 ?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.
?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.Abstract
Background
The aim of this work was to improve oral bioavailability. The uptake of a series of quaternary ammonium salt didodecyl dimethylammonium bromide (DMAB)-modified nanoparticles (with uniform sizes ranging from 50 nm to 300 nm) into heterogeneous human epithelial colorectal adenocarcinoma cells (Caco-2) and human colon adenocarcinoma cells (HT-29) was investigated.
Methods
Coumarin-6 (C6) loaded poly (lactide-co-glycolide) (PLGA) nanoparticles were prepared with DMAB using the emulsion solvent diffusion method. The physicochemical properties and cellular uptake of these nanoparticles were studied. Deserno’s model was applied to explain the experimental observations.
Results
The results showed that the surface modification of PLGA nanoparticles with DMAB notably improved the cellular uptake. The cellular uptake was size-dependent and had an optimal particle size of 100 nm. The experimental data was integrated numerically, and was in agreement with the theoretical model.
Conclusion
These results indicated that the interactions between the charged nanoparticles and the cells resulted from various forces (eg, electrostatic forces, hydrophobic forces, bending and stretching forces, and limited receptor-mediated endocytosis), and the uptake of the nanoparticles occurred as a result of competition.
Introduction
As nanotechnology has developed, polymeric nanoparticles have come to be considered the most promising candidate for the oral administration of drugs.Citation1,Citation2 Nanoparticles (NPs) not only effectively protect drugs from gastrointestinal interferences (chemical and enzymatic degradation), but also circumvent the liver first-pass metabolism caused by cytochrome P450/P-glycoprotein.Citation3–Citation5 Various novel biomaterials (eg, didodecyl dimethylammonium bromide (DMAB), vitamin E d-alpha tocopheryl polyethylene glycol-1000 succinate, and montmorillonite) have been used for oral administration.Citation6–Citation8 In combination with biodegradable materials such as coumarin-6 (C6) loaded poly (lactide-co-glycolide) (PLGA), these biomaterials have been used to prepare small nanoparticles (<300 nm). The surface properties of these nanoparticles were significantly enhanced at increased electric potentials and hydrophobicities, thereby strengthening the adhesion of the nanoparticles to the gastrointestinal cells. The uptake of the nanoparticles was improved, which increased the oral bioavailability and the therapeutic efficiency.Citation9,Citation10
In efforts to find effective therapy formulations, heterogeneous human epithelial colorectal adenocarcinoma cells (Caco-2) and human colon adenocarcinoma cells (HT-29) are often used to simulate the gastrointestinal tract in in vitro screening studies. In vitro studies focus on the cellular uptake value of nanoparticles, which provides information on the effectiveness of the nanoparticle preparation. A number of systematic studies evaluating cellular uptake have been reported in the literature;Citation11–Citation13 they found that the cellular uptake was affected by various nanoparticle characteristics, including the nanoparticles’ size, surface chemical structure, and shape.Citation14 The phenomenon of particle size-dependent uptake has attracted attention from many researchers.Citation9,Citation15,Citation16 However, the results of their studies differ. Most agreed that the smallest wrapping time depended on the particle size; through studies of the receptor-mediated endocytosis of viruses, the optimal size was determined to be approximately 50 nm.Citation17 Other studies found that the maximum uptake of nanoparticles occurred for 100 nm particles.Citation9,Citation18 These results indicated that the interactions between nanoparticles and cells could vary dramatically, resulting in different optimal sizes for cellular uptake. When the ectogenic nanosize particles touch cells, the cell membranes curve, allowing the cell to phagocytize or fuse with the drug-loaded nanoparticles.Citation11 The penetration of the particles into the cells depended on the adhesion strength, the membrane tension, and the bending energy. The efficiency of this penetration affected the final uptake.Citation19,Citation20 According to the theory of dissipative particle dynamics, Smith et al found that cell membranes could not fully wrap around the particle if the adhesion strength was below a threshold value.Citation21 These well-defined models successfully captured some of the features of the particle-uptake process, and may be applied here to investigate physical models for cellular uptake.
