Abstract
Chlamydia trachomatis is a bacterial sexually transmitted infection affecting millions of people worldwide. Previous vaccination attempts have employed the recombinant major outer membrane protein (MOMP) of C. trachomatis nonetheless, with limited success, perhaps, due to stability, degradation, and delivery issues. In this study we cloned C. trachomatis recombinant MOMP DNA (DMOMP) and encapsulated it in chitosan nanoparticles (DMCNP) using the complex coacervation technique. Physiochemical characterizations of DMCNP included transmission and scanning electron microcopy, Fourier transform infrared and ultraviolet-visible spectroscopy, and zeta potential. Encapsulated DMOMP was 167–250 nm, with a uniform spherical shape and homogenous morphology, and an encapsulation efficiency > 90%. A slow release pattern of encapsulated DMOMP, especially in acidic solution, was observed over 7 days. The zeta potential of DMCNP was ~8.80 mV, which indicated that it was highly stable. Toxicity studies of DMCNP (25–400 μg/mL) to Cos-7 cells using the MTT assay revealed minimal toxicity over 24–72 hours with >90% viable cells. Ultra-violet visible (UV-vis) spectra indicated encapsulated DMOMP protection by chitosan, whereas agarose gel electrophoresis verified its protection from enzymatic degradation. Expression of MOMP protein in DMCNP-transfected Cos-7 cells was demonstrated via Western blotting and immunofluorescence microscopy. Significantly, intramuscular injection of BALB/c mice with DMCNP confirmed the delivery of encapsulated DMOMP, and expression of the MOMP gene transcript in thigh muscles and spleens. Our data show that encapsulation of DMOMP in biodegradable chitosan nanoparticles imparts stability and protection from enzymatic digestion, and enhances delivery and expression of DMOMP in vitro and in mice. Further investigations of the nanoencapsulated DMCNP vaccine formulation against C. trachomatis in mice are warranted.
Introduction
Chlamydia trachomatis is the leading cause of bacterial sexually transmitted infections in both developed and developing countries,Citation1 with an estimated 90 million reported new cases annually.Citation2 Prolonged infection with C. trachomatis often leads to other complications such as pelvic inflammatory disease (PID), ectopic pregnancy, infertility, and chronic abdominal pain.Citation3 In addition, enhancement of the human immunodeficiency virus (HIV) transmission can occur due to the presence of a C. trachomatis infection.Citation4C. trachomatis infections and associated complications amass expenses in excess of US$10 billion annually,Citation5 and are a significant socioeconomic burden.
Antibiotic regimens are effective for treatment of C. trachomatis infections, albeit with limitations, because once infection has ensued and becomes chronic, treatment of the bacteria may prove futile leading to reinfection. Hence, development of a vaccine formulation is a more promising and effective approach for controlling C. trachomatis. Attempts at vaccine formulation have been underway, predating the 1970s where whole organisms were tested against different serovars.Citation6 However, protection gained from the whole organism vaccine was short-lived and strain specific.Citation6 Thus, as yet there is still no approved vaccine against C. trachomatis.
Progression in C. trachomatis analysis led to the characterization of several surface exposed proteins in particular its major outer membrane protein (MOMP). Determination of MOMP as a structurallyCitation7 and immunodominantCitation8 protein of C. trachomatis put it at the forefront of being the best understood and most desirable vaccine candidate. MOMP is a 40 kDa cysteine rich protein with numerous immunogenic B- and T-cell epitopes and protective antigens,Citation7,Citation9–Citation13 thus making it ideally suited and attractive as a vaccine candidate. The native form of MOMP reportedly elicited a protective immune response to a C. trachomatis genital challenged infection in mice, which was similar to that elicited by live elementary bodies.Citation14 However, a native MOMP vaccine is not practical because of the cost associated with its mass production. Therefore, recombinant MOMP (rMOMP) has been widely employed in vaccine studies but protection attained in efficacy studies is not as robust as that of native MOMP.Citation15 Another barrier faced in the use of MOMP in vaccine formulation is its rapid degradation via proteases, which often leads to the poor cellular uptake of MOMP and thus a reduction in its immunogenic capacity.Citation16,Citation17 As a result of this instability of MOMP, vaccine formulations against C. trachomatis also now target DNA-based systems.
DNA vaccines, like their protein counterpart, possess the ability to induce both cellular and humoral immune responses as demonstrated for a variety of pathogens.Citation18,Citation19 Long-term persistence of the presented immunogen is also achieved via DNA vaccination. Other benefits of a DNA-based vaccine include its ability to polarize T-cell help, especially to a Th1 immunological response, as well as ease in the burden of production, compared with a protein-based vaccine. DNA vaccines are also beneficial through the extension in shelf-life gained (storage and shipping capacity), as well as through vaccine stability, providing yet again a less expensive means of manufacturing.
A major hurdle in development of a vaccine against C. trachomatis is an effective delivery system for either a protein-or DNA-based immunogen. Although several delivery systems have been employed in C. trachomatis vaccine development projects,Citation9,Citation13,Citation20 they have not been successful in rendering complete protection against this pathogen. An immunogen, coupled with an effective vaccine delivery system, appear to be paramount in potentially achieving complete protective immunity against C. trachomatis. To this end, nanoparticles have emerged as novel delivery vehicles for vaccination against many pathogens,Citation21,Citation22 including C. trachomatis.Citation20,Citation23 Notably amongst these nanoparticles is chitosan, a cationic polymer with favorable biological properties.Citation24
Chitosan is generated from the deacetylation of chitin, the structural component in the exoskeleton of crustaceans. The deacetylated chitosan backbone of glucosamine units has a high density of amine groups, permitting strong electrostatic interactions with proteins and genes, which carry overall negative charge at neutral pH conditions.Citation25,Citation26 Chitosan is relatively non-toxic and biodegradableCitation27 with a high charge density.Citation28 This along with its low level of toxicity, high encapsulate capacity,Citation21 mucoadhesiveness, and drug penetration enhancement capacity across mucosal barriers,Citation29 make it very attractive in the realm of vaccine delivery development. Chitosan can be degraded into N-acetyl-glucosamine by general body lysozymes, which subsequently is excreted as carbon dioxide via the glycoprotein synthetic pathway.Citation30
In the present study, we formulated a DNA vaccine encoding for MOMP of C. trachomatis (DMOMP) and encapsulated it in chitosan nanoparticles (DMCNP) using the complex coacervation technique. The DMCNP was subjected to physiochemical characterizations including Fourier transform infrared and ultra-violet (UV) spectrophotometry to verify encapsulation; and zeta potential and electrophoresis mobility analyses for stability determinations followed by transmission and scanning electron microscopy for morphology and size. Next, we investigated the toxicity of DMCNP on Cos-7 cells, and the in vitro release of encapsulated DMOMP from DMCNP. Expression of MOMP protein in Cos-7 cells transfected with DMCNP was verified by Western blotting and immunofluorescence microscopy. Finally, in vivo expression of the MOMP gene transcript in muscle tissues and spleens of mice injected with DMCNP was accomplished by reverse transcription polymerase chain reaction (RT-PCR). Here we present and discuss our results in the context of chitosan nanoparticles as an effective delivery system for DMOMP.
