Abstract
The low transfection efficiency of chitosan is one of its drawbacks as a gene delivery carrier. Low molecular weight chitosan may help to form small-sized polymer-DNA or small interfering RNA (siRNA) complexes. Folate conjugation may improve gene transfection efficiency because of the promoted uptake of folate receptor-bearing cells. In the present study, chitosan was conjugated with folate and investigated for its efficacy as a delivery vector for siRNA in vitro. We demonstrate that the molecular weight of chitosan has a major influence on its biological and physicochemical properties, and very low molecular weight chitosan (below 10 kDa) has difficulty in forming stable complexes with siRNA. In this study, chitosan 25 kDa and 50 kDa completely absorbed siRNA and formed nanoparticles (≤220 nm) at a chitosan to siRNA weight ratio of 50:1. The introduction of a folate ligand onto chitosan decreased nanoparticle toxicity. Compared with chitosan-siRNA, folate-chitosan-siRNA nanoparticles improved gene silencing transfection efficiency. Therefore, folate-chitosan shows potential as a viable candidate vector for safe and efficient siRNA delivery.
Introduction
Gene transfer technology has developed at a rapid pace in recent years. Projected medical applications run the gene therapy gamut from hereditary to infectious diseases, cancers, and arthritis. Different vector systems have been tested in gene transfer studies for potential clinical use. Viral vectors, such as adenoviruses and retroviruses, are very effective in terms of transfection efficiency, but have limitations in vivo, in particular because of safety concerns. Nonviral gene transfer systems offer several potential advantages over virus vectors. They are noninfectious, relatively nonimmunogenic, have low acute toxicity, can accommodate large small interfering RNA (siRNA)/DNA plasmids, and can be produced on a large scale. Numerous nonviral gene transfer systems have been proposed, including cationic liposomes and polymers.Citation1–Citation3
The discovery of siRNAs has given new therapeutic vision to the treatment of incurable diseases and genetic disorders. RNA interference is a sequence-specific gene-silencing mechanism, leading to target protein downregulation in the cytoplasm of mammalian cells.Citation4 Short, double-stranded RNAs are cleaved into 21–25-base pair fragments by the RNAse III-like enzyme, Dicer, and incorporated into RNA-induced silencing complexes, thereby mediating targeted mRNA degradation.Citation4 Chemically synthesized siRNAs and short-hairpin RNA expression plasmids, which are sequence-specific in targeting mRNAs, are common methods that mimic Dicer cleavage.Citation5 However, siRNAs are susceptible to nuclease destruction and cannot penetrate the cell membrane because of their highly charged backbone. An effective delivery system, in which siRNA are incorporated in carriers, could protect and transport siRNA to the cytoplasm of target cells.Citation1–Citation3
Chitosan and its derivatives have attracted attention as favorable nonviral vectors involved in the delivery of plasmid DNA in the field of siRNA delivery in vitroCitation6–Citation9 and in vivoCitation10 because of their low toxicity, high biocompatibility, and biodegradability. The main drawback of chitosan as a gene carrier is its typically low transfection efficiency. It has been demonstrated that transfection efficiency is closely related to the molecular weight of polymers.Citation11 Chitosan (10–150 kDa), with a specific degree of deacetylation, allows maximum transgenic expression in vitro.Citation12 Another strategy for improving transfection is to take advantage of folate uptake by cells to promote targeting and internalization, thereby enhancing transfection efficiency. Folate-mediated transfection has been shown to facilitate DNA internalization into cells via membrane receptors both in vitro and in vivo.Citation13 Folate receptors are overexpressed on activated (but not resting) macrophages that cause or contribute to inflammatory diseases, such as rheumatoid arthritis. Malignant cells also found to overexpress folate receptors include ovarian cancer and other malignancies.Citation14 Folate receptor distribution is largely restricted to malignant tissues, and folate receptors are now considered to be selective drug targets for the treatment of cancer and inflammatory diseases.Citation15
In the present study, we labeled low molecular weight chitosans with folic acid (FA). The potential of folate-conjugated chitosan as a carrier for siRNA delivery was then explored in vitro. The experimental data reveal that the functional group on folate increased the transfection efficiency of the chitosan-siRNA complex in HeLa and OV-3 cell lines, which are known to have high folate receptor expression.
Materials and methods
Materials
Chitosan (molecular weight 53.4 kDa, 85% deacetylation) was purchased from Wako Chemicals USA Inc (Richmond, VA). Dulbecco’s Modified Eagle’s Medium, fetal bovine serum, 0.25% trypsin-ethylenediamine tetra-acetic acid (EDTA) solution, penicillin-streptomycin mixture, agarose, ethidium bromide, and Lipofectamine® reagent were sourced from Invitrogen Canada Inc (Burlington, ON). FA and other chemicals, if not otherwise stated, were obtained from Sigma-Aldrich Chemical Co, (St Louis, MO). Sjogren syndrome antigen (SSB, GenBank accession number NM_009278)-targeted siRNA was provided by Merck and Co, Inc, (West Point, PA). SSB siRNA was selected as a validated siRNA to evaluate our delivery strategies.
Low molecular weight chitosan and folate-chitosan synthesis
Chitosan of different low molecular weights (2, 5, 10, and 25 kDa) was prepared as described elsewhere.Citation16 Chitosan of different molecular weights was further deacetylated by treatment with concentrated NaOH solution (50%) to achieve approximately 96% deacetylation, according to a previously reported procedure.Citation17 Folate-chitosan conjugation with poly(ethylene glycol) (PEG) was done according to the method reported by Cho et al,Citation18 with some modifications. Experimental details are given in the Supplementary information section.
Instrumentation
1H-NMR spectra were recorded using a Bruker ARX-400 400 MHz spectrometer. A DCl/D2O (1:100, v/v) mixture served as the solvent for the chitosan spectra. Ultraviolet-visible spectra were measured using a Hewlett Packard 8452A photodiode array spectrometer. The folate incorporation level was 1.1% with respect to chitosan amino groups, as determined by ultraviolet spectroscopy using FA as the standard. The extinction coefficient (λ363 nm) of folate was ascertained to be 6165 M−1 cm−1 in phosphate buffer 0.1 M (pH 7.4).