Recent studies of DMAB-modified PLGA nanoparticles have illustrated their improved potential as an oral drug delivery system.Citation22,Citation23 Compared to poly vinyl alcohol (PVA), DMAB has become known as a more suitable stabilizer because of its smaller size and positive zeta potential.Citation24,Citation25 The current study used PLGA nanoparticles stabilized with the cationic surfactant DMAB to imitate the size-dependent cellular uptake phenomenon. The results showed that this dependency was generated, and that these particles had the potential to increase the oral bioavailability of drugs such as paclitaxel. A fluorescent marker, C6, was encapsulated in the nanoparticles to allow visualization of the cellular uptake.Citation26 It was found that the cell uptake of modified polymeric nanoparticles in Caco-2 and HT-29 cells was size dependent; the maximum uptake was observed for particles of 103 nm. However, these results differed from results obtained using receptor-mediated endocytosis. To explain this phenomenon, this study proposed a new model for the size-dependent uptake based on the theory of dynamics. This model explained the results very well. This physical model could be helpful in preparing nanoparticles for specific applications and for solving other problems in this area.
Methods
Materials
The PLGA (50:50, molecular weight: 10,000 Da) was purchased from the Shandong Institute of Medical Instruments (Jinan, China). Fluorescence marker C6 and triton® X-100 were obtained from Sigma-Aldrich (St Louis, MO). DMAB, propylene carbonate, and PVA (molecular weight: 30000–70000 Da) were also purchased from Sigma-Aldrich. Ethyl acetate (analytical grade) and acetonitrile (HPLC grade) were purchased from Merck KGaA (Darmastadt, Germany). Distilled water (Milli-Q, 18 M Ω; Millipore, Billerica, MA) was used in the synthesis. All other materials were analytical grade. Fetal bovine serum (FBS), RPMI-1640 medium, penicillin-streptomycin (10000 U/mL), trypsin-EDTA solution (0.5% trypsin, 5.3 Mm EDTA tetra-sodium), and Hank’s balanced salt solution (HBSS) were obtained from Gibco BRL (Galthersberg, MD). The BCA™ Protein Assay Kit was purchased from the Beyotime Institute of Biotechnology (Nanjing, China).
Preparation of nanoparticles
Nanoparticles loaded with C6 were prepared by the emulsification-diffusion method. In detail, 200 mg of PLGA (50:50) and 10 mg of C6 were dissolved in 10 mL of ethyl acetate or propylene carbonate. Then, the oil phase was slowly added into a 20 mL aqueous solution containing 0.5% DMAB, and emulsified twice using a high speed homogenizer (Fluko, Shanghai, China) at 15,000 g for 10 min, for 5 min each time. The resulting oil-in-water emulsion solution was slowly poured into 100 mL of water under moderate magnetic stirring. The organic solvents were removed through rotary-evaporation or dialysis. The formed nanoparticles were collected by centrifugation (J2-MC, Backman, 34,000 rpm, 30 min, 4°C) and washed with distilled water three times. The nanoparticle pellet was re-suspended in solution for further study. Blank nanoparticles were prepared by the same method. Nanoparticles, blank or loaded with C6, were prepared by the solvent emulsification-diffusion-evaporation method. Briefly, 200 mg of PLGA block copolymer (50:50, 10,000 Da) and 10 mg of C6 in 10 mL of oil phase solution (ethyl acetate) was poured in 200 mL of aqueous solution containing 5% (w/v) PVA and emulsified using a microfluidizer (M-110Y; Microfluidics, Newton, MA) with circulating pressure of 18,000 psi, ten cycles for each sample. The resulting oil-in-water emulsion was then evaporated to remove the ethyl acetate. The formed nanoparticles were collected by centrifugation (J2-MC, Backman, 10,000 rpm, 30 min, 4°C) and washed with distilled water three times. The nanoparticle pellet was re-suspended in solution for further study. Untrapped C6 was removed by centrifugation. The nanoparticle samples were freeze-dried. Three or four milligrams of C6-loaded nanoparticles were dissolved in 10 mL of DMF solution. The drug concentration was determined using a fluorescence spectrofluorometer (FP6500; JASCO, Japan) (λex 430 nm, λem 485 nm). Drug loading content and drug loading efficiency were calculated. Mean hydrodynamic diameters and mean size distributions were determined at 25°C by means of a dynamic-light scattering technique with particle size analyzer (Zetasizer Nano-S90; Malvern Instruments, Malvern, UK). The polydispersity index, a dimensionless measure of size distribution, was also recorded. Zeta potential is an indicator of surface charge and particle stability in dispersion; it was measured using a zeta potential analyzer (90 Plus; Brookhaven Instruments, Huntsville, NY). Transmission electron microscopy (TEM) images were taken using a JEM 1230 electron microscope (JEOL, Tokyo, Japan) at an acceleration voltage of 80 kV.