Methods and materials
Materials
The phCMV1 vector, containing the intron A from the human CMV IE (immediate-early) gene was purchased from Genlantis, (San Diego, CA, USA). Restriction enzymes (BamH I, Not I and Pst I) were purchased from New England Bio-labs (Ipswich, MA, USA); donkey anti-goat IgG (H+L) was purchased from Invitrogen (Carlsbad, CA, USA). Medium molecular weight chitosan and chitosanase (Streptomyces griseus) were obtained from Sigma-Aldrich (St Louis, MO, USA). Polyclonal antibodies against C. trachomatis (MOMP) were purchased from Fitzgerald Industries International (Acton, MA, USA), with FITC rabbit anti-goat antibodies IgG (H+L) being purchased from Southern Biotech (Birmingham, AL, USA). Plasmid DNA preparation kits, as well as DNA gel extraction kits were purchased from Qiagen (Valencia, CA, USA). Amaxa Cell Line Nucleofector Kit was purchased from Lonza (Walkersville, MD, USA).
Plasmid phCMV1-MOMP construction for DNA vaccine
The MOMP gene of C. trachomatis was amplified from the plasmid pET-MOMPCitation9 by polymerase chain reaction (PCR) and cloned into the phCMV1 vector. Primers used for PCR amplification were phCMV1 MOMP BamH I Dir (for forward) and phCMV1 MOMP Not I Rev (for reverse) with the respective sequences: 5′GGATCCACCATGGCTTCCTCCTTGCATGCTCTGCCTG3′, and 5′CCACCGCGGCCGCTCATTAAGTAAGTCGACGGAACTGAGCA3′. Phusion high-fidelity DNA polymerase protocol was used to amplify the MOMP gene with the optimized forward primer containing a Kozak consensus sequence and unique cloning restriction enzyme site (BamH I). The reverse primer also contained Not I unique and two different stop codons used to ensure the expression of only the full length MOMP protein. The PCR product was analyzed by gel electrophoresis for the correct size and then subjected to unique restriction enzyme digestion using BamH I and Not I. Likewise, the phCMV1 vector was subjected to the same restriction enzyme digestion. Both the MOMP gene and phCMV1 vector were purified by low melting agarose electrophoresis, quantified prior to ligation, and then used to transform competent Escherichia coli MC1061. Transformed bacteria were selected on solid media containing kanamycin, clones were screened using previous cloning restriction enzymes and Pst I, and then confirmed by sequencing. A recombinant clone with the appropriate sequenced MOMP gene (DMOMP) was scaled using Qiagen Endo-free Giga kit (Valencia, CA, USA) and used for the entire study.
Chitosan purification
Chitosan preparation and purification were achieved following previous methodsCitation21 with modifications. Chitosan flakes (1 g) were dissolved in 15 mL of 1 M NaOH and heated for 2 hours at 50°C with continuous stirring. The resulting solution was filtered, washed with deionized water and dried overnight at 40°C. Dried chitosan flakes were dissolved in 0.1 M acetic acid, filtered using a Buchner filtration unit (Fischer Scientific, Houston, TX, USA), adjusted to pH 8.0, ultimately resulting in the formation of purified chitosan precipitates. Chitosan precipitates were collected by centrifugation, vacuum-dried for 20 hours at room temperature, and then dissolved in 1% acetic acid to obtain a 2% chitosan solution.
Preparation of DMOMP in chitosan nanoparticles
DMOMP was prepared in chitosan nanoparticles using the complex coacervation methodCitation21 with minor modification. The chitosan solution consisted of 0.2 mL of 2% chitosan, in combination with 0.8 mL of 5 mM sodium acetate (pH 5.5). The DNA solution consisted of 0.4 mL of DMOMP and 3.6 mL of 45 mM sodium sulfate. Both solutions were heated to 55°C for 15 minutes in a water bath. A ratio of 1:3 chitosan to DNA solution was obtained by adding the chitosan solution drop-wise to the DNA with gentle agitation for 15–25 seconds thus resulting in a spontaneous nanoparticle formation (DMCNP). Phosphate buffered saline (PBS) was encapsulated in chitosan nanoparticles (CNP) to serve as a negative control. Both DMCNP and CNP were stored at 4°C until used.
Encapsulation efficiency and in vitro release analyses
Encapsulation efficiency of DMCNP was determined by sucrose gradient ultracentrifugation as describedCitation21 using the formula: Encapsulation Efficiency = A − B/A × 100 where (A) is the total sum of DNA loaded and, (B) is free DNA.
The analysis of DMOMP released from DMCNP was evaluated using similar intestinal fluid (SIF, pH 7.0) and similar gastrointestinal fluid (SGF, pH 2.0) essentially as published.Citation21 In addition, samples were incubated in PBS (pH 7.4) and incubated at 37°C. Samples were collected at varying time-intervals (day 0.08, 1, 2, 4, 5, and 7) and the released DNA samples were measured using NanoDrop ND1000 (Thermo Scientific, Rockford, IL, USA) at 260 nm.
The released products from each time-point were precipitated with 5 M NaCl and amplified by PCR as described above and amplicons subjected to agarose gel electrophoresis for verification of the specific DMOMP product. The resulting products were confirmed as DMOMP in reference to the positive DMOMP clone with an expected 1154 base pair (bp) size.
Protection assessment of DMCNP
The integrity of protection of encapsulated DMOMP in chitosan nanoparticles was validated using previously described methodsCitation21,Citation31 with some modification. Both DMCNP and DMOMP (1 mg/mL) were subjected to enzymatic digestion with Nde I and Sal I in combination with, or without chitosanase for 2 hours at 37°C according to the manufacturer’s protocol. For DNAase 1 analysis, DMOMP or DMCNP suspensions were incubated in DNAase 1 for 15 minutes at 37°C and the reaction was stopped by the addition of iodoacetic acid (a DNAase 1 inhibitor). All samples were analyzed on a 0.8% agarose gel for the presence or absence of DNA or nanoparticles. Corresponding bands were visualized with a ChemiImager gel documentation apparatus (Alpha Innotech Corp, San Leandro, CA, USA).
