Abstract
Carbon nanotubes have been explored as heat-delivery vehicles for thermal ablation of tumors. To use single-walled carbon nanotubes (SWNT) as a “molecular heater” for hyperthermia therapy in cancer, stable dispersibility and smart-delivery potential will be needed, as well as lack of toxicity. This paper reports the preparation of a model complex comprising DNA-treated SWNT and anti-human IgG antibody and the specific binding ability of this model complex with the targeted protein, ie, human IgG. Treatment with double-stranded DNA enabled stable dispersibility of a complex composed of SWNT and the antibody under physiological conditions. Quartz crystal microbalance results suggest that there was one immobilized IgG molecule to every 21,700 carbon atoms in the complex containing DNA-treated SWNT and the antibody. The DNA-SWNT antibody complex showed good selectivity for binding to the targeted protein. Binding analysis revealed that treatment with DNA did not interfere with binding affinity or capacity between the immobilized antibody and the targeted protein. The results of this study demonstrate that the DNA-SWNT antibody complex is a useful tool for use as a smart “molecular heater” platform applicable to various types of antibodies targeting a specific antigen.
Introduction
Carbon nanotubes have been intensively investigated for biological and biomedical applications. Carbon nanotubes release substantial vibrational energy after exposure to near-infrared radiation. This release of energy produces heating localized within a tissue, which can potentially be used for phototherapy in the treatment of cancer.Citation1–Citation5
Our hypothesis was that single-walled carbon nanotubes (SWNT) could be used as an exothermic molecular device for hyperthermic cancer therapy. To achieve good performance for an SWNT-based molecular heating device, two main issues should be considered, ie, achieving stability and good dispersibility of SWNT in body fluids and selective attachment of the SWNT to targeted tumor cells. In a previous study,Citation6 we prepared DNA-SWNT by treatment with double-stranded DNA. The DNA-SWNT was shown to improve the dispersibility of SWNT in vitro (culture medium) and in vivo (subcutaneous mouse tissue) significantly when compared with acid-treated SWNT. This result suggests that functionalization with DNA is an efficient approach to improve the dispersibility of SWNT in body fluids.
To achieve selective targeting of cells, conjugation of DNA-SWNT with certain ligands specifically recognizing tumor cells would be the most efficient approach. Targeting ligands, including folic acid, peptides, and antibodies, have been used for carbon nanotube-based drug delivery systems to develop a smart-delivery nanocarrier.Citation7,Citation8 Several researchers have studied specific antibodies attached to the side wall of the SWNT. Chakravarty et alCitation9 prepared an anti-CD22-targeted SWNT construct and demonstrated its selective binding to tumor cells. The SWNT was solubilized by interaction with biotinylated poly(ethylene glycol) (PEG), and coupled with an antibody-avidin conjugate. McDevitt et alCitation10 synthesized SWNT antibody constructs from covalently attached amino-terminated PEG and thiolated anti-CD 20 antibody and evaluated their specific ability to target a Daudi tumor cell line.
Covalent attachment of antibody directly to the side wall of SWNT is a very feasible procedure. However, this may cause structural changes in antibody molecules, which would alter their specific binding ability. How to design antibodies for attachment to SWNT may be a critical factor in binding of the SWNT antibody complex to targeted cells. Because DNA-SWNT was found to improve the dispersibility of SWNT significantly,Citation6 a complex based on DNA-SWNT with an antibody attached is a potential model for a stable and smart exothermic device. However, the effect of DNA coating on the specific binding ability of the DNA-SWNT antibody complex to targeted cells is not clear.
The purpose of this study was to prepare a model complex composed of DNA-SWNT and an antibody, to clarify the relationship between them quantitatively, and to investigate the specificity, affinity, and capacity of antibody binding to the target protein. If the antibody shows strong and specific binding affinity to the targeted protein, even when immobilized on DNA-coated SWNT, the DNA-SWNT would be a suitable platform for a complex with antibodies. For this purpose, we designed a simple model made of DNA-SWNT and antibody raised against human IgG as a target protein. Using this system, key factors such as immobilization methods and the binding properties of antibodies can be evaluated. In this study, we prepared a DNA-SWNT antibody model complex using biotinylated IgG and an intermediate complex (streptavidin covalently bound to DNA-SWNT), estimated the binding ratio of SWNT and antibody, and evaluated the effect of DNA coating on the specificity, affinity, and capacity of binding of the DNA-SWNT antibody complex to the targeted protein.