Cellular uptake of C6-labeled nanoparticles
Caco-2 cells of passages 25–30 were obtained from the Institute of Biochemistry and Cell Biology of Chinese Academy of Sciences (Shanghai, China). Caco-2 cells were cultured in MEM medium with 1% no-essential amino acid and L-glutamine, and 1% penicillin-streptomycin solution supplemented with 10% FBS at 37°C, 5% CO2, and 95% humidity. Another human colon adenocarcinoma cell line, HT-29, was also obtained from the Institute of Biochemistry and Cell Biology of Chinese Academy of Sciences (Shanghai, China). These cells were cultured in RPMI-1640 medium containing 10% FBS and 1% penicillin-streptomycin at 37°C, 5% CO2, and 95% relative humidity. The medium was replenished every other day. Quantitative studies of nanoparticle uptake were determined using the methods reported.Citation9 Caco-2 and HT-29 cells were used to simulate the gastrointestinal (GI) barrier for oral chemotherapy. Caco-2 or HT-29 cells were seeded at 5 × 104 cells on 96-well black plates (Costar; Corning Inc, Corning, NY) and incubated until the cells reached 80% confluence. The medium was changed with HBSS (pH 7.4) and incubated for 30 min. After equilibration, the culture medium was changed with 100 μL of medium made up of specified C6-loaded nanoparticle suspension (500 μg/mL in HBSS). The samples were incubated with the cells for 4 hours at 37°C. Then, the cells were washed using phosphate-buffered saline (PBS, pH 7.4) to eliminate free particles that were not taken up by the cells. After washing three times, 0.5% triton® X-100 in a 0.2 N NaOH solution was added to destroy the cell membrane completely. Quantitative measurement was then performed by the definite quantitative determination of each cell lysate with a fluorescence microplate reader (Tecan, Männedorf, Switzerland, λex 430 nm, λem 485 nm). For each sample, the negative control and protein amount were also determined.
Confocal laser scanning microscopy
Caco-2 and HT-29 cells were seeded on glass bottom culture dishes and incubated at 37°C, 5% CO2, and 95% relative humidity. After 80% confluence, the medium was removed and replaced by HBSS. Thirty minutes later, the solution was replaced with the C6 nanoparticle suspension (size: 56 nm and 97 nm, 500 μg/mL in HBSS). The cells were incubated with particles at 37°C for 4 hours. Then, the cells were washed three times with a PBS (pH 7.4) solution to eliminate free particles that were not taken up by the cells. Cells were then fixed with 4% paraformaldehyde in PBS for 30 min. Then samples were examined with a confocal laser scanning microscope (LSM 510; Carl Zeiss Microscopy GmbH, Jena, Germany). The excitation wavelengths were 430 nm (C6, green).