Chitosan and folate-chitosan-siRNA complexes
Chitosan and folate-chitosan of different molecular weights were dissolved overnight with low heating (below 45°C) in 20 mM acetic acid solution, adjusted to final concentrations of 1, 5, or 10 mg/mL in 5 mM acetic acetate buffer (pH 5.5) and then passed through a 0.45 μm filter.
Chitosan-siRNA or folate-chitosan-siRNA complexes at weight ratios of 10:1, 50:1, or 100:1 were generated at room temperature by adding 50 μg (50 μL of 1 mg/mL), 250 μg (50 μL of 5 mg/mL), or 500 μg (50 μL of 10 mg/mL) of chitosan or folate-chitosan in 5 mM acetate buffer (pH 5.5) to 5 μg of siRNA in 50 μL of 4.3 mM sodium sulfite buffer (pH 5.5). The solution was vortexed immediately at maximum speed for one minute. The complex solutions produced were used for the following experiments without further modification.
Particle size
Particle size was measured by Zetasizer Nano ZS90 (Malvern Instruments Ltd, Worcestershire, UK). Next, 100 μL of chitosan-siRNA or folate-chitosan-siRNA complexes at weight ratios of 10:1, 50:1, or 100:1 were obtained as indicated earlier. The final volume of 100 μL was processed with disposable, low-volume polystyrene (100 μL) cuvettes. Particle size was measured in triplicate.
Zeta potential
The zeta potential of chitosan-siRNA or folate-chitosan-siRNA complexes was quantified by Zetasizer Nano ZS90 based on the principle of phase analysis light scattering, and 100 μL of chitosan-siRNA or folate-chitosan-siRNA complexes at weight ratios of 10:1, 50:1, or 100:1 were generated as indicated earlier. The solution was then diluted to 1 mL with mixed buffer (half volume of 5 mM acetate buffer and half volume of 4.3 mM sodium sulfite buffer). Measurements were repeated three times in disposable zeta potential capillary cells.
siRNA integrity after synthesis
siRNA binding to chitosan or folate-chitosan was measured by 2% agarose gel electrophoresis in Tris-borate EDTA buffer (4.45 mM Tris-base, 1 mM sodium EDTA, 4.45 mM boric acid) at pH 8.0. Chitosan or folate-chitosan-siRNA complexes were loaded (20 μL of sample containing 0.5 μg of siRNA and 3.5 μL of 50% glycol) on the gel, and electrophoresed at 150 V for 30 minutes. The gel was stained with ethidium bromide (0.5 μg/mL) and photographed in a MultiImage™ light cabinet (Alpha Innotech Corporation, San Leandro, CA).
Cell culture and transfection protocol
HeLa cervical carcinoma (folate receptor-positive), OV-3 ovarian carcinoma (folate receptor-positive), and MG-63 osteosarcoma (folate receptor-negative) cells were obtained from the American Type Culture Collection (Rockville, MD). They were grown in Dulbecco’s Modified Eagle’s Medium, Ham’s F12/Dulbecco’s Modified Eagle’s Medium, and RPMI 1640, respectively. The cells were cultured in an atmosphere of 5% CO2 and 95% O2 at 37°C. They were seeded, 24 hours prior to transfection, in 24-well plates at a density of 50,000 cells/well in 1 mL of their usual culture medium supplemented with 10% fetal bovine serum and 1% penicillin-streptomycin. On the day of transfection, the culture medium was replaced by 500 μL of complete medium containing free siRNA, chitosan-siRNA, or folate-chitosan complexes and siRNA equivalent to 5 μg. After 6 hours of incubation, the cells were incubated in 1 mL of complete medium until 60 hours after transfection. Chitosan or folate-chitosan nanoparticle at weight ratios of 50:1 were incubated for 30 minutes at room temperature and tested without further modification.
Cell viability
The cells were seeded and transfected with free siRNA, chitosan-siRNA, or folate-chitosan-siRNA complexes, followed by 24 hours of incubation. Cell viability was then evaluated by 3-[4, 5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide (MTT) colorimetric assay based on MTT reduction by mitochondria in viable cells to water-insoluble formazan. Absorbance was measured at 570 nm using an EL800 universal microplate reader (Molecular Devices Corporation, Menlo Park, CA). Cell viability was expressed as the percentage of absorbance relative to the controls, which comprised cells not exposed to chitosan materials. Experiments were performed in triplicate. IC50, representing the concentration at which 50% of cell growth is inhibited, was calculated and expressed as the mean ± standard error of the mean.
Evaluation of transfection efficiency
Total RNA was extracted from cells using TRIzol reagent (Invitrogen Canada Inc) according to the manufacturer’s specifications. RNA was quantified using a RiboGreen RNA quantitation kit (Molecular Probes, Eugene, OR). For real-time reverse transcription-polymerase chain reaction (RT-PCR) analysis, 1 μg of total RNA was reverse-transcripted with a reverse transcription kit (Qiagen Inc, Mississauga, ON), as detailed in the manufacturer’s guidelines. One-fifth of the reverse transcriptase reaction was analyzed by real-time quantitative PCR. The following primers were used: SSB, forward 5′-CCA AAA TCT GTC ATC AAA TTG AGT ATT-3′, reverse 5′-CCA GCC TTC ATC CAG TTT TAT CT-3′;Citation19 β -actin housekeeping gene, forward 5′-AGC CTC GCC TTT GCC GA-3′, reverse 5′-CTG GTG CCT GGG GCG-3′.Citation20 SSB mRNA expression in cultured cells was expressed as the fold change relative to control conditions (nonstimulated cells), according to the ΔΔCT method detailed in the guidelines supplied by the manufacturer (Stratagene, La Jolla, CA). ΔCT values were first calculated by subtracting CT values for the β-actin housekeeping gene from each sample. ΔΔCT values were then computed by subtracting ΔCT values of the controls from the ΔCT values of each treatment. Fold changes compared with the controls were then determined by raising 2 to the power of ΔΔCT. Each PCR was performed in triplicate on two separate occasions in at least three independent experiments.