Materials and methods
Preparation of DNA-SWNT
Preparation of the acid-treated SWNT (average diameter 1 nm, average length 800 nm, CG100, South West Nanotechnology, Norman, OK) was carried out according to a method described previously.Citation6 Acid-treated SWNT 10 mg and DNA 100 mg (derived from salmon testis, average 300 bp, Maruha-Nichiro Holdings, Tokyo, Japan) were sonicated (90 W, type 1510, Branson, Danbury, CT) in 10 mL of deionized water for one hour. The resulting aqueous solution of DNA-SWNT (1 mg/mL, based on SWNT amount) was subjected to preparation as an antibody complex.
Preparation of DNA-SWNT antibody complex
The DNA-SWNT solution (containing 10 mg of SWNT) was supplemented with 2 mL of N-hydroxysuccinimide (0.87 M, Nacalai Tesque, Kyoto, Japan) and 2 mL of 1-ethyl-3(3-dimethylaminopropyl) carbodiimide hydrochloride (EDC, 0.52 M, Wako Chemicals, Osaka, Japan), and gently stirred at room temperature. After mixing for 3 hours, 2 mg of streptavidin (Sigma-Aldrich, St Louis, MO) was added to the solution and stirred for 3 hours at 5°C, and then 1 mL of 2-aminoethanol solution (5 mM, Nacalai Tesque) was added to mask the unreacted carboxyl group on the DNA-SWNT, with stirring for one further hour at 5°C. The resulting solution was filtered using a polytetrafluoroethylene membrane (0.2 μm pore size) to collect the DNA-SWNT streptavidin complex.
The filtered DNA-SWNT streptavidin complex was adjusted to a concentration of 1 mg/mL (based on amount of SWNT) in phosphate-buffered solution (containing 0.01% n-dodecyl-β-D-maltoside, DDM, Dojindo, Kumamoto, Japan) using ultraviolet absorbance. The biotinylated anti-human IgG (200 μg, generated in a donkey, having minimal cross-reactivity with other species, Rockland Immunochemicals, Gilbertsville, PA) was added to 2 mL of the DNA-SWNT streptavidin solution and gently stirred for 2 hours at 5°C. After filtration using a polytetrafluoroethylene membrane (0.2 μm pore size), the DNA-SWNT antibody complex was dispersed in phosphate-buffered solution containing DDM (final concentration 500 mg/mL). A schematic diagram for preparation of the DNA-SWNT antibody complex is shown in .
Figure 1 Procedure for immobilization of streptavidin on the DNA-SWNT surface, and binding of biotinylated IgG to immobilized streptavidin to prepare the DNA-SWNT antibody complex. (A) Suspension of DNA-SWNT antibody complex.
Abbreviations: Ab, antibody; SWNT, single-walled carbon nanotubes; NHS, N-hydroxysuccinimide; EDC, 1-ethyl-3(3-dimethylaminopropyl) carbodiimide hydrochloride.
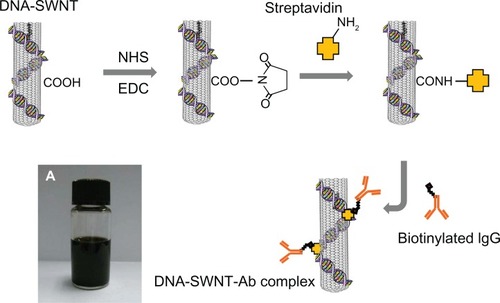
Preparation of SWNT antibody complex
The SWNT antibody complex was prepared from acid-treated SWNT. The preparation was carried out using the same procedure as described for preparation of the DNA-SWNT antibody complex.
Measurement of binding ratio using quartz crystal microbalance
A gold-coated quartz sensor with a fundamental frequency of 27 mHz (Affinix C4, Initium, Chigasaki, Japan) was used. For the quartz crystal microbalance (QCM) system used in this study, 1 Hz of frequency decrease was calibrated as an increase of 0.03 ng.Citation11,Citation12 A mixture of the DNA-SWNT, N-hydroxysuccinimide, and water-soluble carbodiimide was prepared according to the method described earlier and stirred for 3 hours. A 2 μL droplet of this solution was carefully placed on the sensor surface and dried for one hour. After rinsing twice with deionized water to remove unbound DNA-SWNT, it was dried again for 30 minutes. The amount of coated DNA-SWNT was determined from the change in frequency before and after coating.