Results and discussion
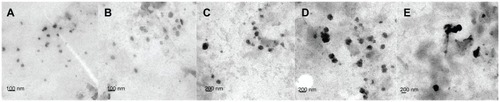
C6-marked nanoparticles were prepared with DMAB by the emulsion-diffusion-evaporation method. PVA was also used to prepare nanoparticles through a previously reported method.Citation23,Citation24 The physical characterization of products was performed using dynamic light scattering, TEM, and zeta potential measurements. By controlling the concentrations of the stabilizer and the emulsive pressure, the researchers obtained a series of DMAB-modified nanoparticles with sizes ranging from 50 nm to 300 nm. The particle sizes, polydispersity indices, and zeta potentials of the C6-loaded nanoparticles are presented in . TEM images of the nanoparticles showed that they possessed excellent uniformity of both shape and size (). The zeta potential varied when different emulsifiers were used in the nanoparticle fabrication process. All of the DMAB-capped nanoparticles in the study showed a positive charge, with zeta potentials of around 58 mV. The nanoparticles created in this study were found to have sizes of 50–300 nm; the size had no effect on the zeta potential. PVA-modified nanoparticles were negatively charged, with a zeta potential of about −1.4 mV. The encapsulation efficiency of the PVA-modified nanoparticles was (30% ± 4.2%). With DMAB as the emulsifier, however, the efficiency reached more than 40%. It was concluded that DMAB’s positive charge may have been the cause of this high encapsulation efficiency. In the present study, the nanoparticles were found to be stable in dispersion, with high absolute zeta potential values. Both the drug-loaded particles and the blank particles were freeze-dried to obtain a fine powder, which was normalized to the same molar concentration for cellular assays.
Table 1 Physicochemical properties of nanoparticles (n = 3)
Figure 1 TEM micrograph of DMAB-modified PLGA nanoparticles with different average sizes: (A) 50 nm; (B) 100 nm; (C) 150 nm; (D) 200 nm; and (E) 300 nm.
Abbreviations: TEM, transmission electron microscopy; DMAB, didodecyl dimethylammonium bromide; PLGA, coumarin-6 loaded poly (lactide-co-glycolide).
C6 was used as a label to evaluate the intestinal permeability of the nanoparticles. The cellular uptake was determined for the two types of nanoparticles using a fluorescence microplate reader. Two human colonic carcinoma cell lines (Caco-2 and HT-29) were used as in vitro models to quantify the cellular uptake of the fluorescent nanoparticles. These cell lines were chosen because of their structural and functional similarities with mature enterocytes. Free C6 was incubated with Caco-2 cells and HT-29 cells for 4 hours as a control. The uptake of NPs by Caco-2 and HT-29 was also examined, using fluorescence spectroscopy, to study the relationship between cellular uptake and the sizes of NPs. This was performed for both PVA- and DMAB-coated nanoparticles.
The cellular uptake efficiency results are displayed in . The control experiments showed that free C6 could not be directly internalized by the cells, so the measured fluorescence directly reflected the uptake of fluorescent nanoparticles (rather than free fluorescence molecules). This result clearly showed that the cellular uptake of DMAB-coated nanoparticles was much higher than that of PVA-coated nanoparticles, which confirmed that DMAB has the potential to improve the oral bioavailability of anticancer drugs such as paclitaxel. The DMAB-modified nanoparticles showed smaller sizes and higher zeta potentials than the PVA-coated nanoparticles, meaning that the DMAB-coated nanoparticles could be entrapped by the cells more easily.
Figure 2 Cellular uptake efficiency of PVA-coated nanoparticles and DMAB-coated nanoparticles with different sizes (A). PLGA nanoparticles were incubated in Caco-2 cells (gray bars) and HT-29 cells (black bars) for 4 hours at a concentration of 500 μg/mL, at 37°C; the uptake rate is given as a function of the size of the nanoparticle (B).
Note: The calculated curve (red and blue solid lines) agreed well with the experimental data (red squares and blue circles, respectively).
Abbreviations: PVA, poly vinyl alcohol; DMAB, didodecyl dimethylammonium bromide; PLGA, coumarin-6 loaded poly (lactide-co-glycolide).
