?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.
?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.Abstract
Background and methods
Applications of the anticancer agent, ellipticine, have been limited by its hydrophobicity and toxicity. An efficient delivery system is required to exploit the enormous potential of this compound. Recently, EAK16-II, an ionic-complementary, self-assembling peptide, has been found to stabilize ellipticine in aqueous solution. Here, the anticancer activity of ellipticine encapsulated in EAK16-II (EAK-EPT) was evaluated in vitro and in vivo.
Results
Our cellular uptake, toxicity, and apoptosis results in an A549 human lung carcinoma cell line indicate that EAK-EPT complexes are significantly more effective than treatment with EAK16-II or ellipticine alone. This is due to the ability of EAK16-II to stabilize ellipticine in a protonated state in well formed nanostructures approximately 200 nm in size. In vivo observations in an A549 nude mouse tumor model show higher antitumor activity and lower cytotoxicity of EAK-EPT complexes than in the control group treated with ellipticine alone. Tumor growth in animals was significantly inhibited after treatment with EAK-EPT complexes, and without any apparent side effects.
Conclusion
The anticancer activity observed in this study coupled with minimal side effects encourages further development of peptide-mediated delivery of anticancer drugs, ellipticine in the present case, for clinical application.
Introduction
Lung cancer is a leading cause of cancer-related mortality in both men and women, and is responsible for well over one million deaths worldwide annually.Citation1 Over 80% of all diagnosed lung cancers are nonsmall cell lung carcinoma,Citation2 which is associated with high rates of proliferation and metastases, as well as a poor prognosis when diagnosed in the advanced stages compared with other cancers.Citation3 Conventional treatment methods for lung cancer are mainly surgery for suitable candidates and chemotherapy for patients with advanced disease, with a mean survival rate of less than one year for most patients with advanced lung cancer.Citation4 Therefore, new agents are continually being investigated in the hope of reducing the death rate attributable to lung cancer.Citation5
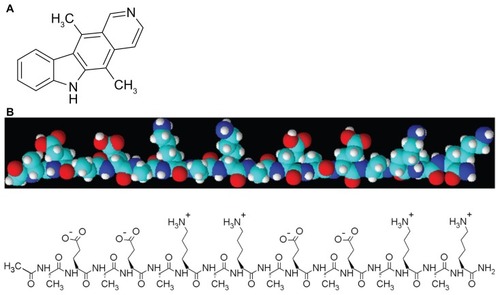
Ellipticine (5,11-dimethyl-6H-pyrido[4,3-b] carbazole), shown in , is a natural product isolated from Ochrosia elliptica and several other species of Ochrosia and Bieekeria vitiensis.Citation6 The main reason for the interest in ellipticine and its derivatives for clinical purposes is their high efficacy against several types of cancer.Citation7 Most ellipticine derivatives are mutagenic to Salmonella typhimurium Ames tester strains, bacteriophage T4, Neurospora crassa, and mammalian cells, and induce prophage lambda in Escherichia coli.Citation8 Likely mechanisms for the antitumor, mutagenic, and cytotoxic activity of ellipticine have been suggested to be intercalation with DNA,Citation7,Citation9 inhibition of topoisomerase II activity,Citation7,Citation10–Citation12 and formation of covalent DNA adducts mediated by cytochrome P450 and peroxidases.Citation8,Citation13,Citation14 However, the clinical application of ellipticine is greatly limited by hydrophobicity and severe side effects.Citation15,Citation16 Thus, appropriate delivery systems are required to stabilize ellipticine in aqueous solution and reduce its toxicity.
Figure 1 Molecular structures of (A) ellipticine and (B) self-assembling EAK16-II peptide (n-AEAEAKAKAEAEAKAK-c).
Many nanoparticle-based delivery systems have been developed for cancer chemotherapy, including liposome, polymeric micelles, albumin-bound nanoparticles, and lipoproteins.Citation17–Citation20 Among the many new nanobiomaterials, a special class of self-assembling peptides has been extensively explored to identify hydrophobic anticancer agents and small interfering RNA.Citation21–Citation23 Self-assembling peptide- based delivery systems have unique repetitive amino acid sequences that allow them to assemble spontaneously into stable nanostructures.Citation24,Citation25 Importantly, these peptides have many advantages over other delivery systems, including biocompatibility, low toxicity, stability, and ease of preparation. Citation26 Previous studies have shown that EAK16-II, an amino acid pairing, ionic-complementary self-assembling peptide, stabilizes ellipticine in aqueous solution.Citation21,Citation27,Citation28 EAK16-II has shown great potential to deliver ellipticine in a protonated state and to induce cytotoxicity in different cancer cells.
In this research, EAK16-II was used as a carrier for ellipticine for extensive in vitro and in vivo experiments, focusing on developing a suitable formulation and determining the cytotoxicity, cellular uptake, and pharmacodynamics of the delivery system. The objective was to clarify whether ellipticine encapsulated in EAK16-II (EAK-EPT) has enhanced anticancer activity in vitro and inhibits tumor growth in vivo while minimizing undesirable toxicity.
Materials and methods
Materials
The EAK16-II peptide (molecular weight 1657.66 g/mol, >95% pure by high-pressure liquid chromatography) was synthesized using the AAPPTEC peptide synthesizer (Apex 396, Louisville, KY) at the University of Waterloo, ON, Canada. Synthesis was started with amidation of the C-terminal and ended with acetylation of the N-terminal. The synthesized peptide was further purified with high-pressure liquid chromatography and characterized by matrix-assisted laser desorption ionization time-of-flight mass spectroscopy. This peptide has a sequence of Ac-AEAEAKAKAEAEAKAK-NH2, where A corresponds to alanine, E to glutamic acid, and K to lysine. The anticancer drug, ellipticine (98% purity), was purchased from EMD-Merck (Mississauga, Canada).