The DNA-SWNT-coated sensor was exposed to 500 μL of HEPES buffer (containing 0.1 M NaCl and 0.01% DDM). When the frequency of the sensor had stabilized, the streptavidin solution was injected (final concentration 0.1 mg/mL) and frequency was monitored as a function of time. The amount of streptavidin bound to DNA-SWNT was determined from this frequency shift. The sensor surface was then exposed to 5 mM 2-aminoethanol solution for one hour to mask the unreacted carboxyl groups of the DNA-SWNT. The sensor was exposed to 500 μL of TE buffer containing 0.15 M NaCl, and the biotinylated anti-human IgG was injected (final concentration 50 μg/mL). The amount of IgG bound to the DNA-SWNT streptavidin complex was determined from the frequency shift. The amounts of streptavidin and IgG bound to the acid-treated SWNT were determined in the same manner as for measurement of the DNA-SWNT antibody.
Detection of binding of SWNT antibody complex to target protein
Ninety-six-well polystyrene microplates (BD Biosciences, Franklin Lakes, NJ) were incubated overnight at 4°C with 100 μL of 10 μg/mL human IgG or rabbit IgG (ChromPure, whole molecule, Jackson ImmunoResearch Laboratories Inc, West Grove, PA). After washing with phosphate-buffered solution, the wells were blocked with 3% bovine serum albumin dissolved in phosphate-buffered solution containing 0.01% DDM for 30 minutes. The solution was then replaced with different concentrations of either biotinylated anti-human IgG antibodies or a complex (DNA-SWNT antibody or SWNT antibody), each of which was dissolved in phosphate-buffered solution containing 1% bovine serum albumin and 0.01% DDM. DNA (1 mg/mL) was also included in the incubation mixture for experiments using the complex or IgG alone. After incubation at 37°C for 60–90 minutes, the wells were washed three times with 0.05% Tween 20 containing phosphate-buffered solution.
To detect the complex, the wells were treated at room temperature for 1.0–1.5 hours with mouse monoclonal anti-streptavidin antibody (1000×, S3E11, Pierce Biotechnology, Rockford, IL) and then goat monoclonal anti-mouse IgG antibody (light chain-specific, cross-reacted minimally with other species) conjugated with horseradish peroxidase (20,000×, Jackson ImmunoResearch Laboratories Inc), both of which were dissolved in phosphate-buffered solution containing 1% bovine serum albumin and 0.01% DDM. The wells were washed thoroughly with 0.05% Tween 20 containing phosphate-buffered solution after each treatment. To detect the biotinylated antibodies without SWNT, streptavidin (1 μg/mL) was first bound to the biotin side chain, and then incubated with the antibodies in the above order. Prior to horseradish peroxidase assay, absorbance at 450 nm was measured as a blank (AbsBlank). Thereafter, horseradish peroxidase assay was performed using 3,3′,5,5′-tetramethylbenzidine (TMB) as a substrate and an excess amount of H2O2 (TMB substrate reagent set, BD Falcon®). The reagents were incubated in the wells for 0–15 minutes, followed by stopping the reaction with 1 M phosphoric acid. Absorbance (AbsTMB) of the reaction mixture at 450 nm was measured, which was dependent on the production of yellow compound, ie, oxidized TMB (diimine form of TMB), under acidic conditions. The difference in value (AbsTMB − AbsBlank) was calculated to obtain the amount of oxidized TMB. The initial production rate of oxidized TMB was calculated on the basis of the Lambert–Beer Law, and the molar extinction coefficient at 450 nm (ε450) of oxidized TMB was reported to be 5.9 ×104 M−1 cm−1.Citation13
Statistics
All values are presented as the mean ± standard error of the mean (n, number of observations). Statistical analysis was performed using the unpaired t-test. A P value less than 0.05 was considered to be statistically significant.
Results
Preparation of DNA-SWNT antibody and SWNT antibody complexes
The DNA-SWNT antibody complex suspension was stable in water and phosphate-buffered solution for at least two weeks at 4°C without aggregation or precipitation (). However, the SWNT antibody suspension (without DNA) formed a small amount of precipitate after being stored for two weeks at 4°C.
