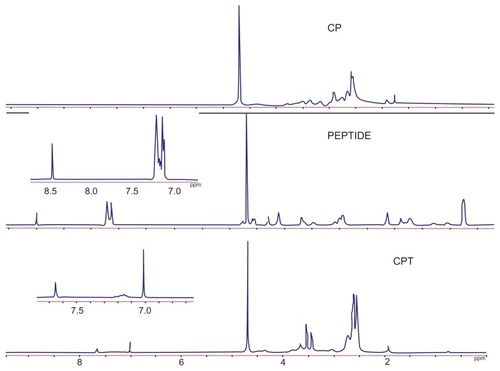
Figure 3 (A) Whole band ultraviolet scanning of chitosan-linked polyethylenimine, peptide, and chitosan/polyethylenimine/peptide complex. (B) The grafting rate of the chitosan/polyethylenimine/peptide complex.
Abbreviations: Abs, absorbance; CP, chitosan-linked polyethylenimine; CPT, chitosan/polyethylenimine/peptide complex.
Biophysical characterization of the polyplexes
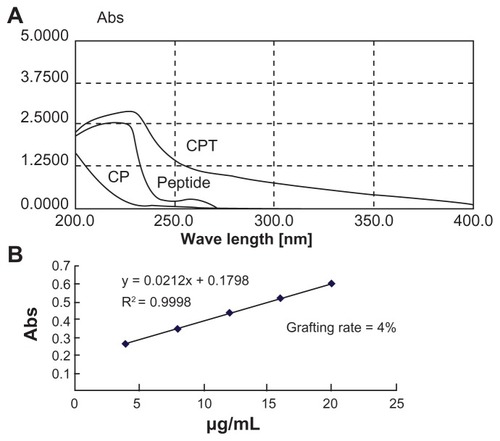
The DNA condensation capacity of CPT was determined by gel retardation experiments. The migration of DNA was completely retarded when the w/w ratio of CPT/DNA was around six (). The particle size and zeta potential of CPT/ DNA were measured by dynamic light scattering (Zetasizer 3000HS; ). It was shown that all three complexes (PEI/DNA, CP/DNA, and CPT/DNA) had small particle sizes under 200 nm and positive zeta potential, which was suitable for cell uptake. However, the zeta potential of CP/DNA and CPT/DNA were only half of that of PEI. For cell uptake via endocytosis, it is required that the particles have a cationic surface for cell binding. However, as Fischer et al reported, toxicity of the nanoparticles increased with increased aggregation and adherence of nanoparticles on the cell surface, which resulted in significant necrosis.Citation12 Therefore, it is not good for the cells if the zeta potential of the nanoparticles is too high. From this viewpoint, CP/DNA and CPT/DNA were more suitable than PEI/DNA for gene delivery. Transmission electron microscopy (JEM-2100; JEOL, Tokyo, Japan) was employed to observe the nanoparticles of CPT/DNA complexes (). The shape of the nanoparticles was spherical and the particle size was around 170–200 nm when the w/w ratio was at 20.
Figure 4 Physicochemical properties of the polyplexes. (A) Agarose gel electrophoresis of the chitosan/polyethylenimine/peptide complex at different weight/weight ratios. Lane 1: marker; lane 2: naked DNA; lane 3: weight/weight = 0.5; lane 4: weight/weight = 1; lane 5: weight/weight = 2; lane 6: weight/weight = 3; and lane 7: weight/weight = 4. (B) zeta potential of the polyplexes formed by pGL3 plasmid with polyethylenimine, chitosan-linked polyethylenimine, and chitosan/polyethylenimine/peptide complex at the gene delivery’s weight/weight ratios and (C) Particle size (measured by dynamic light scattering) (D) Morphology of the chitosan/polyethylenimine/peptide/DNA complex (weight/weight = 20) as observed by transmission electron microscope (×65,000). Bar: 200 nm.
Abbreviations: CP, chitosan-linked polyethylenimine; CPT, chitosan/polyethylenimine/peptide complex; PEI, polyethylenimine.