Stability studies
Stability assessment of DMCNP was established following previously described methods.Citation21 DMCNP was added to individual micro-centrifuge tubes and adjusted to various pH values (2, 4, 6, 8, 9, 10, 10.5, and 12) followed by incubation at 37°C on a shaker for 30 minutes. All samples were analyzed by agarose gel electrophoresis and visualized using the ChemiImager™ gel documentation system (Genetic Technologies, Inc, Miami FL, USA).
Determination of zeta potential
The stability of DMCNP and CNP was measured by dynamic light scattering using a Zetasizer Nano-ZS (Malvern Instruments, Malvern, UK). Samples of DMCNP and CNP were suspended in filtered distilled water, sonicated, and placed in a disposable cuvette for zeta potential measurements. Nanoparticles were measured for several cycles and comparisons were made based upon their potential and conductivity. Each sample was measured three times and is reported as the mean of triplicate samples.
Morphology and size characterizations
Morphological analysis and approximation of nanoparticle sizes were confirmed using transmission electron microscopy (TEM) and scanning electron microscopy (SEM), respectively. TEM samples were prepared and visualized as reported.Citation21 Similar preparations were made for SEM visualization with a single drop being placed on a glass slide mounted on a SEM stub.
Fourier transform infrared spectra and ultra violet visualization analyses
Fourier transform infrared spectra (FTIR) were recorded for DMCNP and CNP nanoparticles in attenuated total reflectance (ATR) mode using an IR spectrophotometer (Thermo Fisher Nicolet 380 FT-IR, Thermo Fisher Scientific, Waltham, MA, USA).Citation23 The spectra were obtained with 64 scans/sample ranging from 4000 to 400 cm−1 and a resolution of 4 cm−1 and the sample chamber was purged with dry N2 gas. UV-vis was conducted to ascertain the encapsulation of DMOMP in chitosan. Nanoparticles were diluted in deionized water and the absorbance and spectral wavelength were used to determine whether absorption occurred on the outside of the nanoparticle.
Cytotoxicity studies
Cos-7 cells were purchased from ATCC (Manassas, VA, USA), propagated, and maintained as described.Citation21 Cytotoxicity of DMOMP and DMCNP to Cos-7 cells was measured using the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT) dye reduction assayCitation32 and the Cell-Titer 96 Cell Proliferation Assay kit (Promega, Madison, WI, USA). Cells were seeded in a 96-well plate at a density of 105 cells/well in 50 μL Minimal Essential Media (MEM) (Life Technologies, Carlsbad, CA, USA) supplemented with 10% Fetal Bovine Serum (FBS) (Life Technologies) and incubated overnight at 37°C under 5% CO2. DMOMP or DMCNP were added in concentrations ranging from 25 to 400 μg/mL and after either 24, 48, or 72 hours, supernatants were removed, cells washed twice with sterile PBS, followed by addition of 15 μL of MTT dye solution to each well and cells further incubated for 3 hours at 37°C under 5% CO2. To stop the reaction, 100 μL of solubilization solution/stop mixture was added to each well and plates incubated for 30 minutes at room temperature. Absorbance at 570 nm was measured using a TECAN Sunrise plate reader (TECAN US Inc, Durham, NC, USA). The percentage of cell viability was obtained using the optical density readings of nanoparticle-treated cells compared to those of normal cells (control), where percent viability = [A]test/[A] control × 100, where [A]test is the absorbance of the test sample and [A]control is the absorbance of the control sample.
Transfection and Western blot analyses
Cos-7 cells (1 × 106 cells/well) were transfected by electroporation with DMOMP and DMCNP at concentrations of 2, 5, and 10 μg using the Amaxa Cell Line Nucleofector kit-R. Transfected cells were incubated for 48 hours at 37°C with 5% CO2, after which they were washed in PBS and fixed using 10% trichloroacetic acid, followed by successive washing with 70%, 90%, and 100% ethanol with a final wash in 1× tris buffered saline and tween 20. Cells were blocked for 30 minutes in blocking buffer (3% dry milk in 1× tris buffered saline), washed in PBS and then incubated for 1 hour with goat anti-C. trachomatis polyclonal antibodies followed by a secondary FITC rabbit anti-Goat IgG (H+L) antibody. For some experiments cells were stained with DAPI combined with anti-fade (Invitrogen) mounting solution. Immunofluorescence of cells was visualized using a Nikon Eclipse Ti-U microscope (Nikon Instruments, Melville, New York, USA).
In addition to transfection of Cos-7 cells via electroporation, cells were also chemically transfected using Lipofectamine™ reagent 2000 (Invitrogen). Cos-7 cells were seeded (4 × 105 cells/well) in a 6-well plate for 24 hours after which they were transfected with DMCNP or phCMV 1 vector in opti-MEM transfection media and incubated at 37°C for 48 hours. Cell lysates were collected using M-Per mammalian protein extraction reagent (Thermo Fisher Scientific); the resulting lysates were run on an SDS-PAGE gel (Bio-Rad Laboratories, Hercules, CA, USA), transferred onto a PVDF membrane and probed using anti-MOMP polyclonal antibodies (Fitzgerald Industries), followed by an Alexa fluor 680 secondary antibody (Life Technologies). The fixed antibody was viewed using the LI-COR Odyssey imaging apparatus.
Animals
Six- to eight-week old BALB/c female mice (Charles Rivers, Raleigh, NC, USA) were used for this study. Animal studies were performed following a protocol approved by the Alabama State University (Montgomery, AL, USA) Institutional Animal Care and Use Committee. Mice were housed under standard pathogen-free environmental conditions at ambient temperatures of 25°C, and provided sterile food and water ad libitum. Mice (6 per group) were injected intramuscularly with DMCNP (80 μg/200 μL of PBS) or with 200 μL of PBS. Mice (2 per group) were sacrificed on day 8 and their thigh muscles and spleens were harvested and stored in RNAlater® (Life Technologies) at −80°C until analyzed.
RNA isolation from mouse tissues
RNA samples were extracted from mouse thigh muscles and spleens using a Qiagen gel extraction kit which included a DNAase-I digestion step according to the manufacturer’s protocol. RNA quantifications were made using NanoDrop ND-1000 spectrophotometer at an absorbance of 260 nm. Complementary DNA (cDNA) was produced from quality RNA using the SuperScript™ II Reverse Transcriptase kit (Life Technologies) following the manufacturer’s protocol. RT-PCR amplification of the MOMP gene transcript was performed using the cDNA as a template, and the same primers as used for cloning.