Measurement of binding ratio of DNA-SWNT antibody complex using QCM
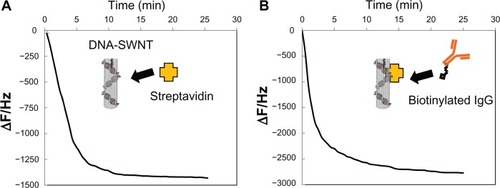
The amounts of immobilized SWNT and DNA-SWNT on the QCM sensor surface were 146.3 ± 14.2 ng and 147.3 ± 14.0 ng, respectively (n = 7). The curve in shows typical frequency changes as a function of time, indicating binding of streptavidin to the DNA-SWNT-immobilized QCM. Addition of streptavidin decreased the frequency (with an increase in mass) in several minutes. The curve in shows the typical frequency change when anti-human IgG antibody was applied to the sensor on which the DNA-SWNT-streptavidin had been immobilized. These consecutive changes in frequency represent the amounts of streptavidin bound to DNA-SWNT and the amounts of IgG then bound to DNA-SWNT-streptavidin. However, the molar ratio for binding of SWNT, streptavidin, and IgG could not be calculated accurately because the molecular weight of SWNT of variable length cannot be estimated. Therefore, we calculated the number of carbon atoms in SWNT immobilized on the QCM sensor using Avogadro’s constant (). The calculated numbers for carbon atoms were 7.30 × 1015 and 7.35 × 1015 for SWNT and DNA-SWNT, respectively. On the basis of the molecular weights of streptavidin (60,000) and IgG (150,000), as well as the QCM results, the binding ratio (carbon atoms of SWNT to molecules of streptavidin to molecules of antibody) for the DNA-SWNT antibody complex was comparable with that for the SWNT antibody complex.
Table 1 Binding ratio of the SWNT antibody and the DNA-SWNT antibody complexes calculated from quartz crystal microbalance data
Figure 2 Typical frequency changes for (A) streptavidin immobilized to DNA-SWNT according to the quartz crystal microbalance sensor, and (B) binding of biotinylated anti-human IgG to the streptavidin-immobilized DNA-SWNT according to the quartz crystal microbalance sensor.
Abbreviation: SWNT, single-walled carbon nanotubes.
Selective binding of DNA-SWNT antibody complex to target protein
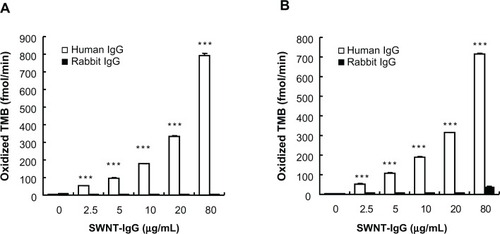
To examine the specificity of binding of the SWNT antibody complex to the target protein, we chose human IgG and rabbit IgG as target molecules and adhered each of these two to the bottom of the wells. Here, we produced an SWNT antibody complex using IgG generated in a donkey against human IgG. shows that the SWNT antibody complex (without DNA) binds selectively to human IgG (the target), but not rabbit IgG, up to a concentration of 30 μg/mL. This indicates that even a complex with a bulky structure did not alter the specificity of binding to the target molecule. When the DNA-SWNT antibody complex was used, specificity of binding was retained (), confirming that treatment with DNA is an effective method of increasing dispersibility without affecting the binding selectivity of IgG.
Figure 3 Selective binding of the SWNT antibody complex to target protein in the presence or absence of DNA. Either human IgG or rabbit IgG was adhered to the bottom of a microplate well. Initial rates of oxidized TMB production are plotted against the concentration of the SWNT antibody complex. (A) SWNT was not treated with DNA. (B) SWNT was treated with DNA, and the buffer contained 1 mg/mL DNA.
Note: ***P < 0.01, compared with the corresponding value for rabbit IgG as the target protein (n = 4).
Abbreviations: TMB, 3,3′,5,5′-tetramethylbenzidine; SWNT, single-walled carbon nanotubes.
Binding affinity and capacity of DNA-SWNT antibody complex to target molecule
To quantify the binding ability of the SWNT antibody complex and compare several binding parameters under different conditions, the binding kinetics of the SWNT antibody (anti-human IgG antibody) complex to its target molecule were examined. shows typical data for binding between the SWNT antibody complex and the target molecule in the absence of DNA.