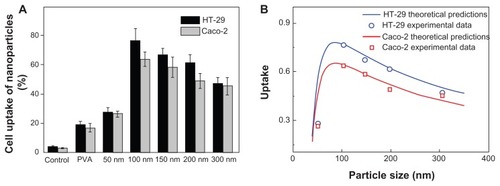
The cellular uptake of DMAB-modified nanoparticles did not increase when the size was smaller than 100 nm. The cellular uptake exhibited size-dependence for the DMAB-coated PLGA nanoparticles in both cell models, and the optimum size was around 100 nm for both. This was different from the maximum cell uptake at a nanoparticle size of 50 nm that was reported for receptor-mediated endocytosis.Citation27 Other investigations showed similar results.Citation9,Citation18 These results indicated that there was another mechanism by which the surfactant-capped nanoparticles interacted with the cell membrane apart from receptor-mediated endocytosis. This study aimed to uncover this mechanism.
Recently, Peetla and Labhasetwar investigated the effects of different cationic surfactants on the biophysical interactions of surfactant-modified nanoparticles with an endothelial cell model membrane.Citation25 Their results indicated that DMAB-modified nanoparticles changed the surface pressure of the membrane to a remarkable extent, and were more aggressive to the cells than either cetyl trimethylammonium bromide- or dodecyl trimethylammonium bromide-modified nanoparticles. These findings clearly suggested that the basic mechanisms of the interactions of nanoparticles with cell membranes were homogeneous. Here, Deserno’s model was extended to analyze this difference in detail.
During the nanoparticle wrapping process, the elastic energy increases with the deformation of the cell membrane. The elastic energy is the sum of the bending and stretching components. The bending energy of the cell membrane is described by HelfrichCitation28 as
where κB and κG are the bending stiffnesses for the mean and Gaussian curvatures, respectively, κ1 and κ2 are the principal curvatures, and κ0 is the spontaneous curvature. The stretching energy of the cell membrane is approximated as the work performed by the surface tension, γ,
where ΔA is the change in the surface area of the cell membrane, and γ = γ0 + ke1ΔA/A0. In this equation, γ0 is the baseline surface tension when the cell is spherical, kel is the elastic expansion modulus, and A0 is the surface area before the cell deformation.Citation29 If wad denotes the adhesion strength induced by driven forces (eg, the electrostatic interaction), and Aad denotes the contact area, the adhesion energy can be expressed as:
The total Gibbs free energy of the system can be written as:
Since this paper does not consider topological changes, the energy due to Gauss curvature is negligible. EquationEquation 4(4) can be rewritten as
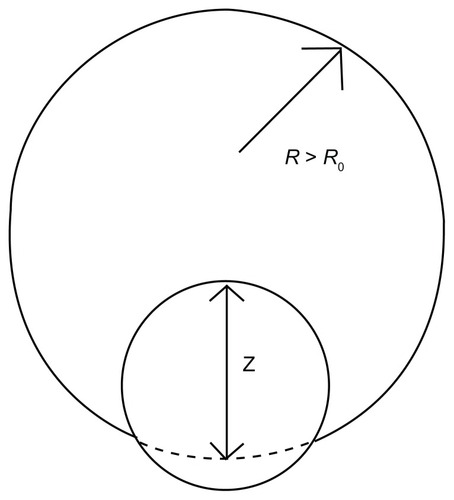
There is a change of the surface area ΔA = πa2z2, and the contact area Aad = 2πa2z, where a is the radius of nanoparticles, and z is the degree of penetration ().Citation30,Citation31 Using the expressions for ΔA and Aad in combination with EquationEquation 5(5) yields:
Figure 3 Schematic view of a nanoparticle adhering to a cell.
Abbreviations: R0, original radius of the cell; R, radius of the deformed cell; z, an index by which to estimate the percentage of the particles taken by the cell.