Results
Purification and expression of DMOMP
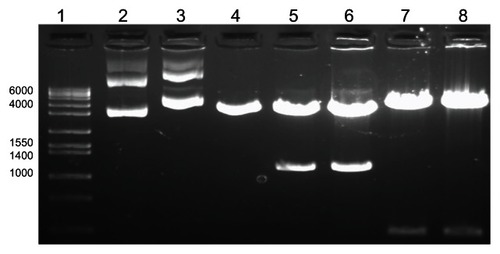
The major outer membrane protein of C. trachomatis was isolated, amplified through PCR, and cloned into the phCMV1 vector resulting in the vaccine construct, DMOMP. The purified DNA clones as analyzed by restriction enzymes digestion on agarose gel electrophoresis are shown in with the native phCMV1 vector (lane 2), native phCMV1 MOMP (lane 3), phCMV1 vector (4239 bp) restricted BamH 1 and Not 1 (lane 4), phCMV1 MOMP positive clones (4212 bp and 1134 bp) restricted BamH 1 and Not 1 (lanes 5 and 6) and phCMV1 MOMP clones (lanes 7 and 8) restricted Pst 1 (4823 bp and 531 bp). Verification of the positive clones was also confirmed by DNA sequencing (Auburn University Genomics and Sequencing laboratory, Auburn, AL, USA).
Figure 1 Agarose gel electrophoresis analysis of DMOMP.
Notes: Molecular weight (1 kb) marker (lane 1), native phCMV1 vector (lane 2), native phCMV1 MOMP (lane 3), phCMV1 vector (4239 bp) restricted BamH 1 and Not 1 (lane 4), phCMV1 MOMP positive clones (4212 bp and 1134 bp) restricted BamH 1 and Not 1 (lanes 5 and 6) and phCMV1 MOMP clones (lanes 7 and 8) restricted Pst 1 (4823 bp and 531 bp).
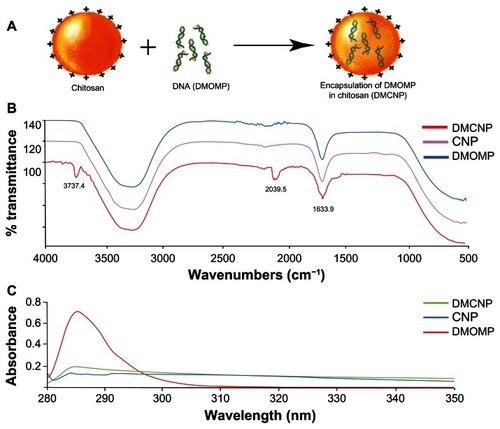
Structural characterization: FT-IR and UV-vis analyses
DMOMP was encapsulated in chitosan nanoparticles as illustrated in . A modified method of the complex coacervation technique was used to prepare DMCNP with an achievement of >90% encapsulation efficiency. To validate encapsulation of DMOMP in chitosan both FTIR and UV-vis analyses were performed. FTIR analysis of DMOMP indicated peaks at 2039.5 cm−1 and 3737.4 cm−1 representative of the chemical components of the hydroxyl and amine groups (). After encapsulation these chemical signature peaks were no longer detectable, almost providing a mirror absorbance image of CNP. These results indicate DMOMP successful encapsulation within the chitosan nanoparticle due to the absence of the aforementioned peaks within the DMCNP profile. UV-vis analysis () also confirmed the successful encapsulation of DMOMP within the chitosan polymer shell as illustrated by the absorbance readings of DNA at a wavelength of ~285. In comparison, no absorbance was observed on the surface of DMCNP or CNP further verifying the successful encapsulation of DMOMP within the chitosan nanoparticle.
Figure 2 Encapsulation of DMOMP (MOMP DNA) in chitosan nanoparticles. (A) A schematic representation of DMOMP-chitosan nanoparticle construction. (B) Fourier Transfer-Infrared spectroscopy (FT-IR) of DMCNP (DMOMP encapsulated in chitosan nanoparticles), CNP (PBS encapsulated in chitosan nanoparticles) and DMOMP. Recording of the FT-IR spectrum was achieved through 32 scans with a sample ranging from 500 to 4000 cm−1 and a resolution of 4 cm−1 at ambient temperature. Chemical signature peaks observed within the DMCNP spectrum at 2039.5 cm−1 are absent from the DMOMP and CNP spectra. (C) Ultra-violet visible spectra of DMCNP, CNP and DMOMP.
Abbreviations: CNP, phosphate buffered saline encapsulated in chitosan nanoparticles; DMOMP, DNA of the major outer membrane protein of C. trachomatis; DMCNP, DMOMP encapsulated in chitosan nanoparticles.
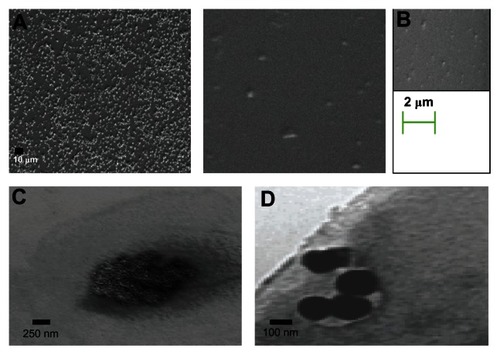
Size and surface morphology (SEM and TEM) of nanoparticles
Particle size is a major determinant in the level of mucosal and epithelial tissue uptake of nanostructures, as well as the intracellular interchange of particles.Citation33 SEM analysis revealed CNP and DMCNP to be spherical structures () with sizes of ~50–75 nm for CNP in comparison to ~167–250 nm for DMCNP, thus indicating an increase in size due to the encapsulation process. TEM was also employed in performing morphological analysis and size determination of the CNP and DMCNP formulations. TEM images revealed uniformity in particle size and a homogenous morphology (), corroborating the SEM acquired images ().
Figure 3 Size and morphological assessments of nanoparticles by SEM and TEM. SEM analyses of CNP (A), DMCNP (B), and TEM analysis of CNP (C) and DMCNP (D).
Note: A drop of the nanoparticles was deposited on a copper grid for TEM, or a glass slide mounted on a stub for SEM.
Abbreviations: CNP, phosphate buffered saline encapsulated in chitosan nanoparticles; DMOMP, DNA of the major outer membrane protein of C. trachomatis DMOMP; DMCNP, DMOMP encapsulated in chitosan nanoparticles.