Figure 4 Binding of the SWNT antibody complex to its specific target. (A) Typical saturation curve. (B) Lineweaver–Burk plots. The data derived from the saturation curve were fitted to the dashed line (see text). The line represents 1/Vmax (obtained from ordinate intercept) and 1/Km (obtained from abscissa intercept). 1/v = 0.007373 (1/[SWNT]) + 0.000812, R2 = 0.999. Vmax,SWNT = 1231, Km,SWNT = 9.09 (μg/mL). (C) Hill plots. Slope of the line represents nH = 0.959.
Abbreviations: TMB, 3,3′,5,5′-tetramethylbenzidine; SWNT, single-walled carbon nanotubes.
![Figure 4 Binding of the SWNT antibody complex to its specific target. (A) Typical saturation curve. (B) Lineweaver–Burk plots. The data derived from the saturation curve were fitted to the dashed line (see text). The line represents 1/Vmax (obtained from ordinate intercept) and 1/Km (obtained from abscissa intercept). 1/v = 0.007373 (1/[SWNT]) + 0.000812, R2 = 0.999. Vmax,SWNT = 1231, Km,SWNT = 9.09 (μg/mL). (C) Hill plots. Slope of the line represents nH = 0.959.Abbreviations: TMB, 3,3′,5,5′-tetramethylbenzidine; SWNT, single-walled carbon nanotubes.](/cms/asset/406fbdc4-6bd0-42e7-afeb-bc7caea80921/dijn_a_34202_f0004_b.jpg)
As shown in , the greater the concentration of the complex that was incubated, the higher the rate of oxidized TMB production that was obtained, suggesting concentration-dependent binding of the complex to its target molecule. The saturation curve obtained as described above appeared to be distorted at higher concentrations. A Lineweaver–Burk plot of the data gave an almost straight line, although it had a nonlinear portion at the two lowest 1/[SWCT] values (). After omitting these values, the affinity constant (Km,SWNT) and maximum reaction (Vmax,SWNT) were estimated (). A Hill plot showed a straight line, aside from the two highest values of log[SWNT] (). The estimated Hill coefficient (nH) was close to unity, indicating that the reaction obeys the Michaelis–Menten equation and occurs between the SWNT antibody complex and the target molecule.
Table 2 Binding affinity and capacity of SWNT-Ab and IgG itself to the target molecule in the absence of DNA
shows typical data for binding between IgG and its target in the absence of DNA. shows that the higher the concentration of IgG incubated, the higher the rate of oxidized TMB obtained. shows the Lineweaver–Burk and Hill plots. After omitting the data for the two highest concentrations of IgG, several parameters (Km,IgG, Vmax,IgG, and nH) were estimated (). The nH value was close to unity, indicating that the reaction also obeys the Michaelis–Menten equation in the absence of DNA.
Figure 5 Binding of IgG to its specific target. (A) Typical saturation curve. (B) Lineweaver–Burk plots. The data derived from the saturation curve were fitted to the dashed line (see text). The line represents 1/Vmax (obtained from ordinate intercept) and 1/Km (obtained from abscissa intercept). 1/v = 0.041700 (1/[IgG]) + 0.000719, R2 = 0.999. Vmax,IgG = 1391, Km,IgG = 58.0 (ng/mL). (C) Hill plots. Slope of the line represents nH = 0.957.
Abbreviation: TMB, 3,3′,5,5′-tetramethylbenzidine.
![Figure 5 Binding of IgG to its specific target. (A) Typical saturation curve. (B) Lineweaver–Burk plots. The data derived from the saturation curve were fitted to the dashed line (see text). The line represents 1/Vmax (obtained from ordinate intercept) and 1/Km (obtained from abscissa intercept). 1/v = 0.041700 (1/[IgG]) + 0.000719, R2 = 0.999. Vmax,IgG = 1391, Km,IgG = 58.0 (ng/mL). (C) Hill plots. Slope of the line represents nH = 0.957.Abbreviation: TMB, 3,3′,5,5′-tetramethylbenzidine.](/cms/asset/0c6fc786-20fe-4ceb-80d5-bc1c7aef3a68/dijn_a_34202_f0005_b.jpg)
Because the Vmax values varied between tests, we performed a pair of assays for the SWNT antibody complex and for IgG alone in each set of experiments, and the relative Vmax(Vmax,SWNT/Vmax,IgG) for the SWNT antibody complex to that for IgG itself was estimated. The relative Vmax,SWNT was close to unity, suggesting that even a complex with a bulky structure did not change the binding capacity ().