Biological characterization of the polyplexes in vitro
Cytotoxicity of the CP copolymer
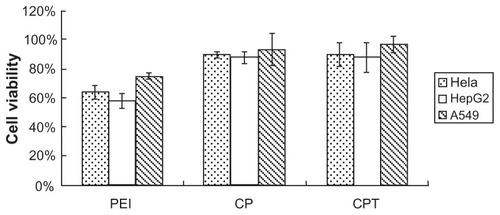
Cytotoxicity was one of the main concerns for the copolymer, particularly for in vivo delivery. shows the results of in vitro cytotoxicity of the copolymer analyzed by methylthiazol tetrazolium assay in three cell lines (HepG2, A549, and HeLa cells). As shown in , no cytotoxicity was detected in the three types of cells treated with CPT/ DNA and CP/DNA. In contrast, the PEI/DNA-treated group was shown to have high cytotoxicity. The cytotoxicity of PEI is probably caused by polymer aggregation on cell surfaces, impairing important membrane functions. The high positive charge of polymers can cause cell membrane damage. The examined zeta potential of PEI/DNA was nearly 40, which was much higher than CP/DNA and CPT/DNA. Also, the cationic polymers may interfere with critical intracellular processes of cells. In particular, the primary amine was reported to disrupt protein kinase C function through disturbance of protein kinase activity.Citation21,Citation22 It has been reported that high molecular weight PEI is significantly more toxic than low molecular weight PEI.Citation8,Citation9 Therefore, the low molecular weight PEI should be chosen to reduce the cytotoxicity of synthetic products.
Figure 5 Cytotoxicity of the complexes in three cell lines: chitosan-linked polyethylenimine/DNA (mass ratio = 10), polyethylenimine/DNA (nitrogen/phosphorus ratio = 10), and chitosan/polyethylenimine/peptide/DNA (mass ratio = 20).
Notes: n = 3; data are expressed as mean ± standard deviation (error bars).
Abbreviations: CP, chitosan-linked polyethylenimine; CPT, chitosan/polyethylenimine/ peptide complex; PEI, polyethylenimine.
Transfection efficiency of polymers in vitro
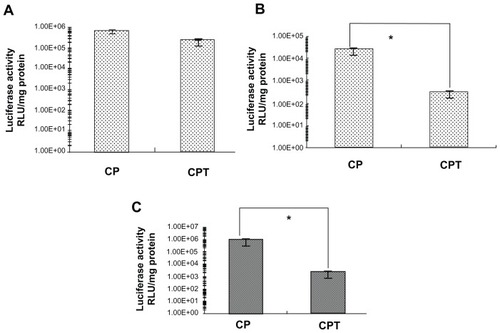
Previous studies have demonstrated that CP/DNA has a high transfection efficiency in several kinds of tumor cells with no significant differences.Citation14 The linking of the targeting peptide with CP may cause changes in the transfection property. shows the transfection efficiency of CP/DNA and CPT/DNA at functional ratios in four cell lines. It was found that the transfection efficiency of CPT/DNA was lower than that of CP/DNA. It is known that a consequential cellular barrier hindering the transfection efficiency of nonviral gene vectors is the insufficient release of endosomally trapped DNA in the cell cytosol. The proton-sponge mechanism of PEI could help the DNA to escape from the endosome and facilitate gene entry into the nucleus.Citation6 In the synthesis and characterization of the copolymer, it was assumed that the reduced PEI in CPT may induce the decline of transfection efficiency compared to CP. The results demonstrated this assumption. However, the transfection efficiency of CPT/DNA was significantly higher in HepG2 cells than in HeLa and A549 cells. The transfection efficiency of CP/ DNA:CPT/DNA in HepG2 cells was 3:1, while in HeLa cells and A549 cells, it was 358:1 and 80:1, respectively. It was suggested that CPT had a receptor-mediated endocytosis through the FQHPSF peptide. HepG2 cells are outstanding model cells of parenchymal cells in the liver, which have a multitude of unknown receptors. FQHPSF peptide was selected from a phage display library to specifically target to HepG2 cells. The receptor of this peptide in HepG2 cells is not very clear.Citation19 When transfected into HepG2 cells, the receptors on HepG2 cells play a role in not allowing a slice of complex to enter into the endosome. The cellular uptake of nontargeting cationic complexes has been proposed to proceed through endocytic routes, remain closely attached to the vesicle membrane of early and late endosomes, and fuse with the lysosomal vesicles, assembling in the perinuclear region. The fusion of endosomes with lysosomes seems to be partially prevented when PEI has been utilized. The DNA is allowed to reside longer in the nonlysosomal compartment, thereby avoiding DNA degradation by lysosomal nucleases and promoting transfection. Nonetheless, for the targeting complexes, the complex may enter into the nucleus directly after cellular uptake. This could illustrate why the transfection efficiency of CPT/DNA did not decrease too much in HepG2 cells, but decreased in other nontarget cells.