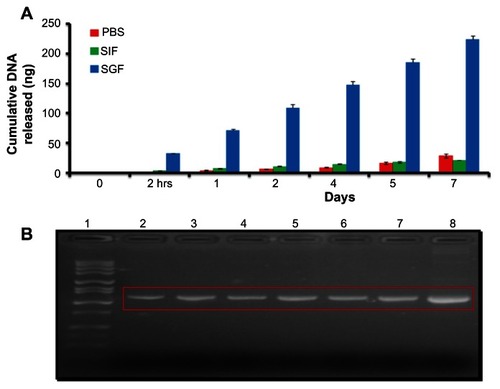
Cumulative in vitro release of DMOMP from DMCNP
Release of DMOMP from DMCNP was assessed through incubation of samples in SGF (pH 2), SIF (pH 7) and PBS (pH 7.4) over 7 days (). When DMCNP was incubated in SGF, DMOMP was released at a more rapid rate, indicating an increased rate of degradation of chitosan due to its presence in an acidic environment. The release pattern of DMOMP from DMCNP was slow and similar for both SIF and PBS mainly because of the neutral pH 7 solutions, in comparison to that of SGF, whereby DMOMP was released more rapidly at pH 2 (). Products released during the 7-day period were amplified and amplicons verified by agarose gel electrophoresis (). The results show that the released products (, lanes 2–6) were indeed DMOMP as validated by the expected size (1154 bp) of the DMOMP positive clone (, lane 6).
Figure 4 Cumulative DNA release and electrophoretic analysis of released DMOMP. (A) Analysis of encapsulated DMOMP released from DMCNP was evaluated using similar intestinal fluid (SIF, pH 7.0) and similar gastrointestinal fluid (SGF, pH 2.0). Samples were collected at each designated time-interval (2 hours, day 1, 2, 4, 5 and 7) and the released DNA measured using NanoDrop at 260 nm. Each bar represents the mean ± standard deviation of triplicate samples. (B) The released products from each time-point were precipitated with 5 M NaCl and amplified by PCR and amplicons subjected to agarose gel electrophoresis for verification of the specific DMOMP product.
Notes: Lanes indicate the sequential collection of amplicons as follows: lane 1 (1 kb marker), lane 2 (2 hours), lane 3 (day 1), lane 4 (day 2), lane 5 (day 4), lane 6 (day 5), lane 7 (day 7) and lane 8 (positive DMOMP clone) with an expected size of 1154 bp.
Abbreviations: DMOMP, DNA of the major outer membrane protein of C. trachomatis; DMCNP, DMOMP encapsulated in chitosan nanoparticles; hrs, hours; PBS, phosphate buffered saline; PCR, polymerase chain reaction; SGF, similar gastrointestinal fluid; SIF, similar intestinal fluid.
Stability studies of DMOMP and DMCNP
Electrophoresis analysis was conducted to assess protection of DMOMP by chitosan nanoparticles. Samples were exposed to chitosanase, DNAase, and restriction enzymes prior to analysis. Electrophoresis analysis of the DMCNP which was not subjected to enzymatic treatments showed no indication of DMOMP released (, lane 2). Following digestion of DMCNP with chitosanase, the released DMOMP was present in the gel indicating degradation of the nanoparticle by the enzyme (, lane 3). When DMCNP was exposed to both chitosanase and DNAase digestion, no DMOMP was seen in the gel, indicating complete degradation of DNA and chitosan (, lane 4). Simultaneous exposure of DMCNP to chitosanase, restriction enzymes, and DNAase revealed the absence of DMOMP (, lane 5). However, when DMCNP was digested with only restriction enzymes, DMOMP was also not present in the gel, verifying that chitosan surrounds DMOMP, thus preventing restriction enzyme access (, lane 6). DMOMP alone was used as a positive control as validation of the respective product sizes (, lane 8). The findings from this study suggest the capacity of chitosan nanoparticles to protect the encapsulated DMOMP from enzyme degradation.
Figure 5 Stability studies of DMOMP in chitosan nanoparticles. (A) Electrophoretic analysis of CNP protection of encapsulated DMOMP after incubation with chitosanase, DNAase I, and restriction enzymes. Both DMCNP and DMOMP (1 mg/mL) were subjected to enzymatic digestion with NdeI and SalI in combination with, or without chitosanase or with DNAase 1. Lane 1, (1 kb molecular marker), lane 2 (trapped nanoparticle in well), lane 3 (released DMOMP in red box), lanes 4 and 5 (degraded released DMOMP), lane 6 (blank), lane 7 (1 kb marker), and lane 8 (DMOMP positive clone). (B) pH stability of encapsulated DMOMP.
Notes: DMCNP was added to individual micro-centrifuge tubes and adjusted to various pH values (2, 4, 6, 8, 9, 10, 10.5 and 12) followed by incubation at 37°C on a shaker for 30 minutes. Lanes 1 and 10 are 1 kb marker. All samples were analyzed by agarose gel electrophoresis and visualized using the ChemiImager gel documentation system.
Abbreviations: CNP, phosphate buffered saline encapsulated in chitosan nanoparticles; DMOMP, DNA of the major outer membrane protein of C. trachomatis; DMCNP, DMOMP encapsulated in chitosan nanoparticles; RE, restriction.
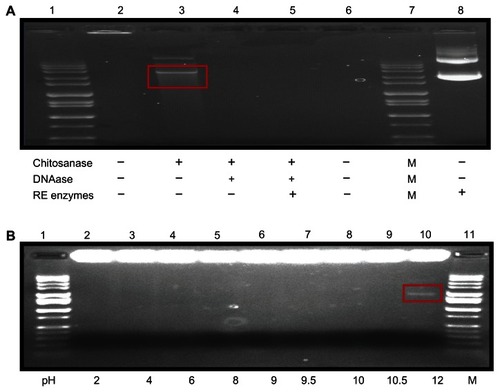
Stability, integrity, and binding aptitude of the DMCNP in different solutions were examined through an electrophoretic mobility analysis using 1.0% agarose gel. DMCNP complex disintegration in various pH solutions is shown in . Stability of the DMCNP was sustained up to pH 10 with no DMOMP being released (). Minor disintegration of DMCNP complexes was observed at pH 10.5 (, lane 9) with a prominent band being observed at pH 12 (, lane 10). These studies show the ability of chitosan to protect DMOMP from degradation up to pH 10 prior to nanoparticle
Zeta potential
Assessment of zeta potential is critical in nanoparticle studies, as it may affect both particle stability and mucoadhesion.Citation34 The zeta potential value for DMCNP was 8.80 mV in comparison to 0.148 mV for CNP alone (). These results indicate an increase in stability of DMCNP when compared to that of CNP.