According to the QCM results and the presumed molecular weight of SWNT, the Km value based on the concentration of IgG (Km,IgG(SWNT)) was calculated to be (3.78 ± 0.56) × 10−8 M, which was 100-fold higher than that of Km,IgG [(3.65 ± 0.20) × 10−10 M] (). Statistical analysis revealed that the pKm,IgG(SWNT) value was significantly smaller than the pKm,IgG value. This result suggests that the streptavidin-SWNT structure greatly attenuated the binding affinity of IgG to the target molecule.
Next, we tested the binding affinity and capacity of the DNA-SWNT antibody complex and IgG itself in the presence of DNA (1 mg/mL, ). After omitting the plots at the two highest concentrations of the SWNT antibody complex and IgG, several parameters, ie, Km,IgG, Vmax,IgG, and nH, were calculated from the Lineweaver–Burk and Hill plots on the basis of the saturation curve (data not shown). A two-molecule reaction appeared to take place, namely between the DNA-SWNT-Ab complex and the target molecule. According to the relative Vmax (Vmax,SWNT/Vmax,IgG), Km,IgG(SWNT), and Km,IgG values, the bulky structure of the DNA-SWNT antibody complex did not alter the binding capacity (Vmax), although it greatly attenuated the binding affinity of IgG to the target molecule because the pKm,IgG(SWNT) value was significantly smaller than the pKm,IgG value.
Table 3 Binding affinity and capacity of DNA-treated SWNT-Ab and IgG itself to the target molecule in the presence of DNA
Comparison of the kinetic parameters for IgG in the presence or absence of DNA shows that an increase in dispersibility by DNA did not change the binding affinity or capacity of the SWNT antibody complex or for IgG ( and ).
Discussion
In this report, we describe a targeted carbon nanotube system using a DNA-SWNT antibody complex to create soluble and stable SWNT that retains specificity for binding to the target protein under physiological conditions. For preparing the model DNA-SWNT antibody complex system, we introduced carboxylic acid groups onto the SWNT surface as attachment sites. The carboxylic acid groups were introduced onto the DNA-SWNT by acid treatment. During this process, carboxylic groups were formed at the ends of the tubes, and were used as conjugation sites to link other ligands to the SWNT surface.Citation14,Citation15 Streptavidin was then attached covalently onto the acid-treated SWNT via an N-hydroxysuccinimide and EDC coupling reaction to yield stable amide linkages. Subsequently, biotinylated IgG was allowed to bind specifically to the DNA-SWNT-streptavidin. Using this concept, various carbon nanotube systems could be designed using specific biotinylated antibodies corresponding to the targeted proteins.Citation16
The streptavidin-biotin complex shows the highest-affinity noncovalent interaction (binding constant, 10−15 M).Citation17 This coupling system is frequently used in biotechnology because of its high affinity constant and stability. The QCM results in this study indicate that approximately one biotinylated IgG molecule bound to one streptavidin molecule immobilized on the SWNT. Although streptavidin has four sites for biotin recognition, three sites were reported not to contribute to further biotin recognition.Citation18 A possible explanation for this is steric hindrance due to the relatively large size of IgG and streptavidin molecules. The relatively large streptavidin molecule (5.4 ×5.8 ×4.8 nm)Citation19 was immobilized on the side wall of the SWNT via a covalent bond, which may have caused structural deformation of the streptavidin molecule. A similar result was reported for gelatin nanoparticles attached to antibodies by the avidin-biotin complex (about 2.4 of the four biotin binding sites in the avidin molecule were available for complexation with biotin).Citation20 Furthermore, segmental mobility of biotinylated IgG would affect the binding efficiency. The biotinylated IgG used in this study for the model complex system has a spacer arm 1.35 nm in length between the biotinyl group and the IgG molecule. A longer spacer arm length would confer a more flexible nature to the biotinylated IgG. There is a possibility that the spacer arm length between the biotinyl group and the IgG molecules is a key factor for binding of the biotinylated IgG molecule to the immobilized streptavidin molecule on the SWNT.