Figure 6 Transfection efficiency of chitosan-linked polyethylenimine/DNA and chitosan/polyethylenimine/peptide/DNA (pGL3 control) complexes in (A) HepG2 cells, (B) A549 cells, and (C) HeLa cells.
Note: *P < 0.05.
Abbreviations: CP, chitosan-linked polyethylenimine; CPT, chitosan/polyethylenimine/peptide complex; RLU, relative light unit.
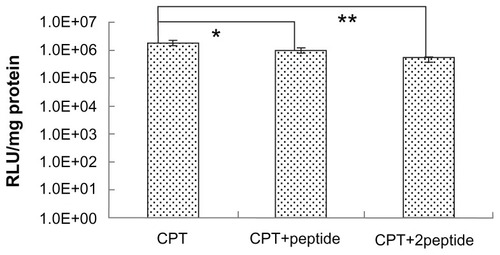
To further demonstrate the target properties caused by the peptide, competitive inhibition experiments were carried out. In this experiment, free peptide was primarily incubated with HepG2 cells before CPT/DNA were added. The receptors on the HepG2 cell surface were occupied by the free peptide, which blocked the receptor mediated internalization of CPT/DNA. It was found that the gene transfection efficiency of the free peptide pretreated group was lower than the untreated group (). The finding that the degree of inhibition was affected by the dose of peptide also confirmed the targeting specificity of CPT. However, although the amount of free peptide was high, the transfection efficiency didn’t decrease to zero, suggesting that the receptor-mediated endocytosis was not the only way for the complex to enter cells. The PEI-induced nonspecific cell uptake also played an important role in transfection in vitro.
Figure 7 The free peptide competitive study. HepG2 cells were incubated in Dulbecco’s modified Eagle medium with different peptide concentrations before the transfection of chitosan/polyethylenimine/peptide/DNA polyplexes at weight/weight ratio of 20. Luciferase expression was detected at 24 hours posttransfection.
Notes: *P < 0.05; **P < 0.01; values are presented as mean ± standard deviation.
Abbreviations: CPT, chitosan/polyethylenimine/peptide complex; RLU, relative light unit; +peptide, 0.4 mg/well of peptide added; +2peptide, 0.8 mg/well of peptide added.
Cellular internalization of CPT/DNA
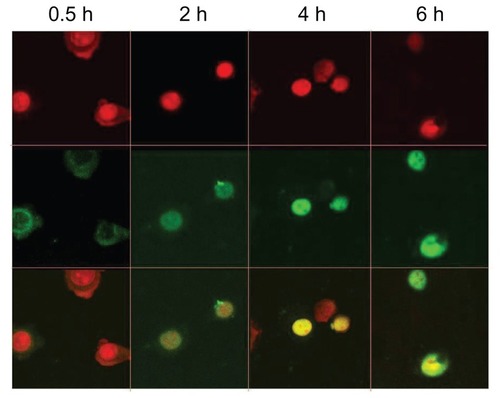
Cellular internalization of CPT/DNA was observed under confocal laser scanning microscopy. shows the time-dependent changes of CPT/DNA internalization. It was observed that the complexes were internalized in 0.5 hours and they were distributed mostly into the cytoplasm. The complexes entered into the nucleus in 2 hours and more DNA was detected in the nucleus after 4 hours. It is supposed that the ability and the time to escape from the endosome and enter the nucleus of vectors are very important for transfection efficiency. Therefore, PEI (25 kDa) could escape from the endosome and enter the nucleus more quickly, and it is difficult for the DNA carried by chitosan to enter the nucleus.Citation23,Citation24 As the amount of PEI grafted on CPT was lower than CP, it was assumed that the time for CPT/DNA to enter into the nucleus ought to be longer than that of CP/DNA. However, it was found that the time for CP to enter the nucleus was 4 hours, which was longer than CPT, suggesting CPT may induce specific internalization in another way to reduce the time of escaping from the endosome.
Figure 8 Intracellular distribution of fluorescein isothiocyanate-labeled DNA plasmid/chitosan/polyethylenimine/peptide complexes was observed with a confocal fluorescence microscope in HepG2 cells at 0.5, 2, 4, and 6 hours postincubation.