FA competition study
Cells were cultured under the same conditions as for the transfection tests. A 1 mg/mL FA solution was prepared. On the day of transfection, the culture medium was replaced by 500 μL of complete medium containing free siRNA, chitosan-siRNA, or folate-chitosan nanoparticles (the amount of siRNA in each well was 5 μg) with or without 20 μg free FA. The cells were subsequently incubated at 37°C with these solutions for 4 hours in the presence of 5% CO2 and with fresh medium for another 48 hours. Transfection efficiency was assessed as described earlier in the RT-PCR assay section.
Statistical analysis
The data are expressed as the mean ± standard error of the mean of at least three independent experiments. All statistics were generated by Prism software (GraphPad Software, San Diego, CA). Statistical significance was analyzed using the unpaired Student’s t-test. P < 0.05 was considered to be statistically significant.
Results
Characterization of chitosan-siRNA complexes
The particle size and surface charge of different molecular weight (2, 5, 10, 25, and 50 kDa) chitosan-siRNA complexes was measured (). The degree of siRNA binding to chitosan was estimated by 2% agarose gel electrophoresis (). The size of the particle complexes was directly proportional to the chitosan to siRNA weight ratio and molecular weight of chitosan. When the weight ratio or molecular weight increased, the size was amplified. The 2 kDa chitosan-siRNA was unstable (). At a chitosan to siRNA weight ratio of 10:1, the 2 kDa chitosan-siRNA complexes were larger in size (606.4 ± 5.8 nm, ), and their zeta potential indicated a negative charge (−4.05 ± 0.55 mV, ). The 2 kDa chitosan did not bind siRNA completely at any weight ratio (lane 3 in ), indicating a weak interaction between siRNA and chitosan at the low molecular weight of 2 kDa. The positive surface charge of other chitosan-siRNA complexes ranged from 12.4 mV to 17.8 mV (). Polydispersity indexes were between 0.22 and 0.41 ().
Figure 1 Characterization of chitosan-siRNA complexes. Size (nm) (A), zeta potential (mV) (B), and PDI (C) of chitosan-siRNA complexes.
Note: The chitosan-siRNA complexes were at a chitosan to siRNA Sjogren syndrome antigen weight ratio of 10:1, 50:1, and 100:1, respectively.
Abbreviations: PDI, polydispersity index; MW, molecular weight; siRNA, small interfering RNA.
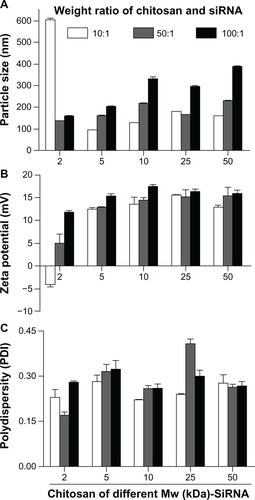
Figure 2 Electrophoresis analysis on 2% agarose gel. Effect of molecular weight of chitosan on siRNA binding efficacy at various chitosan to siRNA Sjogren syndrome antigen weight ratios of (A) 10:1, (B) 50:1, and (C) 100:1.
Notes: Lane 1, ladder 100 base pairs; lane 2, free siRNA (0.5 μg/lane); lane 3, 2 kDa chitosan-siRNA complexes; lane 4, 5 kDa chitosan-siRNA complexes; lane 5, 10 kDa chitosan-siRNA complexes; lane 6, 25 kDa chitosan-siRNA complexes; lane 7, 50 kDa chitosan-siRNA complexes.
Abbreviation: siRNA, small interfering RNA.
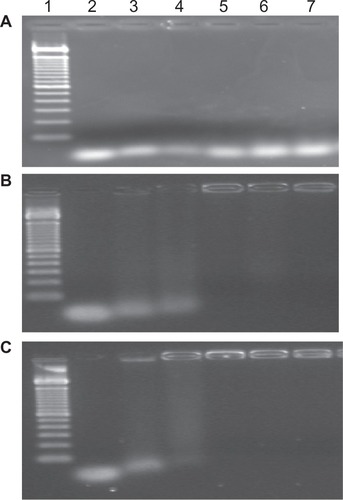
For chitosan-siRNA complexes (5, 10, 25 and 50 kDa) at a chitosan to siRNA weight ratio of 10:1, the migration behavior was virtually the same as that of naked siRNA (), indicating that this weight ratio is unsuitable for protecting siRNA. At a chitosan to siRNA weight ratio of 50:1, the particle size varied between 161.1 and 216.1 nm (). At a 100:1 weight ratio, the particle size increased with the molecular weight of chitosan (between 201.5 and 338.5 nm, ). However, only siRNA binding to 25 kDa or 50 kDa chitosan at a weight ratio of 50:1 could be observed with one peak of size distribution (results not reported).
Cytotoxicity of chitosan and chitosan-siRNA complexes
The cytotoxicity of the chitosan samples was examined by MTT assay using HeLa cells (). The dependence of cell viability on polymer composition after 24 hours of cell incubation with different concentrations (0 to 3 mg/mL) of the polymer solution is shown in . IC50 values for chitosan of different molecular weights were 0.21 mg/mL (2 kDa chitosan), 0.42 mg/mL (5 kDa chitosan), 2.2 mg/mL (10 kDa chitosan), 2.2 mg/mL (25 kDa chitosan), and 1.5 mg/mL (50 kDa chitosan).
Figure 3 (A) In vitro cytotoxicity of the chitosans (polymer alone) of different molecular weights on HeLa cells as measured by MTT assay. (B) In vitro cytotoxicity of chitosan-siRNA complexes.
Notes: The chitosan-siRNA complexes were at a chitosan to siRNA Sjogren syndrome antigen weight ratio of 10:1, 50:1, and 100:1, respectively.
Abbreviations: MW, molecular weight; siRNA, small interfering RNA.