A549 nonsmall lung carcinoma cells were cultured in high glucose Dulbecco’s Modified Eagle’s Medium (DMEM; Gibco-Invitrogen Corporation, Carlsbad, CA) with 10% heat-inactivated fetal bovine serum and 1% antibiotics (streptomycin + penicillin).
MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) was purchased from Sigma-Aldrich (St Louis, MO) for the in vitro cytotoxicity experiments. A terminal deoxynucleotidyl transferase (TUNEL) apoptosis assay kit was purchased from Roche Diagnostics GmbH (Mannheim, Germany).
Five-week-old male Balb/c nude mice were obtained from the B and K Universal Group Limited (Shanghai, China). The mice were maintained under a 12-hour light/dark cycle at 25°C and a humidity of 60% ± 10%. All experiments were performed in accordance with institutional and governmental regulations on the use of experimental animals.
Formation of EAK-EPT complexes
EAK16-II solutions were made by dissolving peptide powder in pure water (with a 0.2 μm filter) at a concentration of 3 mg/mL (1.81 mM) followed by 10 minutes of bath sonication. A certain amount of ellipticine powder was weighed in a tube of EAK16-II solution to reach a concentration of 1.5 mg/mL (6.1 mM), followed by stirring for 24 hours. Initially the solution was very cloudy. To make a clear complex, the pH of the sample was changed to acidic with 1 M HCl until the solution became clear. The pH was thereafter adjusted with 1 M NaOH for the animal study to a pH of 5.6–5.8. The EAK-EPT complexes were prepared at a mass ratio of 2:1.
The ellipticine control sample was added to 5% ethanol/filtered water (with a 0.2 μm filter) to a concentration of 1.5 mg/mL followed by stirring for 24 hours. To make a clear complex, the pH of the ellipticine control sample was changed to acidic with 1 M HCl until the solution became clear. The pH was adjusted with 1 M NaOH to pH 5.6–5.8 thereafter for the animal study.
Atomic force microscopy
The self-assembled peptide and complex nanostructures were imaged by atomic force microscopy (PicoScan; Molecular Imaging, Phoenix, AZ) on a mica substrate. The samples were prepared according to the following procedure: a 10 μL sample of the peptide solution (15 minutes after preparation) was deposited on the freshly cleaved mica substrate that was fixed onto an atomic force microscopy sample plate. The solution was incubated on the mica surface for 10 minutes to allow the peptide nanostructures to adhere onto the surface, and then rinsed with water twice. Surplus moisture on the surface was collected with tissue paper. The sample plate was then covered with a Petri dish to avoid possible contamination and left to dry overnight. Atomic force microscopic images were taken at room temperature using the tapping mode. All images were acquired using a 225 μm silicon single-crystal cantilever (type NCL-16; Molecular Imaging) with a typical tip radius of 10 nm and a frequency of 175 kHz. A scanner with a maximum scan size of 6 × 6 μm2 was used. Data sets were subjected to third-order flattening. The atomic force microscopic images underwent further dimension analysis with free Gwyddion scanning probe microscopy data analysis software (Czech Metrology Institute, Brno, Czech Republic).
Dynamic light scattering
The hydrodynamic diameters of the peptide assemblies and peptide-drug complexes were obtained on a Zetasizer Nano ZS (Malvern Instruments, Worcestershire, UK). Appropriate settings, viscosity, refractive index, and dispersant solvent were set for each measurement at 25°C. A small 45 μL volume of the sample was transferred from the vial to a quartz microcell with a 3 mm light path (Hellma Canada Ltd, Concord, Canada). The scattered light intensities of the samples were collected at an angle of 173°. This was known as backscatter detection, ie, the relationship between the size of a particle and its scattered light intensity obtained with the multimodal Contin algorithm, which was provided in the Dispersion Technology Software 5.1 package (Malvern Instruments).
Cytotoxicity assay
A549 cells were plated onto 96-well plates (5 × 103 cells per well) in DMEM with 10% fetal bovine serum in the presence of EAK16-II (0.01, 0.1, 1.0, 10.0, and 100.0 μg/mL), ellipticine (0.005, 0.05, 0.5, 5.0, and 50.0 μg/mL), or EAK-EPT complexes (in which concentrations of EAK are 0.01, 0.1, 1.0, 10.0, and 100.0 μg/mL, and concentrations of ellipticine are 0.005, 0.05, 0.5, 5.0, and 50.0 μg/mL). Because 5% ethanol was added into ellipticine solution (1.5 mg/mL) during preparation, ethanol was treated as a control, and the concentration of ethanol in cytotoxicity assay was calculated according to its ratio in EPT solution (1.7 × 10−4, 1.7 × 10−3, 1.7 × 10−2, 1.7 × 10−1, 1.7 μL/mL). DMEM medium served as the blank and untreated cells served as controls. The treatments were diluted four-fold in culture medium. Cells were incubated for 24 and 48 hours at 37°C with 5% CO2, 95% air, and complete humidity prior to MTT assay. The MTT solution was prepared at 5 mg/mL in phosphate-buffered saline and was filtered through a 0.2 μm filter followed by dilution in DMEM. MTT (20 μL/well) was added to each well. Cells were further incubated with MTT for 4 hours. The insoluble formazan was dissolved in dimethyl sulfoxide (180 μL/well) solution. The plate was further incubated for 5 minutes at room temperature. Cell viability was assessed by measuring absorbance at 570 nm using a 96-plate reader (Bio-Rad, Hercules, CA). Cell survival was expressed as a percentage of the absorbance value determined for control cultures using the following calculation:
where As is absorbance of sample, Ac is absorbance of control, and Ab is absorbance of blank.