Notes: The upper panel shows the nucleus (red) and the middle panel shows the fluorescein isothiocyanate plasmid (green). The lower panel shows the overlay of the top two panels.
CPT-induced effective antitumor activity in vivo
Tumor targeting studies have found that ineffective in vivo therapy was always caused by the low amount of therapeutic gene accumulated in the tumor site. In a previous study, the CCL22 gene was transferred to H22 cells by CP/DNA and then the cells were inoculated to tumor-bearing ICR mice. The tumor growth inhibiting effect was observed. In comparison with both the PEI/DNA and control group, the tumor growth rate was significantly inhibited in the CP/DNA group.Citation14 In the present study, CPT was used as a vector in the in vivo therapy experiment by using an ascites tumor model. As there were several kinds of cells in ascites, including tumor cells, lymph cell, and blood cells, it is difficult to induce therapeutic effects by nonspecific gene transfection. Here, to demonstrate the target property of CPT, another therapeutic gene, IL-12, was utilized to be transfected by CPT. IL-12 is a multifunctional cytokine that stimulates both innate and adaptive immunity, acting as a consequential regulator of cell-mediated immune responses.Citation25 It was originally characterized in the early 1990s as a cytokine with the potent ability to activate T and natural killer cells.Citation26 Through upregulation of class I and II major histocompatibility molecules, and generating T helper type I immune responses, IL-12 facilitates the presentation of tumor antigens. These effects are tightly associated with the ability of IL-12 to induce expression of interferon-γ,Citation27 and the endogenous production of interferon-γ is required for the antitumor effects of IL-12 administration in most tumor studies. The promising data obtained by the administration of IL-12 recombinant protein in preclinical animal models of cancer raised hopes that recombinant IL-12 could be a powerful therapeutic agent.Citation28 However, clinical trials revealed a modest clinical response that was confined by the development of an adaptive response that downregulated IL-12 activity and the severe toxicity that occurred when high doses of this cytokine were employed.Citation29 Gene therapy can significantly increase cytokine expression in the target organ without excessively elevating systemic cytokine levels, which leads to an increased efficacy/toxicity ratio.
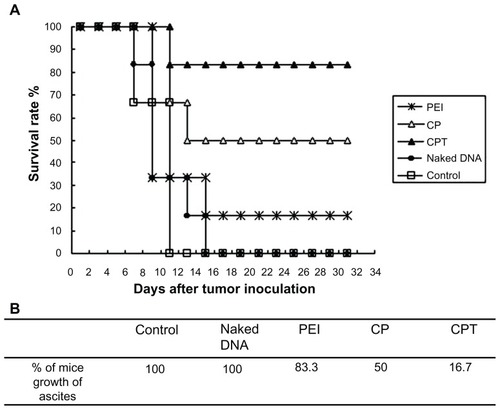
Hence, CPT – which was proven to have long-term expression and target properties – was used in the present study for the gene transfection to achieve in vivo therapeutic effects. shows that the tumor-bearing mice administered with CPT/IL-12 could survive more than 30 days while all of the mice in the control group died within 12 days. Only 16.7% of mice grew ascites in the CPT group, while the number of ascites in the control and naked DNA groups was 100%. The inhibition effect of the ascites tumor by CP and PEI complexes was lower than CPT. This result further confirmed the target property of CPT.
Figure 9 Tumor growth inhibition in mice after receiving phosphate buffered saline, naked DNA (interleukin-12 plasmid), polyethylenimine (25 kDa)/DNA (interleukin-12), chitosan-linked polyethylenimine/DNA (interleukin-12), and chitosan/polyethylenimine/peptide/DNA (interleukin-12) complexes. Imprinting control region mice were inoculated with H22 cells in the abdominal cavity. When tumors grew within 3 days, the mice were randomly assigned to one of the five groups and given the appropriate therapy. Tumor growth was monitored by measuring weight with a caliper every 2 days. (A) Survival rate of tumor-bearing mice. (B) Percentage of mice that experienced ascites growth with the treatment of different gene-carried nonviral vectors.
Abbreviations: CP, chitosan-linked polyethylenimine; CPT, chitosan/polyethylenimine/peptide complex; PEI, polyethylenimine.