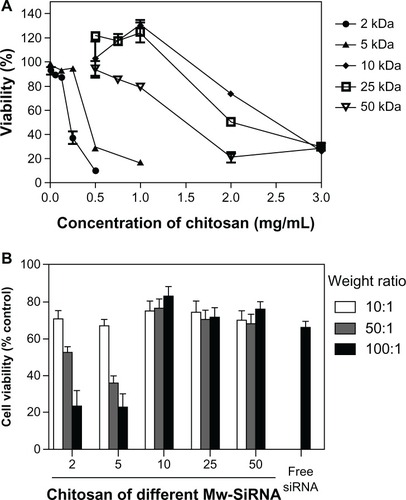
To investigate the cytotoxicity of the chitosan-siRNA complexes, cell viability was also examined by MTT assay after 24 hours of incubation (). Cells not treated with chitosan-siRNA complexes were considered as controls, with cell viability of 100%. shows the effects of molecular weight of chitosan and the chitosan to siRNA weight ratio on cell viability. The results show at least 70% average cell viability for chitosan-siRNA complexes formulated with 10, 25, or 50 kDa chitosan at various weight ratios of 10:1, 50:1, or 100:1 (containing 5 μg of siRNA). The amount of viable cells decreased slightly upon contact with free siRNA (5 μg/well in 24-well plates, 65.9%) in comparison with untreated cells. It was also observed that 5 μg of siRNA-Lipofectamine 2000 dramatically reduced cellular viability to below 40% (results not included). Viability declined significantly when the cells were incubated with 2 kDa or 5 kDa chitosan-siRNA at weight ratios of 50:1 (52.7% and 36.2% for 2 kDa and 5 kDa chitosan, respectively) and 100:1 (23.3% and 22.8% for 2 kDa and 5 kDa chitosan, respectively).
Synthesis and characterization of folate-chitosan-siRNA complexes
The feasibility of folate-mediated chitosan targeting was investigated after coupling of folate with chitosan using PEG as a spacer. Based on the size and electrophoretic behavior of different molecular weight (2, 5, 10, 25, and 50 kDa) chitosan-siRNA complexes, only 25 kDa and 50 kDa molecular weight chitosan was conjugated with folate. The folate content of chitosan-PEG-folate was assessed by ultraviolet spectroscopy and found to be 1.1 mol% for 50 kDa chitosan-PEG-folate and 0.9 mol% for 25 kDa chitosan-PEG-folate with respect to chitosan glucosamine units, which corresponds to the fact that there are four and 1.3 folate molecules on each of the 50 kDa chitosan-PEG-folate and 25 kDa chitosan-PEG-folate chains, respectively (). We selected chitosan-siRNA and FA-chitosan-siRNA complexes at a weight ratio of 50:1. The ability of 25 kDa or 50 kDa folate-chitosan to interact with siRNA was investigated by agarose gel electrophoresis (). Complete siRNA binding to 25 kDa or 50 kDa chitosan and folate-chitosan vectors was observed ().
Figure 4 (A) 1H NMR spectrum of folate-chitosan. Solvent DCl/D2O (1:100). (B) Electrophoresis analysis of chitosan or folate-chitosan/siRNA complexes at a weight ratio of 50:1 on 2% agarose gel. Lane 1, free siRNA Sjogren syndrome antigen (0.5 μg/lane); lane 2, 25 kDa chitosan-siRNA; lane 3, 25 kDa folate-chitosan-siRNA; lane 4, 50 kDa chitosan-siRNA; lane 5, 50 kDa folate-chitosan-siRNA. (C) Time course of nanoparticle size (nM). (D) Time course of zeta potential (mV).
Abbreviations: PEG, poly(ethylene glycol); siRNA, small interfering RNA.
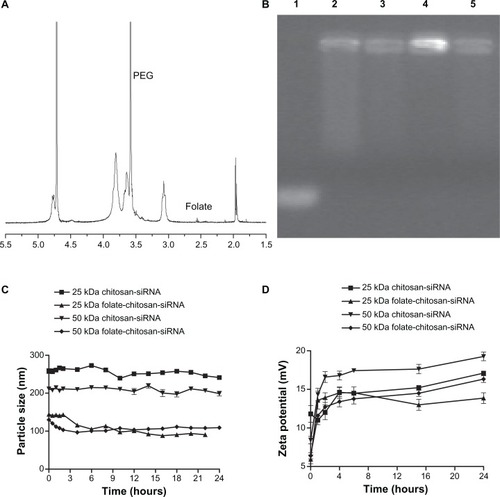
Introduction of the folate ligand onto chitosan decreased the size of the complex (258.0 ± 6.6 nm versus 143.3 ± 0.7 nm for 25 kDa chitosan-siRNA and folate-chitosan-siRNA, respectively). The 50 kDa chitosan-siRNA complex sizes decreased from 207.7 ± 2.6 nm to 120.5 ± 1.9 nm in comparison with 50 kDa chitosan-siRNA and folate-chitosan-siRNA, respectively. The nanoparticles remained stable in size for at least 24 hours (). Their zeta potential increased slightly within 24 hours, ranging from 11.02 ± 0.59 mV to 14.33 ± 0.80 mV (one hour after complex formation) and from 13.87 ± 0.69 mV to 19.27 ± 0.56 mV (24 hours after complex formation), respectively ().
In vitro cytotoxicity of folate-chitosan-siRNA nanoparticles
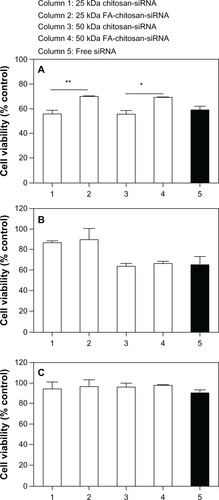
Cell viability was quantified by MTT assay after 24 hours of incubation with chitosan-siRNA or folate-chitosan complexes. Folate receptor-positive (HeLa and OV-3) and folate receptor-negative (MG-63) cells were tested. As shown in , over 50% of average HeLa cell viability was noted for free siRNA-treated cells in comparison with nontreated cells (considered as 100%). Cell viability with chitosan (25 kDa or 50 kDa)-siRNA complexes was not significantly different from that of free siRNA-treated cells, ie, 54%–65%. Compared with free SSB siRNA and chitosan-SSB siRNA, folate-chitosan-SSB siRNA was less cytotoxic and assigned more than 68% viability, which was significantly different from chitosan-SSB siRNA-treated cells (P < 0.01 for 25 kDa polymer-siRNA and P < 0.05 for 50 kDa polymer-siRNA, respectively). An average cell viability of 65% with free siRNA was observed in OV-3 cells (), with an average viability of over 87% for OV-3 cells treated with 25 kDa chitosan-siRNA complexes, which increased slightly to 89% for 25 kDa folate-chitosan-siRNA-treated cells. We noted that 50 kDa chitosan and folate-chitosan-siRNA complexes were likely to decrease the viability of OV-3 cells (64%–69%), but no significant viability difference in OV-3 cells was apparent between 25 kDa chitosan-siRNA and folate-chitosan-siRNA or between 50 kDa chitosan-siRNA and folate-chitosan-siRNA. Average MG-63 cell viability was over 85% with free siRNA-chitosan and folate-chitosan-siRNA complexes at various polymer and siRNA weight ratios. However, there was no significant difference in viability between siRNA-chitosan and folate-chitosan-siRNA complexes ().