Fluorescence microscopy
The cellular uptake of EAK-EPT complexes was studied by imaging the cells under a fluorescence microscope (Axio Observer, Carl Zeiss Ltd, Canada). The procedure used for preparation of the cell samples was as follows: A549 cells were cultured in DMEM medium on a 6-well plate at a concentration of 1.2 × 105 cells per well. After growth overnight, the cells were treated with EAK-EPT complexes (5:1 mass ratio, 0.5:0.1 mg/mL) or the control and incubated for 2 hours. The culture medium was discarded and the cells were washed with phosphate-buffered saline, followed by fixing in 4% (v/v) paraformaldehyde in phosphate-buffered saline for 30 minutes at 4°C. After removing the paraformaldehyde, the cells were washed with phosphate-buffered saline three times before applying Fluoroshield™ (Sigma-Aldrich) mounting medium with or without 4′,6-diamidino-2-phenylindole (DAPI).
Flow cytometry or fluorescence-activated cell sorting
Cellular uptake of EAK-EPT complexes was also studied by flow cytometry (FACSVantage™ SE cell sorter; BD Biosciences, Franklin Lakes, NJ). The procedure used for preparation of the cell samples was as follows: A549 cells were cultured in DMEM medium on a 24-well plate at a concentration of 3.0 × 104 cells per well. After growth overnight, the cells were treated with EAK-EPT complexes (5:1 mass ratio, 0.5:0.1 mg/mL) or the control and incubated for 2 hours. The culture medium was discarded and the cells were washed with phosphate-buffered saline followed by addition of trypsin-ethylenediamine tetraacetic acid to detach the cells from the plate, and the cells were then suspended in 4% paraformaldehyde.
Apoptosis assays
Apoptosis was measured in A549 cells using the TUNEL assay according to the manufacturer’s instructions. Cells were plated onto culture slides at a density of 15,000 cells/cm2 in 10% fetal bovine serum and DMEM. After growth overnight, the cells were treated with EAK16-II 40 μg/mL, ellipticine 20 μg/mL, or EAK-EPT complexes (2:1 mass ratio, 40:20 μg/mL) for 48 hours. The cells were then fixed in 4% paraformaldehyde. After fixation, the slides were rinsed with phosphate-buffered saline. The slides were then incubated in 3% H2O2/methanol for 10 minutes and rinsed in phosphate-buffered saline. Sections were then permeabilized by incubating the slides in 1% Triton X-100/1% sodium citrate for 2 minutes at 4°C. After permeabilization, 50 μL of the TUNEL reaction mixture was added and incubated for 60 minutes at 37°C in a humidified atmosphere in the dark. After washing with phosphate-buffered saline, the slides were incubated with 50 μL of Converter-POD solution in a humidified chamber for 30 minutes at 37°C. After incubation, the slides were rinsed three times in phosphate-buffered saline and incubated with 50–100 μL of DAB substrate for 10 minutes at 15°C–25°C. The slides prepared for quantifying the extent of apoptosis in individual tests were then mounted with coverslips. TUNEL-positive cells were considered apoptotic and analyzed under a light microscope (Leica, Solms, Germany). For quantification, five randomized fields were counted by a blinded observer at a magnification of 200×.
In vivo experiments
In order to generate tumors, Balb/c nude mice (5 weeks of age) were injected subcutaneously in the right flank with 200 μL of cell suspension containing 5 × 106 A549 cells. When tumor volumes had reached 100–200 mm3, the mice were randomly divided into four groups, ie, a control group (n = 6), an EAK16-II group (n = 6), an ellipticine group (n = 6), and an EAK-EPT group (n = 6). Mice in the EAK16-II group were injected intraperitoneally with EAK16-II 22.5 mg/kg once a day for a total of 14 days. Mice in the ellipticine group were injected intraperitoneally with ellipticine 11.25 mg/kg once a day for a total of 14 days. The EAK-EPT group was injected intraperitoneally with EAK-EPT complexes (2:1 mass ratio) at an ellipticine concentration of 11.25 mg/kg once a day for a total of 14 days. Tumor size was measured using a Vernier caliper, and the tumor volume was calculated using the formula: V = 0.5 × longest diameter × shortest diameter.Citation2 Mouse body weight was also monitored. After 2 weeks, all of the mice were sacrificed and the tumors were removed for further analysis.
Statistical analysis
The results are expressed as the mean ± standard deviation. Statistical significance was determined by one-way analysis of variance. Differences were considered significant if P was< 0.05.
Results and discussion
The self-assembling EAK16-II amino acid-pairing peptide has recently been used as a new delivery vehicle for hydrophobic therapeutic agents. The role of EAK16-II in stabilization of ellipticine is associated with its simple sequence and unique structure. EAK16-II contains 16 amino acids in sequence, but is comprised of only three different amino acids, ie, glutamic acid (E), lysine (K), and alanine (A). These three amino acids are arranged in a special manner with hydrophobic (A) and hydrophilic residues (K or E) alternating in the sequence, rendering the peptide amphiphilic ().Citation24,Citation29 The EAK16-II peptide has been found to form stable β-sheet-rich nanostructures in aqueous solution. In a β-sheet arrangement, all hydrophilic residues of the peptide are arranged on one side and the hydrophobic residues are on the other side. This unusual amphiphilic property allows the peptide to interact with the ellipticine, which is hydrophobic. Protonation of ellipticine is usually found at higher peptide concentrations, as a result of relatively low pH (<5, pKa of ellipticine is about 6) in solution,Citation24,Citation30 and protonated ellipticine can be stabilized by ionic interaction with the negatively charged residues (glutamic acid E in this case) of the peptide. Thus, this self-assembling EAK16-II peptide can stabilize hydrophobic ellipticine in aqueous solution.