Table 1 Zeta potential and encapsulation efficiency of nanoparticles
Cytotoxicity studies
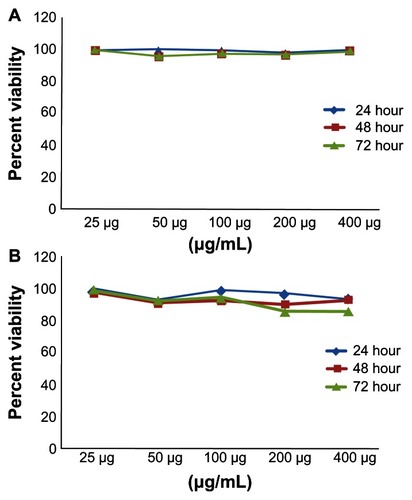
The cytotoxicity of DMCNP and DMOMP was assessed in Cos-7 cells by the MTT assay. Minimal cell death (>90% viable cells) was observed at all concentrations tested from 25 to 400 μg/mL for both DMCNP () and DMOMP () and at all examined time-points. Overall no differences were seen on the viability of Cos-7 cells after their exposure to DMCNP and DMOMP.
Figure 6 Cytotoxicity analyses of (A) CNP and (B) DMCNP to Cos-7 cells.
Notes: Cos-7 cells were seeded in a 96-well plate at a density of 105 cells/well/ 50 μL in the presence or absence of CNP or DMCNP in concentrations ranging from 25 to 400 μg/mL and incubated at 37°C for 24, 48, or 72 hours. The Cell-Titer 96 cell Proliferation Assay kit was used to determine cell viability. Absorbance was read at 570 nm and % cell viability was calculated by using the optical density readings compared to normal cells as indicated in the Materials and method section.
Abbreviations: CNP, phosphate buffered saline encapsulated in chitosan nanoparticles; DMCNP, DNA of the major outer membrane protein of C. trachomatis (DMOMP) encapsulated in chitosan nanoparticles.
Transfection studies: gene delivery and protein expression studies
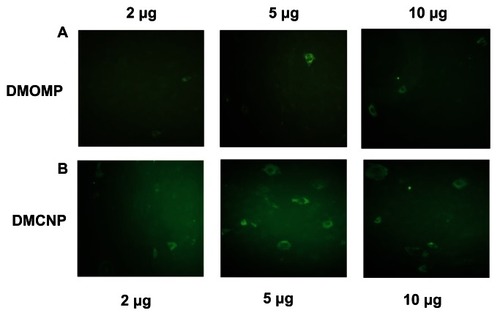
Qualitative expression of protein upon transfection of Cos-7 cells with DMOMP and DMCNP was evaluated by immunofluorescence microscopy (). A level of increase in MOMP protein expression was detected in cells transfected with DMCNP (, row B) in comparison to DMOMP (, row A). Protein expression was greater for the 5 and 10 μg concentrations, than the 2 μg concentration, suggesting the ability of chitosan to enhance the level of MOMP protein expression in Cos-7 cells.
Figure 7 In vitro expression of MOMP protein in transfected Cos-7 cells. Cos-7 cells (1 × 106 cells/well) were transfected by electroporation with DMOMP (A) and DMCNP (B) at concentrations of 2, 5 and 10 μg.
Notes: Transfected cells were incubated for 48 hrs at 37°C, fixed and blocked prior to incubation with goat anti-C. trachomatis polyclonal antibodies followed by a secondary FITC rabbit anti-Goat IgG (H+L) antibody. Immunofluorescence of cells were visualized using a Nikon Eclipse Ti-U microscope.
Abbreviations: DMOMP, DNA of the major outer membrane protein of C. trachomatis; DMCNP, DMOMP encapsulated in chitosan nanoparticles; MOMP, major outer membrane protein of C. trachomatis.
Expression in Cos-7 cells was also evaluated through immunofluorescence microscopy using DAPI nuclei stain () and bright-field () microscopy. DMCNP-transfected Cos-7 cells were probed with anti-MOMP polyclonal antibodies and as shown in , MOMP protein expression was clearly observed in the Cos-7 cells exposed to DMCNP as depicted by the green fluorescence cells but not in those exposed to CNP (data not shown). As confirmation of the immunofluorescence microscopy results we next performed Western blot with protein lysates collected from DMCNP transfected Cos-7 cells (). , lane 2 reveals a band ~48 kDa of the MOMP protein, indicating the successful expression of the full MOMP protein in these transfected cells. Our results provide further evidence for the successful encapsulation, and subsequent release of encapsulated DMOMP from the chitosan nanoparticle.
Figure 8 Expression of MOMP at the protein and gene transcript levels. (A) Cos- 7 cells were transfected as described in , immunostained (positive MOMP fluorescence cells) and then mounted with DAPI (blue nuclei stain) combined with an anti-fade mounting solution. (B) Bright-field visualization of Cos-7 cell monolayer showing the MOMP expressed protein. Red circle (positive MOMP fluorescence cells) shows expression of the MOMP protein. (C) Confirmation of expressed MOMP protein by western blot. Cos-7 cells (4 × 105 cells/well) were transfected with DMCNP or phCMV1 vector using Lipofectamine and incubated at 37°C for 48 hours. Cell lysates were collected, run on an SDS-PAGE gel, transferred onto a PVDF membrane and probed using anti-MOMP polyclonal antibodies followed by an Alexa fluor 680 secondary antibody. The bound antibody was viewed using LI-COR Odyssey imaging apparatus. (D) In vitro expression of MOMP gene transcript.
Notes: RNA samples were extracted from mouse thigh muscles and spleens reversed transcribed to cDNA and then subjected to RT-PCR amplification of the MOMP gene transcript using MOMP specific primers. Lane 1 (MW marker), lanes 2 and 7 (DMOMP positive clones), lane 3 (thigh muscle of DMCNP mice), lane 4 (thigh muscle of PBS mice), lane 5 (spleen from DMCNP mice), and lane 6 (spleen from PBS mice). Red rectangle shows positive MOMP gene transcripts.
Abbreviations: DMCNP, DMOMP encapsulated in chitosan nanoparticles; DMOMP, DNA of the major outer membrane protein of C. trachomatis; MOMP, major outer membrane protein of C. trachomatis; PVDF, polyvinylidene difluoride.