Figure 5 Cell viability. (A) HeLa, (B) OV-3, and (C) MG-63 line cells were transfected with either 5 μg free siRNA SSB or with chitosan-siRNA or folate-chitosan-siRNA complexes containing an equivalent of 5 μg of siRNA SSB.
Notes: Viability was measured with the MTT assay and compared with nontreated cells, which are considered as 100%. Statistical significance was assessed by unpaired Student’s t-test, *P < 0.05; **P < 0.01. Column 1, 25 kDa chitosan-siRNA; column 2, 25 kDa folate-chitosan-siRNA; column 3, 50 kDa chitosan-siRNA; column 4, 50 kDa folate-chitosan-siRNA; column 5, free siRNA.
Abbreviations: FA, folic acid; SBB, Sjogren syndrome antigen; siRNA, small interfering RNA.
In vitro gene silencing of folate-chitosan-siRNA nanoparticles
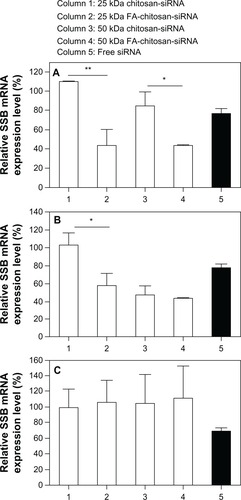
To investigate the gene silencing efficiency of various chitosan and folate-chitosan-siRNA systems, HeLa and OV-3 (folate receptor-positive) and MG-63 (folate receptor-negative) cells were transfected with chitosan-siRNA and folate-chitosan-SSB complexes, respectively. The transfection efficiency of polymer-siRNA complexes was analyzed relative to SSB mRNA expression by real-time RT-PCR. HeLa (), OV-3 (), and MG-63 () cells treated with free SSB siRNA achieved approximately 25% knockdown of SSB mRNA expression compared with nontreated cells. No change in SSB mRNA expression was evident with 25 kDa chitosan-siRNA compared with nontransfected cells (negative controls) in all three cell lines. However, silencing efficiency was dependent on and increased with the molecular weight of chitosan. 50 kDa chitosan-siRNA nanoparticles exerted a 15% gene-silencing effect in HeLa cells (), whereas OV-3 cells transfected with 50 kDa chitosan-siRNA nanoparticles generated 53% gene silencing. MG-63 cells displayed no SSB knockdown with 25 kDa or 50 kDa chitosan-siRNA transfection. Folate conjugation improved the gene silencing efficiency of chitosan. HeLa cells transfected with 25 kDa or 50 kDa folate-chitosan-siRNA showed significantly reduced transcription of SSB mRNA (57% knockdown of SSB mRNA expression) compared with 25 kDa (P < 0.01) and 50 kDa (P < 0.05) chitosan-siRNA, respectively. Compared with normal nontreated cells, 25 kDa and 50 kDa folate-chitosan-SSB siRNA, respectively, inhibited 42% and 57% of SSB gene expression in iOV-3 cells; 25 kDa or 50 kDa folate-chitosan-SSB siRNA manifested no SSB mRNA inhibitory effect in folate receptor-negative MG-63 cells. Chitosan-siRNA complexes containing mismatched control siRNA showed no SSB inhibition, confirming gene silencing efficiency (data not included).
Figure 6 Transfection efficiency in vitro. 5 μg free siRNA SSB or chitosan-siRNA or folate-chitosan-siRNA complexes containing an equivalent of 5 μg of siRNA SSB incubated with (A) HeLa, (B) OV-3, and (C) MG-63 line cells.
Notes: The weight ratio of chitosan or folate-chitosan to siRNA SSB was 50:1. SSB gene expression is measured with real-time RT-PCR and compared with nontreated cells, which are considered as 100%. Statistical significance was assessed by the unpaired Student’s t-test, *P < 0.05; **P < 0.01; P < 0.001. Column 1, 25 kDa chitosan-siRNA; column 2, 25 kDa folate-chitosan-siRNA; column 3, 50 kDa chitosan-siRNA; column 4, 50 kDa folate-chitosan-siRNA; column 5, Free siRNA.
Abbreviations: RT-PCR, reverse transcription-polymerase chain reaction; SBB, Sjogren syndrome antigen; siRNA, small interfering RNA.
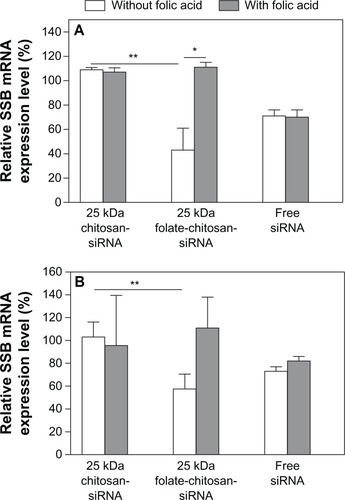
Competition analysis of folate-chitosan-siRNA complexes with free FA
As indicated in , HeLa cells transfected with 25 kDa folate-chitosan-siRNA showed significantly (P < 0.01) reduced transcription of SSB mRNA (57% knockdown of SSB mRNA expression) compared with 25 kDa chitosan-siRNA. In OV-3 cells (), 25 kDa folate-chitosan-SSB siRNA inhibited 42% of SSB gene expression in comparison with normal nontreated cells. Folate-chitosan-siRNA transfection efficiency decreased in the presence of free FA in these cells, suggesting that free FA molecules prevent cellular uptake of the complexes by competitive binding to folate receptors on cell surfaces.