Nanostructure of EAK-EPT complexes
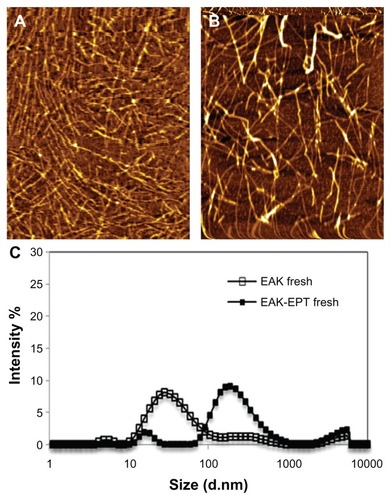
The EAK16-II peptide has been shown to self-assemble into a fiber nanostructure with a high β-sheet secondary structure. As atomic force microscopic images show, EAK16-II forms fibers approximately 9.27 ± 0.82 nm wide with a height of about 0.532 ± 0.036 nm on a mica surface, whereas dynamic light scattering data show the approximate hydrodynamic diameter to be 33.09 ± 0.76 nm for EAK16-II nanoparticles. The differences in size and morphology indicated by these two measurements are due to the different environments, ie, a mica surface and an aqueous solution. Estimation of the hydrodynamic diameter by dynamic light scattering considers the aggregates to be spherical shapes. The nanostructure of the EAK-EPT complexes is shown by atomic force microscopic imaging, forming slightly thicker nanofibers on a mica surface. However, data from dynamic light scattering show stable particles of 194.2 ± 8.94 nm (). The increase in hydrodynamic diameter of EAK-EPT complex particles compared with EAK16-II alone occurs because ellipticine particles are trapped between the peptide nanofibers. The zeta potential of the peptide-drug complex measured using dynamic light scattering shows stable particles in solution (zeta potential 62.83 ± 0.93 mV) at a pH of approximately 3.7–3.8. Ellipticine has a pKa of about 6, and can be protonated in a weakly acidic environment. Fresh EAK16-II in pure water has a pH of about 4.6, which can cause protonation of ellipticine. The higher pH may lead to deprotonation of ellipticine and accordingly a lower zeta potential.Citation27 To study the interaction between EAK-EPT and plasma proteins under physiological conditions, the EAK-EPT complexes were incubated in bovine albumin serum, which is the most abundant protein in plasma. Over time, the zeta potential of EAK-EPT preincubated with bovine albumin serum decreased to −0.51 ± 0.08 mV with a stable particle size of <100 nm. It was observed that ellipticine transformed from a protonated form to a crystalline form, while EAK-EPT interacted with bovine albumin serum molecules.
Antiproliferation induced by EAK-EPT complexes against A549 cells
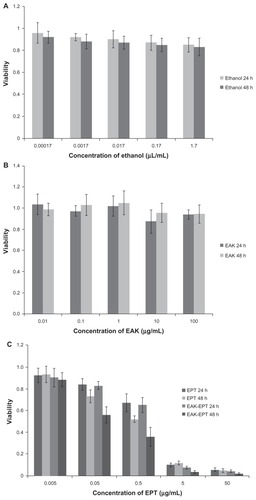
As an antiproliferative drug, ellipticine is a natural alkaloid, first isolated from an Australian evergreen tree of the apocynaceae family in 1959.Citation31 Later, it was found that ellipticine and its derivatives possess anticancer activity against various tumors, including lung cancer. Here, A549 nonsmall cell lung carcinoma cells were used to study the antitumor efficiency of EAK-EPT complexes using the MTT assay technique. We used 5% ethanol to increase the solubility of ellipticine, which is a hydrophobic agent. The 5% ethanol control had little effect on the viability of the A549 cells (). It was found that sole use of EAK16- II had no ability to inhibit A549 cell growth at 24–48 hours (), while the ellipticine and EAK-EPT complex groups obviously inhibited proliferation of A549 cells. With increasing incubation time and dose increments, the antiproliferative effect of ellipticine and EAK-EPT complexes on the A549 cells became more and more marked. The results indicate that cell viability for the group treated with EAK-EPT complexes was significantly lower than that for the ellipticine control group, particularly after 48 hours of incubation (P < 0.05, ). It was concluded that the self-assembling peptide EAK16-II could control the release of ellipticine effectively, and prolonged the inhibitory effect of ellipticine on growth of A549 lung cancer cells in vitro, without inducing toxicity. This advantage for drug delivery systems will be explored in further in vivo experiments.
Figure 3 (A) Cellular toxicity of ethanol controls at 24 and 48 hours. (B) Cellular toxicity of EAK at 24 and 48 hours. (C) Cellular toxicity of ellipticine controls and EAK-EPT complexes for two incubation times of 24 and 48 hours.
Abbreviations: EAK, self-assembling EAK16-II peptide; EPT, ellipticine.