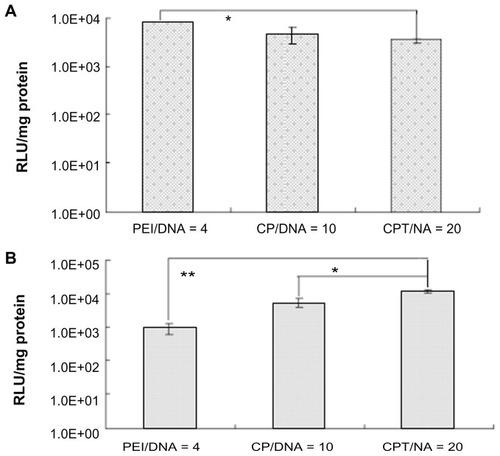
In the in vitro studies, the PEI and CP groups were proven to have much higher transfection efficiency than the CPT group (). This raised the question of why CPT could increase its transfection efficiency in vivo. To simulate the in vivo environment, the ascitic fluid was used – which was obtained from tumor-bearing mice with the cells in the fluid totally removed – to culture H22 cells. Then, the in vitro transfection experiment was performed. It was found that the highest transfection efficiency was observed in the CPT group while the lowest efficiency was observed in the PEI group (). Under physiological conditions, systemic delivery is hindered by complex instability. The physiological salt concentration often promotes aggregation of cationic complexes, which leads to vascular blockage. Additionally, cationic complexes readily bind with serum proteins such as serum albumin, and the protein binding would inhibit cellular uptake, promote aggregation, and possibly encourage phagocytosis. Although the composition of ascites was not very clear, it may decrease the transfection efficiency of PEI significantly. As mentioned above, CPT has a receptor-mediated endocytosis effect. Therefore, its transfection efficiency would not be influenced much. This could explain why CPT/IL-12 exhibited much higher tumor inhibition effect in vivo than other vectors.
Figure 10 Transfection efficiency of chitosan-linked polyethylenimine/DNA, polyethylenimine/DNA, and chitosan/polyethylenimine/peptide/DNA (pGL3 control) complexes in H22 cells in vitro using (A) serum-free Dulbecco’s modified Eagle medium and (B) Dulbecco’s modified Eagle medium with ascites extract.
Abbreviations: CP/DNA, chitosan-linked polyethylenimine/DNA complex; CPT/DNA, chitosan/polyethylenimine/peptide/DNA complex; PEI/DNA, polyethylenimine/DNA complex; RLU, relative light unit.
Conclusion
In this research, a new polymeric gene vector, CPT, was successfully synthesized by crosslinking low molecular weight PEI with chitosan and further coupling the peptide to the polymer for targeting hepatic cells. The new gene vector was characterized in terms of its chemical structure and composition, biophysical parameters, and in vitro and in vivo gene transfection efficiency. The gene vector not only showed the proper particle size and zeta potential and low cytotoxicity, it was also proved to target specificity to receptors on hepatic cells. The delivery of therapeutic IL-12 gene mediated by the new gene vector and the therapeutic efficiency were also studied in mice ascites model. The in vivo study results demonstrated that the new gene vector significantly enhanced the antitumor effect in tumor-bearing mice as compared to PEI (25 kDa), indicating that this new polymeric gene vector is a potential candidate for cancer gene therapy.
Acknowledgments
This work was financially supported by the National Natural Science Foundation of China (30873173, 30973648, and 81001410), Zhejiang Provincial Natural Science Foundation of China (R2090176), Fundamental Research Funds for the Central Universities, and National Basic Research Program of China (2009CB930300).
Disclosure
The authors report no conflicts of interest in this work.