The minimization of the free energy requires that z satisfies
According to recent work, it is reasonable to take z as an index to estimate the percentage of the particles taken by the cell. So, z = 0 and z = 2 represent no uptake and complete uptake, respectively.Citation9 Taking wad/κB=1400 μm−2,γ0/κB=1.25×10−3 nm−2,kel/κB=5 nm−2, and R0 = 5 μm, the theoretical model is integrated numerically and in agreement with the experimental data (). The uptake rate as a function of the nanoparticle diameter is shown in . There existed a critical size (98 nm) below which the uptake rate increased with increasing particle size. The uptake decreased with increasing size when the radius exceeded the critical size.
The following is a possible model for the cellular uptake process: generally, when the nanoparticle is small, the membrane deformation results predominantly from the bending energy. In this case, whether nanoparticles can enter the cell is decided mainly by the driven energy and the bending energy of the membrane. The bending energy of the membrane is larger when wrapping a small particle, which induces the low uptake ratio for small particles. When the particle exceeds the critical size, the cell membrane’s main energy costs are in the stretching energy. In that case, the uptake of the big nanoparticles is difficult because big particles cause higher stretching energies in the membrane. The relation between the size of the nanoparticles and the uptake rate depends on the competition between the bending energy and the stretching energy. This model is consistent with the experimental observations.Citation15 The researchers conducted additional theoretical analysis and discussed the results in detail.
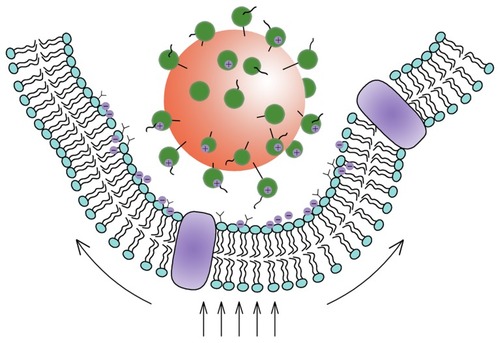
Based on the aforementioned analysis, it would be expected that the major force for inducing the cellular uptake of nanoparticles prepared using DMAB would be complicated. The interactions between the nanoparticles and the cell membrane included electrostatic interactions, and receptor-mediated endocytosis, which should agree well with the improved Deserno’s model presented here. This conclusion was supported by the cell uptake of PVA nanoparticles, as well as other experimental results reported in previous investigations.Citation16,Citation32,Citation33 In the absence of a driving force, such as electrostatic interactions, the cell uptake was always limited, indicating that in some cases the receptor-mediated endocytosis was not the main driving force for the uptake. The uptake of nanoparticles was ultimately shown to be size-dependent. This uptake process was influenced by various forces, including electrostatic forces, hydrophobic forces, bending and stretching forces, and limited receptor-mediated endocytosis (). The improved uptake of DMAB-modified PLGA nanoparticles over PVA-modified particles is easy to understand in light of the additional electrostatic forces present in the DMAB/PLGA system.
Figure 4 Schematic view of the interaction between nanoparticles and cells.
Note: The Deserno’s model showed that the uptake of the nanoparticles occurred as a result of the competition between the bending energy and the stretching energy.
The Caco-2 and HT-29 cell lines were both derived from human colon carcinomas. These cell lines have been investigated as feasible candidates for simulating the GI barrier in vitro. The significantly enhanced uptake into both of these cell types indicated that the DMAB-capped nanoparticles should develop a strong interaction with the GI tract mucus/epithelial surface, making it possible to achieve the oral delivery of anticancer drugs, proteins, peptides, and nucleic acids. Finally, the aggregates of the green C6 found in the confocal laser scanning microscopy experiments confirmed that the nanoparticles had been internalized by the cells ().
Figure 5 Confocal microscopic images of (A) Caco-2 cells and (B) HT-29 cells incubating with DMAB-modified C6-loaded nanoparticles of (a) 100 nm and (c) 50 nm for 4 hours at the concentration of 500 μg/mL, at 37°C. The close-ups of entrapped fluorescent nanoparticles (green) could be observed directly in the right panels [(b) and (d)].