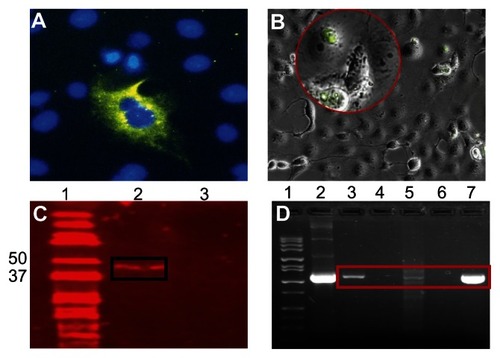
Confirmation of DMOMP expression in vivo was investigated by intramuscular administration of DMCNP to mice. RNA was extracted from thigh muscles and spleens at day 8 and subjected to amplification to determine expression of the MOMP gene transcript. As shown in (lane 3), the expression level of the MOMP gene transcript was elevated in the thigh muscle of the DMCNP group in comparison to that of the PBS group (, lane 4). Spleens from DMCNP mice also showed expression of the MOMP gene (, lane 5) as compared to the PBS group (, lane 6), albeit at lower levels. DMOMP clones were run as positive controls to confirm the correct MOMP gene size ( lanes 2 and 7). Overall these results revealed the expression of the MOMP protein in vitro and its gene transcripts in vivo, suggesting the enhanced delivery potential of chitosan nanoparticles for DMOMP.
Discussion
Presently, one of the foremost problems associated with C. trachomatis vaccine development is the necessity for the induction of strong Th1 immunological responses, as well as delivery of the vaccine candidate. Previously we demonstrated the ability of MOMP to induce a balanced Th1/Th2 serological immune response,Citation9 and the capacity of chitosan to enhance delivery and expression of a nano-encapsulated RSV DNA vaccine.Citation21 In the present study, we developed a DNA vaccine candidate by cloning the immune-dominant MOMP gene of C. trachomatis into the phCMV1 vector and encapsulating it in chitosan nanoparticles. Our results demonstrate the successful encapsulation and characterization of DMCNP as well as the in vitro and in vivo expression of the MOMP protein and gene transcripts.
In our present study, DMCNPs were produced through the complex coacervation technique due to the mild conditions of the technique, and its successive high encapsulation efficiency rate. Encapsulation of plasmid in chitosan in the generation of a nanoparticle is reliant on various parameters including the N/P ratio (the ratios of moles of the amine groups of cationic polymers to those of the phosphate ones of DNA), polymer charge density, polymer structure, molecular weight of chitosan, degree of deacetylation, and pH value.Citation35,Citation36 This technique allowed us to obtain an encapsulation efficiency of 90% or greater which may be due to the addition of sodium acetate to ensure the maintenance of a relatively constant pH, a key parameter in the formation of the encapsulated DMOMP. High rates of encapsulation are achieved when chitosan possessing high levels of deacetylation and increased density of amino acids interacts with phosphate groups present in DNA.Citation37 Similar efficiencies have been reported by other investigators who used the complex coacervation technique for encapsulation.Citation21,Citation32,Citation36,Citation38
The release profiles of encapsulated DMOMP from chitosan nanoparticles were pH dependent as revealed by the results obtained in SGF (pH 2), SIF (pH 7) and PBS (pH 7). Release profile of any biomaterial from a nanoparticle is an important attribute in nanovaccine development as it can influence the immunological response and immunization protocol.Citation23 Within acidic solutions, the amino acid groups of chitosan are protonated with the resultant soluble polysaccharide being charged.Citation39 Studies by Hu et alCitation40 demonstrated that the release of a gene from the nanoparticle depends greatly on the swelling of the nanoparticle and pH values of the releasing solution. Consistent with previous studies our results indicate that the release pattern of encapsulated DMOMP was biphasicCitation21 with an initial burst preceding a controlled release which was also pH dependent.Citation40
The initial release of DMOMP observed in all three pH solutions could be attributed to the release of molecules weakly associated with the chitosan nanoparticles located at, or near the nanoparticle surface, whereas the second phase of the release profile may correspond to DNA molecules more proficiently entrapped within the nanoparticle. In our study, different release profiles were obtained for DMCNP and were dependent on its presence in either an alkaline or acidic solution. At pH values of 7 to 7.4, chitosan nanoparticles are comparatively more swollen than those at a pH < 4.0,Citation40 resulting in a slower release of encapsulated DNA due to slower degradation of the nanoparticles. The release of encapsulated DMOMP in SIF and PBS solutions was less due to chitosan inability to solubilize in neutral solutions. In comparison, DMCNP exposed to SGF released its entrapped DMOMP at a more rapid pace due to excessive degradation of the chitosan nanoparticle in a more acidic solution.
Here our analysis demonstrates the ability of chitosan to protect encapsulated DMOMP from degradation, corroborating previous reports.Citation21,Citation35,Citation41 Our results clearly indicate that release of DMOMP was enzyme-dependent and that our chitosan nanoparticle exhibits protective properties promoting its potential as an effective and efficient transport system capable of safe delivery and protection of DMOMP from degradation by biological enzymes. Within cells, chitosan is degraded by lysozyme and N-acetyl-glucose amidase, both of which are present in the endosomal vesicle. The presence of these two enzymes indicates that degradation and the ensuing release of the plasmid will begin immediately following endocytosis.Citation41
In our study DMCNP nanoparticles ranged from 167–250 nm in diameter and their shapes were spherical with a homologous morphology. It is known that nanoparticles smaller than 10 μm are phagocytized by antigen presenting cells at the mucosal surfaces, leading to a potency increase, or potentiating immune responses.Citation42,Citation43 According to Boyoglu et al,Citation21 the physical state and stability of the nanoparticle can be considered key parameters for efficient and effective peptide delivery. Thus, DMCNP produced through our modified complex coacervation technique possessed the required size necessary to facilitate uptake by APCs, which is necessary for induction of immune responses.
We employed the MTT test to assess the overall effects of DMOMP and DMCNP on the metabolic activity of Cos-7 cells. Our results overall showed no cytotoxic effects of DMOMP or DMCNP for Cos-7 cells as cell viabilities remained greater than 90%. Taking into consideration that proliferating cells are metabolically more active than nonproliferating cells, results obtained can also be interpreted as the possible impact of DMOMP and DMCNP solutions on cellular proliferation. This is perhaps indicative that polymers may favorably manipulate both cellular lysosomal and mitochondrial activity.Citation44
Since its initial usage as a vector-based delivery system for the transport of genes into the cellular matrix,Citation45 chitosan and its derivatives have revealed immense promise for encapsulation of DNA. In our study we demonstrated that chitosan not only protected, but also enhanced expression of DMOMP. Previous studies have shown that chitosan possesses capabilities of opening tight cellular junctions through varying effects upon F-actin filaments.Citation46 This, along with the small size of chitosan nanparticles being engulfed by endocytosis into endocytic vesicles, allows it entry into targeted cells via transferring cytosis,Citation47 which may explain the enhanced effect of DMCNP on encapsulated DMOMP delivery into both Cos-7 cell monolayers and animal tissues within our study. Also, as cell culture substrates, chitosan and its derivatives may allow activation of varying endocytic pathways thus increasing nanoparticle internalization or plasmid tranfection.Citation48 Saka and BozkirCitation49 reported increased levels of transfection efficiency in vitro using chitosan-based nanoparticle formulations, thereby corroborating our results in the present study.