Figure 7 Folic acid competition study. 5 μg free siRNA SSB or chitosan-siRNA or folate-chitosan-siRNA complexes containing an equivalent of 5 μg of siRNA SSB incubated with (A) HeLa and (B) OV-3 line cells with or without 20 μg free folic acid.
Notes: SSB gene expression is measured with real-time RT-PCR and compared with nontreated cells, which are considered as 100%. Statistical significance was assessed by the unpaired Student’s t-test, *P < 0.05; **P < 0.01.
Abbreviations: RT-PCR, reverse transcription-polymerase chain reaction; SBB, Sjogren syndrome antigen; siRNA, small interfering RNA.
Discussion
siRNA silencing technology has been exploited in a wide range of biological studies, but has also become one of the most challenging therapeutic strategies. However, because of its poor delivery and susceptibility to degradation by nuclease, siRNA-based approaches need a protective delivery system. A number of polymer formulations have been proposed in the literature as potential carriers.Citation1–Citation3 Polymer molecular weight, charge density, the ratio of protonatable polymer amine groups to nucleic acid phosphate groups, and the ionic strength of the medium can affect electrostatic binding between siRNA and cationic polymers. Research over the years has revealed that chitosan is one of the more desirable polymeric carriers of siRNA because of its natural biocompatibility, biodegradability, nontoxicity, and high nuclease resistance. The effects of different chitosan (114 kDa or more)-siRNA complexes on transfection activity have been investigated previously.Citation6–Citation9 Rojanarata et alCitation7 reported that chitosan-thiamine pyrophosphate-mediated siRNA enhanced green fluorescent protein gene silencing efficiency, depending on the molecular weight and weight ratio of chitosan and siRNA. Chitosan-thiamine pyrophosphate-siRNA complexes with the lowest chitosan molecular weight (20 kDa) at a weight ratio of 80 showed the strongest inhibition of gene expression.Citation7 The results of our study are in good agreement with reports in the literature.
Complexes prepared from 2 kDa and 5 kDa chitosan were the largest in size and their migration trace for siRNA could be observed. Moreover, 2 kDa or 5 kDa chitosan alone (or chitosan-siRNA complex) showed more toxicity than the other chitosan preparations. The 10, 25, and 50 kDa chitosan-siRNA formed nanosized particles (≤220 nm), but only complete binding of siRNA with 25 kDa or 50 kDa chitosan could be seen and showed delayed siRNA migration without the presence of a trailing band. Our results are consistent with those of a previous study which demonstrated a weak interaction between siRNA and chitosan at a very low molecular weight of about 10 kDa.Citation6 Higher molecular weight chitosan molecules are long and flexible, but have a higher degree of deacetylation (meaning increased positive charge), which enhances electrostatic interaction with siRNA, thus synergistically reducing the size of the complexes and enhancing their stability. As a favorable nonviral vector involved in the delivery of plasmid DNA, 5 kDa and 10 kDa chitosan can form stable chitosan-DNA nanoparticles (data not included). Differences in complexation between chitosan-plasmid DNA and chitosan-siRNA need to be investigated further because of a divergence in size and charge density between plasmid DNA and siRNA.
Taken together, our findings show that binding of siRNA to chitosan was dependent on both the molecular weight of chitosan and the chitosan to siRNA weight ratio. The 25 kDa and 50 kDa chitosan-siRNA nanoparticles formed at a molecular ratio of 50:1 were more stable. We chose 25 kDa and 50 kDa chitosan to synthesize folate-chitosan.
Interestingly, size and particle surface charge were decreased after introduction of the folate ligand onto chitosan. A positive zeta potential of the polymer is necessary to ensure the uptake of complexes by cells carrying a negative charge. It has been reported by our group that the size of folate-chitosan-DNA is indirectly proportional to the charge ratio and zeta potential of folate-chitosan-DNA.Citation22 Due to its short length (21–25 base pairs) and linearity, siRNA binds to folate-chitosan in a manner that is different from plasmid DNA. The weak interaction of siRNA with folate-chitosan contributes to the formation of smaller folate-chitosan-siRNA complexes.
Until recently, folate-conjugated nonviral systems with cationic liposomes and polymer were used to condense siRNA to nanoparticles. No study has attempted to illustrate the ability of folate-conjugated chitosan as a siRNA delivery system. Due to its cationic characteristics, chitosan is able to form complexes with negatively-charged DNA or siRNA by electrostatic interaction. In the present work, simple complexation was used to prepare chitosan and folate-chitosan-siRNA complexes. Our electrophoresis analysis results show that introduction of the folate ligand did not affect siRNA-binding ability. However, sizes of the folate-chitosan-siRNA complexes were decreased in comparison with chitosan-siRNA. As reported by Chan et al,Citation21 the water solubility of folate-PEG-chitosan (255 kDa) was slightly lower than that of PEG-chitosan, owing to the introduction of hydrophobic groups in folate, inducing the precipitation of these conjugates.
In general, chitosan and folate-chitosan are nontoxic as nonviral gene delivery vectors in vitro. Folate-chitosan-DNA nanoparticles have lower cytotoxicity than commercial transfection agents, such as Lipofectamine 2000.Citation22 Decreased cytotoxicity has also been reported after grafting folate-PEG onto polyethylenimine (PEI), which is considered to have high cell toxicity.Citation23 In this study, average cell viability with 25 kDa or 50 kDa chitosan-siRNA was 54%–65% compared with nontreated cells. It increased slightly when the cells were incubated with folate-chitosan-siRNA. These results are in agreement with our previous study,Citation24 which revealed that folate-chitosan-interleukin-1 receptor antagonist plasmid DNA has a better toxicity profile than naked DNA or chitosan-DNA.