Note: Scale bars = 20 μm.
Abbreviations: DMAB, didodecyl dimethylammonium bromide; C6, coumarin-6.
![Figure 5 Confocal microscopic images of (A) Caco-2 cells and (B) HT-29 cells incubating with DMAB-modified C6-loaded nanoparticles of (a) 100 nm and (c) 50 nm for 4 hours at the concentration of 500 μg/mL, at 37°C. The close-ups of entrapped fluorescent nanoparticles (green) could be observed directly in the right panels [(b) and (d)].Note: Scale bars = 20 μm.Abbreviations: DMAB, didodecyl dimethylammonium bromide; C6, coumarin-6.](/cms/asset/4edf75be-d6d2-4b7f-b1c7-a4e7f46b5150/dijn_a_32188_f0005_c.jpg)
Conclusion
In summary, DMAB-modified polymeric nanoparticles could enhance the drug oral bioavailability in vitro for cell models. It was concluded that the cell uptake of modified polymeric nanoparticles was size-dependent in Caco-2 cells and HT-29 cells. The cellular uptake rose to maximum levels for nanoparticles with a size of 100 nm, which differed from the results for uptake via receptor-mediated endocytosis. The results discussed previously suggested that the uptake of nanoparticles prepared with DMAB resulted from the surface potential-induced, passive dynamic behavior of cells. The results also fitted well with a dynamic model. Future research should also address the interactions among the nanoparticles and cell membrane.
Acknowledgment
The suppor t f rom the Nat ional Basic Research Program of China (973 Program, 2009 CB930300) is acknowledged.
Disclosure
The authors report no conflicts of interest in this work.
References
- AnkolaDDViswanadBBhardwajVRamaraoPKumarMNDevelopment of potent oral nanoparticulate formulation of coenzyme Q10 for treatment of hypertension: can the simple nutritional supplements be used as first line therapeutic agents for prophylaxis/therapy?Eur J Pharm Biopharm200767236136917452099
- MittalGSahanaDKBhardwajVRavi KumarMNEstradiol loaded PLGA nanoparticles for oral administration: effect of polymer molecular weight and copolymer composition on release behavior in vitro and in vivoJ Control Release20071191778517349712
- BalaIHariharanSKumarMNPLGA nanoparticles in drug delivery: the state of the artCrit Rev Ther Drug Carrier Syst200421538742215719481
- DongYFengSSPoly(d,l-lactide-co-glycolide)/montmorillonite nanoparticles for oral delivery of anticancer drugsBiomaterials200526306068607615894372
- WinKYFengSSIn vitro and in vivo studies on vitamin E TPGS-emulsified poly(D,L-lactic-co-glycolic acid) nanoparticles for paclitaxel formulationBiomaterials200627102285229116313953
- SahanaDKMittalGBhardwajVKumarMNPLGA nanoparticles for oral delivery of hydrophobic drugs: influence of organic solvent on nanoparticle formation and release behavior in vitro and in vivo using estradiol as a model drugJ Pharm Sci20089741530154217722098
- FengSSZengWTeng LimYVitamin E TPGS-emulsified poly(lactic-co-glycolic acid) nanoparticles for cardiovascular restenosis treatmentNanomedicine (Lond)20072333334417716178
- FengSSMeiLAnithaPGanCWZhouWPoly(lactide)-vitamin E derivative/montmorillonite nanoparticle formulations for the oral delivery of DocetaxelBiomaterials200930193297330619299012
- WinKYFengSSEffects of particle size and surface coating on cellular uptake of polymeric nanoparticles for oral delivery of anticancer drugsBiomaterials200526152713272215585275
- LlabotJMSalmanHMillottiGBernkop-SchnurchAAllemandiDManuel IracheJBioadhesive properties of poly(anhydride) nanoparticles coated with different molecular weights chitosanJ Microencapsul20112841920945969