It was demonstrated that this self-assembling peptide system can be used to control the release of EAK-EPT complexes and enhance their inhibitory effect on A549 lung cancer cells. The higher efficacy of EAK-EPT complexes against cancer cells compared with EAK16-II alone can be explained by the molecular state of the ellipticine in the EAK-EPT complexes. The protonation of ellipticine is usually found at a higher peptide concentration, related to the relatively low pH of the solution (<5, the pKa of ellipticine is about 6),Citation24,Citation30 and protonated ellipticine can be stabilized by ionic interaction with negatively charged residues, glutamic acid (E) in the case of this peptide. Protonated ellipticine has a positive charge, which can interact with a negatively charged cell membrane surface, leading to accumulation of ellipticine at the cell membrane. In addition, such a small molecule with hydrophobic characteristics is expected to cross the cell membrane easily into the cytoplasm.Citation21
Cellular uptake of the peptide-drug complex
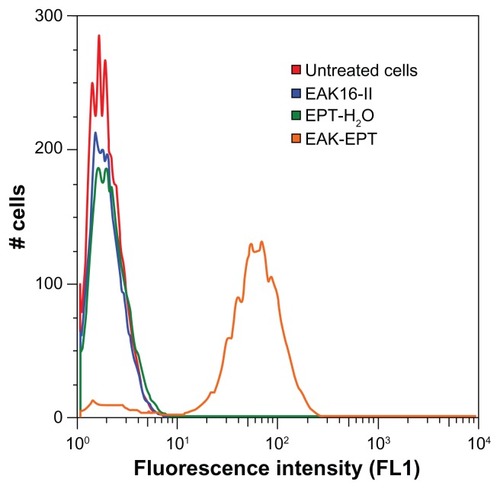
We used flow cytometry and fluorescence microscopy to determine whether ellipticine permeates the cell membrane and which organelle the drug localizes in. The flow cytometry results reveal a greater uptake of EAK-EPT (5:1 mass ratio, 0.5–0.1 mg/mL) into A549 cells compared with the ellipticine control, EAK16-II control, and untreated cells. The fluorescence properties of ellipticine have been extensively studied and reported in our previous publications. There are three different molecular states of ellipticine, ie, neutral, crystalline, and protonated, with corresponding emission fluorescence peaks at about 390–440 nm, 468 nm, and 520 nm, respectively.Citation27 In the current study, the focus is on EAK-EPT complexes, where ellipticine was in a protonated state and is considered as green fluorescence. Both the flow cytometry and fluorescence microscopy used here were capable of detecting green fluorescence emitted by ellipticine at 520 nm.
As shown in , the fluorescent intensity detected in the FL1 (FITC) channel increased dramatically between control samples and EAK-EPT complexes. This result is significant because cellular uptake was much greater for EAK-EPT complexes than for the ellipticine control sample, showing clearly that ellipticine cannot penetrate into the nuclei without the assistance of the peptide. It is worth noting that the exposure time and excitation laser intensity was kept constant for all measurements. The excitation wavelength was at 488 nm, and the emission recorded was on the 530/30 nm filter. Ellipticine is a green fluorescence species that emits light at approximately 520 nm, and can be detected in the FL1 channel. Since all other factors were constant during the experiment and only the amount of ellipticine in the cells was varied, comparison between FL1 fluorescence intensities according to the amount of ellipticine uptaken by the cells for different treatments is reasonable. This was also visualized by fluorescence microscopy.
Figure 4 Flow cytometry results for ellipticine uptake by A549 cells (red), untreated cells (blue), cells treated with EAK16-II control (green), cells treated with EPT-H2O (orange), and cell treated with EAK-EPT complex at 5:1 mass ratio.
Abbreviations: EAK, self-assembling EAK16-II peptide; EPT, ellipticine.
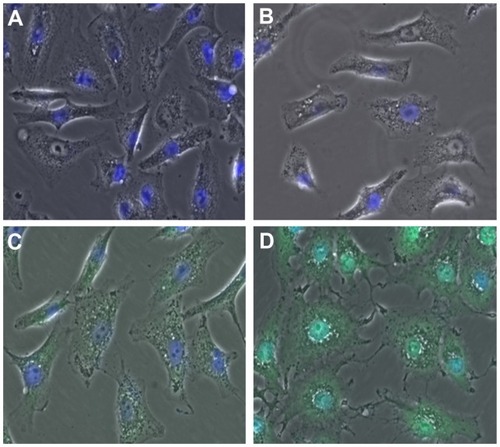
Fluorescence microscopy is a powerful technique for visualizing drug uptake by cells and in particular drug delivery to specific organelles. Given that the literature has reported interaction of ellipticine with DNA at the nucleus level, localization of ellipticine in nuclei should be observable by microscopy. For this purpose, the cells were first stained with DAPI, a standard nucleus dye. In this experiment, the cells were incubated for 2 hours with the following treatments: EAK16-II control, ellipticine control, and EAK-EPT complexes (the same as in the fluorescence-activated cell sorting experiment). Green spots are clearly observable inside the cells for EAK-EPT treated cells, but only blue nuclei appear in the cells treated with the control. When the images from both green and blue filters are overlapped, samples treated with EAK-EPT complexes and stained with DAPI showed an intense cyan color in the nuclei (), a finding significantly different from that in cells treated with ellipticine alone. For the ellipticine control, the nuclei were strongly blue (DAPI) and only slight green uptake was detected in the cytoplasm. This result confirms the fluorescence-activated cell sorting results and indicates cellular uptake of ellipticine by nuclei when it is associated with the EAK16-II peptide ().
Figure 5 Overlap (DAPI + green) images for (A) untreated cells, (B) EAK control cells, (C) ellipticine control cells, and (D) EAK-EPT complex cells. The overlapping blue and green color (cyan) is clearly visible in the nuclei for D.
Abbreviations: DAPI, 4′,6-diamidino-2-phenylindole; EAK, self-assembling EAK16-II peptide; EPT, ellipticine.
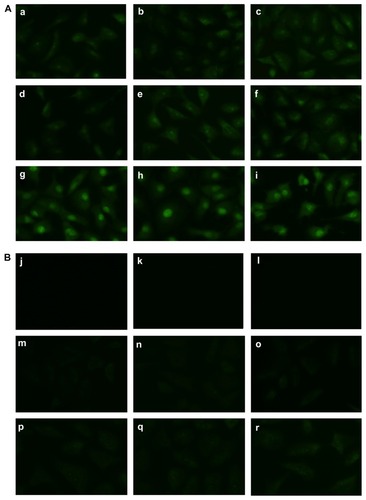
To clarify the uptake mechanism better, a time study of the treatment was conducted over 5 seconds to 2 hours of incubation time for both the ellipticine control and the EAK-EPT complexes. The results indicate the presence of green fluorescence in the cells from the very lowest incubation time (5 seconds) to the longest (2 hours). The images show increasing uptake of ellipticine over time, with ellipticine moving toward the cytoplasm and finally the drug accumulating in nuclei (). However, the uptake of ellipticine alone is not as clear as that of EAK-EPT complexes (). A very small amount of ellipticine is visible in the cytoplasm, but not in the nuclei, even at longer incubation times (). This result illustrates the efficiency of the EAK16-II peptide in delivering the hydrophobic compound, ellipticine, to the nuclei.