DNA was used as an effective solubilization substance to modify SWNT.Citation21 In a previous study,Citation6 we found that DNA functionalization was an effective method to improve the stability and solubility of acid-treated SWNT in culture medium, although the mechanism of interaction between anionic-charged DNA and acid-treated SWNT was not clear. Nakashima et alCitation22 suggested that π-π interaction between the side wall of the SWNT and the nucleic acid bases of DNA with a partially deformed structure generated by sonication is an important factor for solubilization of SWNT. Zheng et alCitation23 indicated that, for the single-stranded molecular CNT-DNA model interaction, π-π stacking interaction forms a helical wrapping on the CNT surface. However, the DNA molecules coated to the acid-treated SWNT surface could be a cause for concern in terms of affecting the specific binding affinity of the DNA-SWNT antibody complex to the targeted protein. A DNA-SWNT antibody complex suspension was stable for at least two weeks, so antibody attachment did not affect the solubilization of DNA-SWNT. To exclude the possibility that the DNA coating (functionalization) causes changes in binding affinity between the immobilized antibody and targeted protein, we carried out a binding analysis for the two types of complexes (DNA-SWNT antibody and SWNT antibody). The binding properties indicated by data for the two types of complexes clearly show that the coated DNA molecules did not interfere with the binding affinity or capacity of the immobilized antibody and target protein (). These findings suggest that treatment with DNA improved the stability and dispersibility of SWNT in body fluid, and that DNA-SWNT could be a smart “molecular heater” vehicle for hyperthermic therapy.
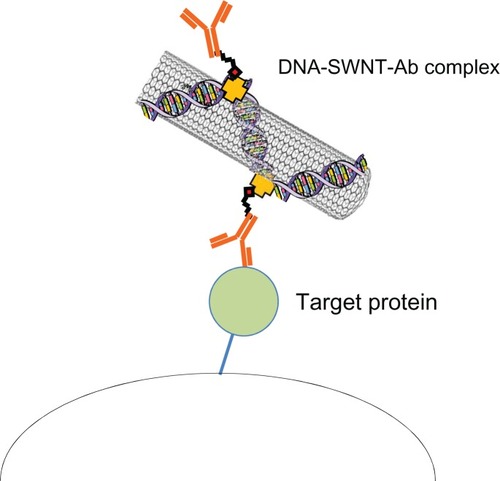
Figure 6 Schematic illustration of the DNA-SWNT antibody complex and binding to its target protein.
Abbreviations: Ab, antibody; SWNT, single-walled carbon nanotubes.
In our previous study, we calculated the total number of carbon atoms in acid-treated SWNT as 37,000 according to the estimated value reported by Kuwahara et al (SWNT of 1.0 nm diameter and 660 nm length has 74,000 carbon atoms).Citation24 From the QCM results for the DNA-SWNT antibody complex, we estimated that one IgG molecule attached to 21,700 carbon atoms (carbon:IgG ratio 18,900 atoms:0.87 molecules, as shown in ). By assuming that the carbon atoms of the DNA-SWNT number 37,000, 1.73 molecules of IgG were immobilized on one model DNA-SWNT antibody complex system. Although there was no difference between the binding affinities of the two types of complexes (with and without DNA treatment) for the target molecule, the affinities of these complexes were lower than that of IgG alone. Although the reason for this difference is unclear, one possible explanation is the segmental mobility of the DNA-SWNT antibody complex. The bulky SWNT attached to the IgG molecule would affect the binding affinity of the complex for the targeted molecule.
In summary, we have demonstrated a scheme for preparing a DNA-SWNT antibody complex containing a targeting IgG moiety. The DNA-SWNT antibody complex showed good specific binding ability for the target protein. Because the complex is based on SWNT with streptavidin attached, variable biotinylated ligands corresponding to the targeted protein can be attached. The results of this study suggest that the DNA-SWNT antibody complex system is a useful and smart “molecular heater” platform applicable for various types of antibodies in response to the corresponding target molecule.
Disclosure
The authors report no conflicts of interest in this work.