References
- Cavazzana-CalvoMHacein-BeySde Saint BasileGGene therapy of human severe combined immunodeficiency (SCID)-X1 diseaseScience2000288546666967210784449
- BoydACGene and stem cell therapyBushAAltonEWFWDaviesJCGriesenbachUJaffeACystic Fibrosis in the 21st CenturyBaselKarger2006221232
- KaplittMGFeiginATangCSafety and tolerability of gene therapy with an adeno-associated virus (AAV) borne GAD gene for Parkinson’s disease: an open label, phase I trialLancet200736995792097210517586305
- YangZRWangHFZhaoJRecent developments in the use of adenoviruses and immunotoxins in cancer gene therapyCancer Gene Ther200714759961517479105
- AndersonWFHuman gene therapyNature1998392667925309579858
- PackDWHoffmanASPunSStaytonPSDesign and development of polymers for gene deliveryNat Rev Drug Discov20054758159316052241
- VermaIMSomiaNGene therapy – promises, problems and prospectsNature199738966482392429305836
- Koping-HoggardMVarumKMIssaMImproved chitosan-mediated gene delivery based on easily dissociated chitosan polyplexes of highly defined chitosan oligomersGene Ther200411191441145215269712
- KimTHKimSIAkaikeTChoCSSynergistic effect of poly(ethylenimine) on the transfection efficiency of galactosylated chitosan/DNA complexesJ Control Release2005105335436615949861
- DensmoreCLOrsonFMXuBAerosol delivery of robust polyethyleneimine-DNA complexes for gene therapy and genetic immunizationMol Ther20001218018810933929
- KichlerALeborgneCDanosODilution of reporter gene with stuffer DNA does not alter the transfection efficiency of polyethyleniminesJ Gene Med20057111459146716041686
- FischerDBieberTLiYElsasserHPKisselTA novel non-viral vector for DNA delivery based on low molecular weight, branched polyethylenimine: effect of molecular weight on transfection efficiency and cytotoxicityPharm Res19991681273127910468031
- KunathKvon HarpeAFischerDKisselTGalactose-PEI-DNA complexes for targeted gene delivery: degree of substitution affects complex size and transfection efficiencyJ Control Release200388115917212586513
- GaoJQZhaoQQLvTFGene-carried chitosan-linked-PEI induced high gene transfection efficiency with low toxicity and significant tumor-suppressive activityInt J Pharm20103871–228629420035848
- ZhaoQQChenJLLvTFN/P ratio significantly influences the transfection efficiency and cytotoxicity of a polyethylenimine/chitosan/ DNA complexBiol Pharm Bull200932470671019336909
- ParkIKParkYHShinBAGalactosylated chitosan-graft-dextran as hepatocyte-targeting DNA carrierJ Control Release20006919710811018549
- ParkMRKimHWHwangCSHighly efficient gene transfer with degradable poly(ester amine) based on poly(ethylene glycol) diacrylate and polyethylenimine in vitro and in vivoJ Gene Med200810219820718064729
- ZhangCGaoSJiangWTargeted minicircle DNA delivery using folate-poly(ethylene glycol)-polyethylenimine as non-viral carrierBiomaterials201031236075608620488533
- BonoiuACMahajanSDDingHNanotechnology approach for drug addiction therapy: gene silencing using delivery of gold nanorod-siRNA nanoplex in dopaminergic neuronsProc Natl Acad Sci U S A2009106145546555019307583
- ZhangBZhangYWangJScreening and identification of a targeting peptide to hepatocarcinoma from a phage display peptide libraryMol Med2007135–624625417622312
- FarhoodHBottegaREpandRMHuangLEffect of cationic cholesterol derivatives on gene transfer and protein kinase C activityBiochim Biophys Acta1992111122392461420259
- KopatzIRemyJSBehrJPA model for non-viral gene delivery: through syndecan adhesion molecules and powered by actinJ Gene Med20046776977615241784
- FengMLeeDLiPIntracellular uptake and release of poly (ethyleneimine)-co-poly(methyl methacrylate) nanoparticle/ pDNA complexes for gene deliveryInt J Pharm20063111–220921416442245
- HashimotoMMorimotoMSaimotoHShigemasaYSatoTLactosylated chitosan for DNA delivery into hepatocytes: the effect of lactosylation on the physicochemical properties and intracellular trafficking of pDNA/chitosan complexesBioconjug Chem200617230931616536460
- ColomboMPTrinchieriGInterleukin-12 in anti-tumor immunity and immunotherapyCytokine Growth Factor Rev200213215516811900991
- ManettiRParronchiPGiudiziMGNatural killer cell stimulatory factor (interleukin 12 [IL-12]) induces T helper type 1 (Th1)-specific immune responses and inhibits the development of IL-4-producing Th cellsJ Exp Med19931774119912048096238
- ManettiRGerosaFGiudiziMGInterleukin 12 induces stable priming for interferon gamma (IFN-gamma) production during differentiation of human T helper (Th) cells and transient IFN-gamma production in established Th2 cell clonesJ Exp Med19941794127312837908322
- Del VecchioMBajettaECanovaSInterleukin-12: biological properties and clinical applicationClin Cancer Res200713164677468517699845
- BerraondoPPrietoJGonzalez-AseguinolazaGAdvances in interleukin- 12 gene therapy for acquired liver diseasesCurr Gene Ther200992627119355864