Figure 6 (A) Time study of cellular uptake of ellipticine from the EAK-EPT complex at (a) 5 seconds, (b) 45 seconds, (c) 1 minute, (d) 2.5 minutes, (e) 5 minutes, (f) 30 minutes, (g) 1 hour, (h) 1.5 hours, and (i) 2 hours. (B) Time study of cellular uptake of ellipticine from the EPT-H2O control at (j) 5 seconds, (k) 45 seconds, (l) 1 minute, (m) 2.5 minutes, (n) 5 minutes, (o) 30 minutes, (p) 1 hour, (q) 1.5 hours, and (r) 2 hours.
Abbreviations: EAK, self-assembling EAK16-II peptide; EPT, ellipticine.
Note that the experimental setup for microscopy was consistent for imaging of all the samples. The cells were fully washed to detach nanoparticles from the surface. However, there was still a chance that the particles on the surface could not be distinguished from particles inside the cells. However, from the observations we made from the fluorescence microscopy images, the green fluorescence intensity differed between the cell cytoplasm and the nucleus. This might indicate uptake of nanoparticles inside the cells. To confirm this assumption, imaging with confocal microscopy is recommended to overcome any resolution limitation by fluorescence microscopy.
Detection of apoptosis in A549 cells by TUNEL assay
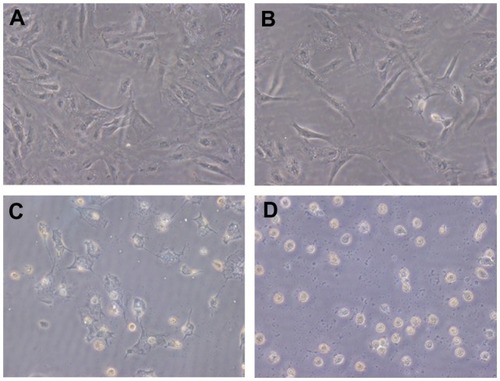
A TUNEL assay was performed to determine whether ellipticine and the EAK-EPT complexes could promote apoptosis of A549 cells. The results of this assay are shown in , where numerous apoptotic cells appear in the ellipticine and EAK-EPT complex groups, but few appear in the control and EAK16-II groups. The apoptotic cell count for the four groups was as follows: EAK-EPT group > ellipticine group > EAK16-II group > control group.
Figure 7 Detection of apoptotic tumor cells by TUNEL assay in the four groups. The magnification of the light microscope was 200×. (A) control, (B) EAK, (C) ellipticine, and (D) EAK-EPT.
Abbreviations: EAK, self-assembling EAK16-II peptide; EPT, ellipticine; TUNEL, terminal deoxynucleotidyl transferase.
TUNEL-positive cells were considered apoptotic and analyzed under a light microscope. The results were calculated as the mean ± standard deviation. The numbers of apoptotic A549 cells are shown in . The results indicate that the numbers of apoptotic cells in the ellipticine and EAK-EPT groups differ significantly from those in the EAK16-II and control groups (P < 0.05). However, the number of tumor cells in the EAK-EPT group is much higher than that in the ellipticine group (P < 0.05).
Table 1 Number of apoptotic tumor cells in four groups (n = 5)
The results of the TUNEL assay show that very few apoptotic cells in the EAK group continue to appear over the time courses observed. In contrast, a large number of apoptotic cells could be seen in the ellipticine and EAK-EPT groups at 48 hours, indicating that cell proliferation was inhibited by ellipticine. However, the number of apoptotic tumor cells in the EAK-EPT group was slightly higher than that in ellipticine group. It is concluded that EAK-EPT had a more inhibitory effect on proliferation of tumor cells than did ellipticine alone.
The results shown above are in accordance with the MTT results. The TUNEL method is generally used for in situ labeling of the DNA strand breaking that occurs in the individual nuclei of apoptotic cells. Cells undergoing apoptosis were identified based on morphologic criteria and cytometry by light microscopy. The greater cell apoptosis seen for the EAK-EPT complexes compared with that for the ellipticine control is probably due to the fact that the EAK16-II peptide can stabilize large amounts of ellipticine in aqueous solution and easily transfer ellipticine into A549 cells. The cellular uptake test for EAK-EPT complexes as shown above indicates that EAK-EPT complexes can penetrate into cells more easily than can ellipticine alone. Thus, this unusual amphiphilic property of EAK16-II allows ellipticine to play an effective role in inducing cell apoptosis.
Antitumor effect of EAK-EPT complexes in vivo
Although a number of EAK-mediated ellipticine delivery systems have been shown to be powerful inhibitors of A549 cell proliferation, in vivo applications for these systems for therapeutic purposes have not been shown. The A549 nude mouse model was used to investigate the potential effects of EAK-EPT complexes on inhibiting the growth of lung cancer cells in vivo. The mice were divided into four groups (ie, model, EAK16-II, ellipticine, and EAK-EPT complexes), each group containing six mice. A549 cells (5 × 106) were subcutaneously injected into the right flanks of the mice in each group. When the tumors reached a size of approximately 100–200 mm3, intraperitoneal treatment was started with EAK16-II, ellipticine, or EAK-EPT. As shown in , inhibition of tumor growth in the nude mice injected with A549 cells increased proportionally in response to time lapse after injection for both the ellipticine and EAK-EPT complex groups, but inhibition was more apparent in the EAK-EPT complex group 14 days after initial injection (). Thus, EAK-EPT complexes obviously exert a stronger antitumor effect than ellipticine alone in nude mice.