- VermaAStellacciFEffect of surface properties on nanoparticle-cell interactionsSmall201061122119844908
- ChaudhuriABattagliaGGolestanianRThe effect of interactions on the cellular uptake of nanoparticlesPhys Biol20118404600221508440
- GaoHShiWFreundLBMechanics of receptor-mediated endocytosisProc Natl Acad Sci U S A2005102279469947415972807
- HariharanSBhardwajVBalaISitterbergJBakowskyURavi KumarMNDesign of estradiol loaded PLGA nanoparticulate formulations: a potential oral delivery system for hormone therapyPharm Res200623118419516267632
- ChithraniBDGhazaniAAChanWCDetermining the size and shape dependence of gold nanoparticle uptake into mammalian cellsNano Lett20066466266816608261
- WangSHLeeCWChiouAWeiPKSize-dependent endocytosis of gold nanoparticles studied by three-dimensional mapping of plasmonic scattering imagesJ Nanobiotechnology2010812334521167077
- LuFWuSHHungYMouCYSize effect on cell uptake in well-suspended, uniform mesoporous silica nanoparticlesSmall20095121408141319296554
- YangJZengRKongTThe effect of surface properties of gold nanoparticles on cellular uptake2007 IEEE NIH Life Sci Syst Appl Workshop200710129295
- WhiteheadKALangerRAndersonDGKnocking down barriers: advances in siRNA deliveryNature Reviews200982129138
- PanyamJLabhasetwarVBiodegradable nanoparticles for drug and gene delivery to cells and tissueAdv Drug Deliv Rev200355332934712628320
- SmithKAJasnowDBalazsACDesigning synthetic vesicles that engulf nanoscopic particlesJ Chem Phys2007127808470317764280
- BhardwajVAnkolaDDGuptaSCSchneiderMLehrCMKumarMNPLGA nanoparticles stabilized with cationic surfactant: safety studies and application in oral delivery of paclitaxel to treat chemical-induced breast cancer in ratPharm Res200926112495250319756974
- SongKCLeeHSChoungYChoKIAhnYChoiEJThe effect of type of organic phase solvents on the particle size of poly(D,L-lactide-co-glycolide) nanoparticlesColloids and Surfaces A: Physicochemical and Engineering Aspects20062761–3162167
- KwonHYLeeJYChoiSWJangYSKimJHPreparation of PLGA nanoparticles containing estrogen by emulsification-diffusion methodColloids and Surfaces A: Physicochemical and Engineering Aspects20011821–3123130
- PeetlaCLabhasetwarVEffect of molecular structure of cationic surfactants on biophysical interactions of surfactant-modified nanoparticles with a model membrane and cellular uptakeLangmuir20092542369237719161268
- SunBRanganathanBFengSSMultifunctional poly(D,L-lactide-co-glycolide)/montmorillonite (PLGA/MMT) nanoparticles decorated by Trastuzumab for targeted chemotherapy of breast cancerBiomaterials200829447548617953985
- HuangJBuLXieJEffects of nanoparticle size on cellular uptake and liver MRI with polyvinylpyrrolidone-coated iron oxide nanoparticlesACS Nano284127151716021043459
- HelfrichWElastic Properties of lipid bilayers – theory and possible experimentsZ Naturforsch C1973C28116937034273690
- HerantMHeinrichVDemboMMechanics of neutrophil phagocytosis: behavior of the cortical tensionJ Cell Sci200511891789179715827090
- DesernoMElastic deformation of a fluid membrane upon colloid bindingPhys Rev E2004693111
- DesernoMGelbartWMAdhesion and wrapping in colloid-vesicle complexesJ Phys Chem B20021062155435552
- DesaiMPLabhasetwarVWalterELevyRJAmidonGLThe mechanism of uptake of biodegradable microparticles in Caco-2 cells is size dependentPharm Res19971411156815739434276
- OhWKKimSChoiMCellular uptake, cytotoxicity, and innate immune response of silica-titania hollow nanoparticles based on size and surface functionalityACS Nano28;4953015313