References
- GannonCJCherukuriPYakobsonBICarbon nanotube-enhanced thermal destruction of cancer cells in a noninvasive radiofrequency fieldCancer20071102654266517960610
- KlingelerRHampelSBuchnerBCarbon nanotube-based biomedical agents for heating, temperature sensoring and drug deliveryInt J Hyperthermia20082449650518923989
- LiuZTabakmanSWelsherKDaiHCarbon nanotubes in biology and medicine: in vitro and in vivo detection, imaging and drug deliveryNano Res200928512020174481
- FisherJWSarkerSBuchananCFPhotothermal response of human and murine cancer cells to multiwalled carbon nanotubes after laser irradiationCancer Res201070110
- MadaniSYNaderiNDissanayakeOTanASeifalianAMA new era of cancer treatment: carbon nanotubes as drug delivery toolsInt J Nanomedicine201162963297922162655
- KawaguchiMOhnoJIrieAFukushimaTYamazakiJNakashimaNDispersion stability and exothermic properties of DNA-functionalized single-walled carbon nanotubesInt J Nanomedicine2011672973621556347
- KamNWSO’ConnellMWisdomJADaiHCarbon nanotubes as multifunctional biological transporters and near-infrared agents for selective cancer cell destructionProc Natl Acad Sci U S A2005102116001160516087878
- DharSLiuZThomaleJDaiHLippardSJTargeted single-wall carbon nanotube-mediated Pt(IV) prodrug delivery using folate as a homing deviceJ Am Chem Soc2008130114671147618661990
- ChakravartyPMarchesRZimmermanNSThermal ablation of tumor cells with antibody-functionalized single-walled carbon nanotubesProc Natl Acad Sci U S A20081058697870218559847
- McDevittMRChattopadhyayDKappelBJTumor targeting with antibody-functionalized radiolabeled carbon nanotubesJ Nucl Med2007481180118917607040
- MarxKAQuartz crystal microbalance: a useful tool for studying thin polymer films and complex biomolecular systems at the solution-surface interfaceBiomacromolecules200341099112012959572
- FurusawaHTakanoHOkahataYTransient kinetic studies of pH-dependent hydrolyses by exo-type carboxypeptidase P on a 27-MHz quartz crystal maicrobalanceAnal Chem2008801005101118211097
- JosephyPDElingTMasonRPThe horseradish peroxidase-catalyzed oxidation of 3, 5, 3′, 5′-tetramethylbenzidine. Free radical and charge-transfer complex intermediatesJ Biol Chem1982257366936756277943
- ParkTJBanejeeSHemraj-BennyTWongSSPurification strategies and purity visualization techniques for single-walled carbon nanotubesJ Mater Chem200616141154
- LiuZGalliFJanssenKGHStable single-walled carbon nanotube-streptavidin complex for biorecognitionJ Phys Chem201011443454352
- JiangKSchadlerLSSiegelRWZhangXZhangHTerronesMProtein immobilization on carbon nanotubes via a two-step process of diimide-activated amidationJ Mater Chem2004143739
- HendricksonWAPahlerASmithJLCrystal structure of core streptavidin determined from multiwavelength anomalous diffraction of synchrotron radiationProc Natl Acad Sci U S A198986219021942928324
- Dupont-FilliardABillonMGuillerezSBidanGComparison by QCM and photometric enzymatic test of the biotin-avidin recognition on a biotinylated polypyrroleTalanta20015598199218968449
- ErdemAPapakonstantinouPMurphyHMcMullanMKaradenizHSharmaSStreptavidin modified carbon nanotube based graphite electrode for label-free sequence specific DNA detectionElectroanalysis201022611617
- BalthasarSMichaelisKDinauerNvon BriesenHKreuterJLangerKPreparation and characterisation of antibody modified gelatin nanoparticles as drug carrier system for uptake in lymphocytesBiomaterials2005262723273215585276
- NoguchiYFujigayaTNiidomeYNakashimaNSingle-walled carbon nanotubes/DNA hybrids in water are highly stableChem Phys Lett2008455249251
- NakashimaNOkuzonoSMurakamiHNakaiTYoshikawaKDNA dissolves single-walled carbon nanotubes in waterChem Lett200332456457
- ZhengMJagotaASemkeEDDNA-assisted dispersion and separation of carbon nanotubesNat Mater2003233834212692536
- KuwaharaSSugaiTShinoharaHDetermining exact molar absorbance coefficient of single-wall carbon nanotubesPhys Chem Chem Phys2009111091109719543607