Figure 8 Size of tumors in the four groups after treatment. (A) control, (B) EAK, (C) ellipticine, and (D) EAK-EPT complexes.
Notes: Results represent the mean ± standard error (n = 6 tumors). *P < 0.05 versus control group; **P < 0.05 versus ellipticine group.
Abbreviations: EAK, self-assembling EAK16-II peptide; EPT, ellipticine.
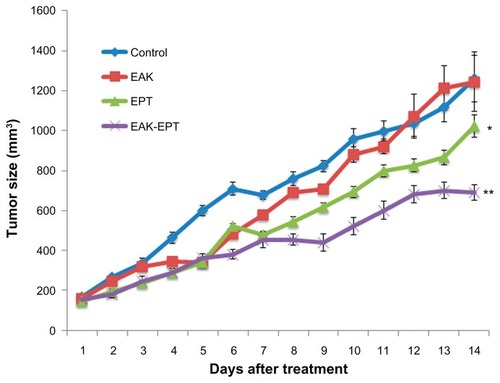
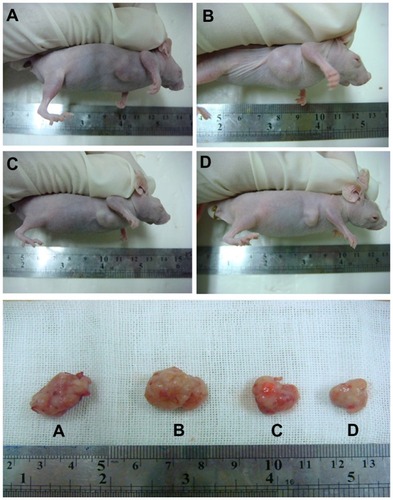
Figure 9 Representative example of tumors from four groups after treatment. On day 14, the tumors were removed. (A) Controls, (B) EAK, (C) ellipticine, and (D) EAK-EPT complexes.
Abbreviations: EAK, self-assembling EAK16-II peptide; EPT, ellipticine.
The in vivo result was in accordance with the MTT assay and cell apoptosis results. The EAK-EPT complexes showed more antitumor activity than ellipticine alone in the A549 lung cancer mouse model, whereas EAK16-II alone had no effect. In our unpublished research, we found in an animal model that EAK-EPT complexes could prolong the residence time of ellipticine and enlarge the area under the curve, which is why EAK-EPT complexes could enhance the therapeutic efficacy of ellipticine in a mouse model of lung cancer. These roles for EAK16-II with regard to ellipticine might be closely related to its structure. The EAK16-II peptide can form stable β-sheet-rich nanostructures in aqueous solution. In a β-sheet arrangement, all hydrophilic residues of the peptide are arranged on one side and the hydrophobic residues are on the other side. This unusual amphiphilic property allows the peptide to interact with the hydrophobic agent ellipticine and protect ellipticine in an aqueous solution. Further, the EPT-EAK nanoparticle complexes may enter cells mainly via endocytotic pathways with rapid uptake in the nucleus, which may enhance the therapeutic effect of ellipticine. Thus, these favorable properties probably facilitate the cellular uptake of ellipticine. After transport, EAK16-II can control the release of ellipticine, protecting it from rapid elimination, and slowing down metabolism of the EAK-EPT complexes in the liver. Therefore, EAK16-II not only conferred stability to ellipticine, but also enabled its controlled release, which would increase the contact time of ellipticine at the site of action and enhance the efficiency of therapy.
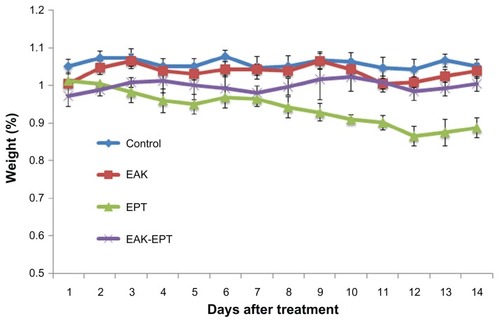
The in vivo toxicity of EAK-EPT complexes was studied by monitoring animal body weight over time. As shown in , compared with the control group, body weight had decreased significantly in the ellipticine group at the end of day 14. However, mouse body weight did not change significantly during treatment with the EAK-EPT complexes. From these results, it can be inferred that the EAK-EPT complex delivery system is safe in animals, with almost no apparent side effects.
Conclusion
EAK16-II, an amino acid-pairing, self-assembling peptide, was used for delivery of the anticancer drug ellipticine. The formulated peptide-drug EAK-EPT complex facilitates the release of ellipticine and has an inhibitory effect on the growth of A549 lung cancer cells in vitro and in vivo. This peptide-drug complex has no apparent toxicity and does not cause weight loss when introduced into the body of the animal. These results suggest that peptide-mediated delivery of hydrophobic anticancer drugs possesses great potential in cancer chemotherapy.
Acknowledgments
YW and PS contributed equally to this work. This project was supported by grant 10410711300 from the Shanghai Committee of Science and Technology, China. This research was financially supported by the Natural Sciences and Engineering Research Council of Canada, Canadian Foundation for Innovation, and the Canada Research Chairs Program for one of the coauthors (PC) and the Alexander Graham Bell Canada Graduate Scholarships for another coauthor (PS). The authors thank Dr Spafford for use of the fluorescence microscopy for uptake experiments, and Tewodros Mamo for assistance with fluorescence imaging at the University of Waterloo, Canada.