The ability of chitosan to deliver DNA and siRNA both in vitro and in vivo has been reported recently.Citation6–Citation10 However, data on folate receptor-targeting delivery of cationic polymer folate-PEG-PEI are available.Citation23 Folate was conjugated to PEI to increase transfection efficiency for targeting purposes because of its low immunogenicity, low toxicity, and high affinity for folate receptors. Folate-PEG-PEI has been demonstrated to be a potentially safe and efficient gene vector, particularly for folate receptor-positive cell gene delivery. In the present study, we determined that folate-chitosan could be complexed with siRNA, and the results with folate-chitosan-siRNA complexes show folate receptor-specific gene transfection. Surface-exposed folate ligands on the complexes were recognized by folate receptors on the membranes of targeted cancer cells, providing active transport of folate-chitosan-siRNA complexes into HeLa and OV-3 cells. We found that folate-conjugated chitosan had greater potential for delivery to targeted cells. The delivery of chitosan complexes to specific cell types has been achieved by conjugating chitosan to various cell-targeted ligands, such as galactose, lactose, and trisaccharides.Citation25 Targeting of tumor cells is promoted by conjugating chitosan with folate.Citation13 Expression of the folate receptor in synovial mononuclear cells and CD14+ cells in patients with rheumatoid arthritis has been reported.Citation25 We have already successfully performed characterization, internalization, and in vitro and in vivo studies of a nonviral gene therapy vector based on chitosan nanoparticles and interleukin-1 receptor antagonist gene therapy in an adjuvant-induced arthritis rat model of rheumatoid arthritis.Citation22,Citation26
Free FA competes with folate-chitosan-siRNA complexes for folate receptor-binding. In HeLa cells, as seen in , SSB mRNA expression was significantly higher (P < 0.05) when transfection was undertaken with 25 kDa folate-chitosan-SSB siRNA complexes in the presence of free FA. Therefore, the transfection ability of HeLa cells was significantly decreased due to free FA in culture medium, whereas no significant difference was found when MG-63 cells (folate receptor-negative) were transfected in the presence of free FA in medium (data not included). These findings indicate that free FA molecules can prevent cellular uptake of the complexes by competitive binding to folate receptors on the cell surface. Folate conjugation improves the transfection efficiency of folate-chitosan for folate receptor-positive cell gene silencing under normal conditions without excess free FA. Because of folate receptor overexpression and activated macrophages in cancer cells, a number of therapeutic agents have been linked with FA for selective delivery to pathological cells, by avoiding normal tissues. Among these therapies, folate-targeted nonviral vectors are considered to be promising treatments for cancer and inflammatory diseases.Citation13–Citation15
In conclusion, siRNA binding to chitosan depends on molecular weight and the chitosan to siRNA weight ratio. Very low molecular weight chitosan (<10 kDa) has difficulty in forming stable complexes with siRNA. Its binding ability is improved when its molecular weight is 25 kDa or more. After addition of a folate ligand to chitosan, the viability and transfection efficiency of folate-chitosan-siRNA complexes in vitro depend on the weight ratios of folate-chitosan to siRNA as well as the type of cell transfected. Higher transfection efficiency and lower toxicity of folate-chitosan are observed in folate receptor-positive (HeLa and OV-3) cells. These findings should encourage further exploration of folate-targeted nonviral gene therapies in cancer and inflammatory diseases, such as arthritis.
Acknowledgements
This work was supported by grants from the National Natural Science Foundation of China (30811120440 and 30911120261), the Shanghai International Collaboration Foundation (08410701800), the Ministry of Science and Technology of China (No.2011DFA30790) and the Canadian Institutes of Health Research (CCM104888). JCF and MB are research scholars of Fonds de la Recherche en Santé du Québec. The authors thank Merck and Co, Inc, for providing the SSB-targeted siRNA used in this study.
Disclosure
The authors report no conflicts of interest in this work.
Supplementary information
Synthesis of low molecular weight chitosans
Chitosans of lower molecular weight (2, 5, 10, and 25 kDa) was prepared by nitrous acid degradation. Briefly, the procedure is explained here for the synthesis of chitosan 25 kDa. Chitosan (molecular weight 53 kDa, 5.0 g) was dissolved in 500 mL of 2% acetic acid solution. Sodium nitrite (NaNO2, 35 mg, 0.5 mmol) in 10 mL of double-distilled water was added in four aliquots. The reaction was conducted at 40°C–50°C for 5–6 hours. After that, the pH of the chitosan solution was adjusted to >10 by adding NaOH solution (5 M). The white precipitate generated was collected by filtration and washed with double-distilled water until the filtrate became neutral (yield 3.3 g, 66%).
The molecular weight of chitosan was determined by gel permeation chromatography with multiple angle light scattering detection. Gel permeation chromatography analyses were carried out on a gel permeation chromatography system consisting of an Agilent 1100 isocratic pump, a Dawn EOS multiangle laser light scattering detector (Wyatt Technology Corporation, Santa Barbara, CA), an Optilab DSP interferometric refractometer (Wyatt Technology), and a TSK-GELPW (Tosoh Biosep, King of Prussia, PA) column eluted with a pH 4.5 acetic acid (0.3 M)/sodium acetate (0.2 M) buffer, injection volume 100 ul, flow rate 0.5 mL per minute, temperature 25.0°C. The dn/dc values of the polymers were measured at 690 nm with the same refractometer used in the off-line mode. Characterizations of the chitosans are listed in Table S1.
Synthesis of chitosan-PEG-folate conjugate
Two chitosans, ie, chitosan 50 K and chitosan 25 K were selectively modified with folate via a PEG 3.4 K spacer.
Table S1 Characterization of the chitosans
The synthesis scheme for chitosan-PEG-folate conjugate is shown in Scheme S1. The conjugate was prepared according to the synthetic procedure reported by Cho et alCitation18 with slight modification. The synthesis was accomplished in three steps, as follows.
Folate-NHS-ester: folic acid (300 mg, 0.67 mmol) was dissolved in 12 mL of dry dimethylsulfoxide to which 93 mg (0.45 mmol) of dicyclohexylcarbodiimide (DCC) and 77 mg (0.67 mmol) of N-hydroxysuccinimide (NHS) were added. The mixture was stirred at room temperature for 6 hours. The white precipitate (a side product of the reaction) was removed by filtration. The filtrate was added to 100 mL of 30% acetone in diethyl ether. The resulting yellow precipitate (folate-NHS-ester) was collected by filtration and washed with acetone and ether. It was used immediately in the next step of synthesis. The yield was 250 mg (83%).
Folate-PEG-COOH: NH2-PEG-COOH (450 mg, 0.13 mmol, molecular weight 3400 Da, Shearwater Polymers, Huntsville, AL) was dissolved in 100 mL of dry pyridine. The folate-NHS-ester (approximately 250 mg, 0.55 mmol) was added to the solution and the reaction mixture was stirred overnight in the dark. Upon completion of the reaction, the pyridine was evaporated in vacuo and the remaining solid was dissolved in 20 mL of deionized water. The product was purified by dialysis against pH 7.4 phosphate buffer (10 mM) for one day and against deionized water for two days. The yield was 450 mg (76%).
Chitosan-PEG-folate: Chitosan (200 mg, 1.0 mmol amino groups) was dissolved in 20 mL of 2% acetic acid aqueous solution. To this solution, folate-PEG-COOH (40 mg, 10 μmol in 5 mL of dimethylsulfoxide and preactivated by reacting with [3-(-dimethylamino)propyl]carbodiimide (EDC, 2.5 mg, 15 μmol) and NHS (5.8 mg, 25 μmol) for 4 hours) was added dropwise. The reaction was continued in the dark for 20 hours with constant stirring. The reaction mixture was then dialyzed using a membrane (molecular weight cutoff, 12–14 kDa) against deionized water for 3 days. The chitosan-PEG-folate conjugate was recovered by freeze-drying. The yield was 210 mg.
The structure of chitosan-PEG-folate conjugate is illustrated in Figure S1.
References
- GaryDJPuriNWonYYPolymer-based siRNA delivery: Perspectives on the fundamental and phenomenological distinctions from polymer-based DNA deliveryJ Control Release2007121647317588702
- ZhangSZhaoBJiangHWangBMaBCationic lipids and polymers mediated vectors for delivery of siRNAJ Control Release200712311017716771
- De FougerrollesARDelivery vehicles for small interfering RNA in vivoHum Gene Ther20081912513218257677
- HammondSMBernsteinEBeachDHannonGJAn RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cellsNature200040429329610749213
- ChenMGrangerAJVanbrocklinMWInhibition of avian leukosis virus replication by vector-based RNA interferenceVirology200736546447217493657
- LiuXHowardKADongMThe influence of polymeric properties on chitosan/siRNA nanoparticle formulation and gene silencingBiomaterials2007281280128817126901
- RojanarataTOpanasopitPTechaarpornkulSNgawhirunpatTRuktanonchaiUChitosan-thiamine pyrophosphate as a novel carrier for siRNA deliveryPharm Res2008252807281418563539
- KatasHAlparHODevelopment and characterisation of chitosan nanoparticles for siRNA deliveryJ Control Release200611521622516959358
- KatasHChenSOsamuyimenAACevherEAlparHOEffect of preparative variables on small interfering RNA loaded poly(D,L-lactide-co-glycolide)-chitosan submicron particles prepared by emulsification diffusion methodJ Microencapsul20082554154818465303
- HowardKARahbekULLiuXRNA interference in vitro and in vivo using a novel chitosan/siRNA nanoparticle systemMol Ther20061447648416829204
- GodbeyWTWuKKMikosAGSize matters: molecular weight affects the efficiency of poly(ethylenimine) as a gene delivery vehicleJ Biomed Mater Res A199945268275
- LavertuMMethotSTran-KhanhNBuschmannMDHigh efficiency gene transfer using chitosan/DNA nanoparticles with specific combinations of molecular weight and degree of deacetylationBiomaterials2006274815482416725196
- SudimackJLeeRJTargeted drug delivery via the folate receptorAdv Drug Deliv Rev20004114716210699311
- LowPSAntonyACFolate receptor-targeted drugs for cancer and inflammatory diseasesAdv Drug Deliv Rev2004561055105815094205
- LeeDLockeyRMohapatraSFolate receptor-mediated cancer cell specific gene delivery using folic acid-conjugated oligochitosansJ Nanosci Nanotechnol200662860286617048492
- AllanGGPeyronMMolecular weight manipulation of chitosan II: prediction and control of extent of depolymerization by nitrous acidCarbohydr Res1995277273282
- MimaSMiyaMIwamotoRYoshikawaSHighly deacetylated chitosan and its propertiesJ Appl Polym Sci19832819091917
- ChoKCJeongJHChungHJJoeCOKimSWParkTGFolate receptor-mediated intracellular delivery of recombinant caspase-3 for inducing apoptosisJ Control Release200510812113116139916
- BolstadAIEikenHGRosenlundBAlarcón-RiquelmeMEJonssonRIncreased salivary gland tissue expression of Fas, Fas ligand, cytotoxic T lymphocyte-associated antigen 4, and programmed cell death 1 in primary Sjögren’s syndromeArthritis Rheum20034817418512528117
- KreuzerKALassULandtOHighly sensitive and specific fluorescence reverse transcription-PCR assay for the pseudogene-free detection of beta-actin transcripts as quantitative referenceClin Chem1999452973009931059
- ChanPKurisawaMChungJEYangYYSynthesis and characterization of chitosan-g-poly(ethylene glycol)-folate as a non-viral carrier for tumor-targeted gene deliveryBiomaterials20072854054916999995
- MansouriSCuieYWinnikFCharacterization of folate-chitosan-DNA nanoparticles for gene therapyBiomaterials2006272060206516202449
- KimSHJeongJHChoKCKimSWParkTGTarget-specific gene silencing by siRNA plasmid DNA complexed with folate-modified poly(ethylenimine)J Control Release200510422323215866348
- MintzerMASimanekEENonviral vectors for gene deliveryChem Rev200910925930219053809
- Nakashima-MatsushitaNHommaTYuSSelective expression of folate receptor beta and its possible role in methotrexate transport in synovial macrophages from patients with rheumatoid arthritisArthritis Rheum1999421609161610446858
- FernandesJCWangHJreyssatyCBone-protective effects of nonviral gene therapy with folate-chitosan DNA nanoparticle containing interleukin-1 receptor antagonist gene in rats with adjuvant-induced arthritisMol Ther2008161243125118500247