Disclosure
The authors report no conflicts of interest in this work.
References
- LiQTainskyMAEpigenetic silencing of IRF7 and/or IRF5 in lung cancer cells leads to increased sensitivity to oncolytic virusesPLoS One20116e2868322194884
- ZhangWChenYWeiHAntiapoptotic activity of autocrine interleukin-22 and therapeutic effects of interleukin-22-small interfering RNA on human lung cancer xenograftsClin Cancer Res2008146432643918927282
- DaraseliaNWangYBudoffAMolecular signature and pathway analysis of human primary squamous and adenocarcinoma lung cancersAm J Cancer Res201229310322206048
- GongHCWangSMayerGSignatures of drug sensitivity in nonsmall cell lung cancerInt J Proteomics872011 [Epub ahead of print.]
- DrobnienėMCicėnienėAZelvienėTPTargeted therapy in patients with non-small cell lung cancer previously treated with chemotherapyMedicina (Kaunas)20114752052522156604
- BykadiGFloraKPCradockJCPoochikianGKDetermination of ellipticine in biological samples by high-performance liquid chromatographyJ Chromatogr19822311371447119056
- AuclairCMultimodal action of antitumor agents on DNA: the ellipticine seriesArch Biochem Biophys19872591143318697
- StiborováMBielerCAWiesslerMFreiEThe anticancer agent ellipticine on activation by cytochrome P450 forms covalent DNA adductsBiochem Pharmacol2001621675168411755121
- SinghMPHillGCPéoc’hDRaynerBImbachJLLownJWHigh-field NMR and restrained molecular modeling studies on a DNA heteroduplex containing a modified apurinic abasic site in the form of covalently linked 9-aminoellipticineBiochemistry19943310271102858068666
- MonnotMMauffretOSimonVDNA-drug recognition and effects on topoisomerase II-mediated cytotoxicity. A three-mode binding model for ellipticine derivativesJ Biol Chem1991266182018291846365
- FosséPRenéBCharraMPaolettiCSaucierJMStimulation of topoisomerase II-mediated DNA cleavage by ellipticine derivatives: structure-activity relationshipMol Pharmacol1992425905951331751
- Froelich-AmmonSJPatchanMWOsheroffNThompsonRBTopoisomerase II binds to ellipticine in the absence or presence of DNA. Characterization of enzyme-drug interactions by fluorescence spectroscopyJ Biol Chem199527014998150047797481
- StiborováMArltVMHendersonCJRole of hepatic cytochromes P450 in bioactivation of the anticancer drug ellipticine: studies with the hepatic NADPH: cytochrome P450 reductase null mouseToxicol Appl Pharmacol2008226258327
- PoljakováJDračínskýMFreiEHudečekJStiborováMThe effect of PH on peroxidase-mediated oxidation of and DNA adduct formation by ellipticineCollect Czechoslov Chem Commun20097111691185
- GarbettNCGravesDEExtending nature’s leads: the anticancer agent ellipticineCurr Med Chem Anticancer Agents2004414917215032720
- ClarysseABrugarolasASiegenthalerPPhase II study of 9-hydroxy-2 N-methylellipticinium acetateEur J Cancer Clin Oncol1984202432476538489
- WangYLiYWangQWuJFangXPharmacokinetics and biodistribution of paclitaxel-loaded pluronic P105/L101 mixed polymeric micellesYakugaku Zasshi200812894195018520140
- PortneyNGOzkanMNano-oncology: drug delivery, imaging, and sensingAnal Bioanal Chem200638462063016440195
- GreenMRManikhasGMOrlovSAbraxane, a novel cremophor-free, albumin-bound particle form of paclitaxel for the treatment of advanced non-small-cell lung cancerAnn Oncol2006171263126816740598
- CorbinIRZhengGMimicking nature’s nanocarrier: synthetic low-density lipoprotein-like nanoparticles for cancer-drug deliveryNanomedicine (Lond)2007237538017716181
- FungSYYangHChenPSequence effect of self-assembling peptides on the complexation and in vitro delivery of the hydrophobic anticancer drug ellipticinePLoS One200834e195618398476
- LawMJafariMChenPPhysicochemical characterization of siRNA-peptide complexesBiotechnol Prog20082495796319194904
- KimSWKimNYChoiYBParkSHYangJMShinSRNA interference in vitro and in vivo using an arginine peptide/siRNA complex systemJ Control Release201014333534320079391
- ZhangSHolmesTLockshinCRichASpontaneous assembly of a self-complementary oligopeptide to form a stable macroscopic membraneProc Natl Acad Sci U S A199390333433387682699
- FungSYOyaizuTYangHThe potential of nanoscale combinations of self-assembling peptides and amino acids of the src tyrosine kinase inhibitor in acute lung injury therapyBiomaterials2011324000400821376387
- ZhangSHolmesTCDiPersioCMHynesROSuXRichASelf- complementary oligopeptide matrices support mammalian cell attachmentBiomaterials199516138513938590765
- FungSYYangHBholaPTSelf-assembling peptide as a potential as a potential carrier for hydrophobic anticancer drug ellipticine: complexation, release and in vitro deliveryAdv Funct Mater2009197483
- BawaRFungSYShiozakiASelf-assembling peptide-based nanoparticles enhance cellular delivery of the hydrophobic anticancer drug ellipticine through caveolae-dependent endocytosisNanomedicine912011 [Epub ahead of print.]
- ZhangSEmerging biological materials through molecular self-assemblyBiotechnol Adv20022032133914550019
- GarbettNCGravesDEExtending nature’s leads: the anticancer agent ellipticineCurr Med Chem Anticancer Agents2004414917215032720
- SaijoNTamuraTNishioKStrategy for the development of novel anticancer drugsCancer Chemother Pharmacol200352Suppl 1S9710112856152