Here in the present study expression was seen not only in Cos-7 cells but also in mice. Expression of the MOMP gene was observed to be localized as well as systemic, respectively in thigh muscles and spleens of mice. Increased levels of gene expression have been demonstrated through chitosan-plasmid usage in comparison to plasmid usage alone.Citation50 Reportedly, Jean and colleagues,Citation50 demonstrated encapsulation of a type 2 diabetes gene for treatment of mice produced results two-fold greater than plasmid administration alone. Our results strongly suggest that DMCNP can improve DMOMP expression with enhanced delivery to targeted tissues.
Conclusion
Our study, to the best of our knowledge, is the first to encapsulate a phCMV1-MOMP DNA construct in chitosan, perform physiochemical characterizations, and assess its expression by both in vitro and in vivo experiments. In our study, DMCNP was generated through the complex coacervation technique with >90 encapsulation efficiency. DMCNP was ~167–250 nm and spherical in shape. Notably, chitosan protected DMOMP from degradation and enhanced its protein expression in Cos-7 cells. More significantly, the MOMP gene transcript was expressed locally and systemically in tissues of mice. Our data accentuate the efficiency of chitosan as a delivery vehicle for the formulation of a C. trachomatis vaccine candidate. In addition, DMCNP has the potential of a promising nano-encapsulated vaccine formulation and warrants further studies in mice.
Acknowledgment
This research was supported by funding from National Science Foundation (NSF) grants NSF-CREST (HRD-1241701) and NSF-HBCU-UP (HRD-1135863). We thank Drs Mamie Coats and Saurabh Dixit for their help with the mice study, Mr Elijah Nyairo for assistance with the FTIR, and Dr Michael Miller, Auburn University, for help with TEM and SEM images. Special thanks go to Eva A Dennis for illustration, and to the Center for NanoBiotechnology Research (CNBR) staff, Yvonne Williams and Lashaundria Lucas for their time and support.
Disclosure
The authors report no conflict of interests in this work.
References
- World Health OrganizationGlobal Prevalence and Incidence of Selected Sexually Transmitted Diseases: Overviews and EstimatesGeneva, SwitzerlandWorld Health Organization1996152
- World Health OrganizationDoRHaR, Prevalence and Incidence of Selected Sexually Transmitted InfectionsGeneva, SwitzerlandWorld Health Organization2011
- WestromLJRJoesoefRReynoldsGHagduAThompsonSEPelvic inflammatory disease and fertility. A cohort study of 1,844 women with laparoscopically verified disease and 657 control women with normal laparoscopic resultsSex Transm Dis199241851921411832
- PlummerFASimonsenJNCameronSJNdinya-AcholaJOKreissJKGakinyaNMCofactors in male–female sexual transmission of human immunodeficiency virus type 1J Infect Dis1991612361237
- BeagleyKWTimmsPChlamydia trachomatis infection: incidence, health costs and prospects for vaccine developmentJ Reprod Immunol200048476810996382
- KarunakaranKPYuHFosterLJBrunhamRCDevelopment of a Chlamydia trachomatis T cell vaccineHum Vaccine20106676680
- CaldwellDKromhoutJSchachterJPurification and partial characterization of the major outer membrane protein of Chlamydia trachomatisInfect Immun198131116111767228399
- ColerRNBhatiaAMaisonneuveJFIdentification and characterization of novel recombinant vaccine antigens for immunization against genital Chlamydia trachomatisFEMS Immunol Med Microbiol20095525827019281568
- SinghSRHulettKPillaiSRDennisVAOhMKScissum-GunnKMucosal immunization with recombinant MOMP genetically linked with modified cholera toxin confers protection against Chlamydia trachomatis infectionVaccine2006241213122416194585
- OrtizLAngevineMKimSKWatkinsDDeMarsRT-cell epitopes in variable segments of Chlamydia trachomatis major outer membrane protein elicit serovar-specific immune responses in infected humansInfect Immun2000681719172310678996
- StaggAJElsleyWAPickettMAWardMEKnightSCPrimary human T-cell responses to the major outer membrane protein of Chlamydia trachomatisImmunology199379198099564
- ZhuSChenJZhengMGongWXueXLiWIdentification of immunodominant linear B-cell epitopes within the major outer membrane protein of Chlamydia trachomatisActa Biochim Biophys Sin201042820043042
- XuWLiuJGongWChenJZhuSZhangLProtective immunity against Chlamydia trachomatis genital infection induced by a vaccine based on the major outer membrane multi-epitope human papillomavirus major capsid protein L1Vaccine2011292672267821324344
- PalSPetersonEMde la MazaLMVaccination with the Chlamydiatrachomatis major outer membrane protein can elicit an immune response as protective as that resulting from inoculation with live bacteriaInfect Immun2005738153816016299310
- SunGPalSWeilandJPetersonEMde la MazaLMProtection against an intranasal challenge by vaccines formulated with native and recombinant preparations of the Chlamydia trachomatis major outer membrane proteinVaccine2009275020502519446590
- EkoFOHeQBrownTA novel recombinant multi-subunit vaccine against ChlamydiaJ Immunol20041733375338215322201
- DaiCWangBMicroencapsulation peptide and protein drugs delivery systemColloid Surf B Biointerfaces200541117120
- TyagiRKGargNKSahuTVaccination strategies against malaria: novel carrier(s) more than a tour de forceJ Control Release201216224225422564369
- KathuriaNKraynyakKACarnathanDBettsMWeinerDBKutzlerMAGeneration of antigen-specific immunity following systemic immunization with DNA vaccine encoding CCL25 Chemokine ImmunoadjuvantHum Vaccin Immunother201281607161923151454
- ChildsTSWebleyWCIn vitro assessment of halobacterial gas vesicles as a Chlamydia vaccine display and delivery systemVaccine2012305942594822846397
- BoyogluSVigKPillaiSRangariVDennisVAKhaziFEnhanced delivery and expression of a nanoencapsulated DNA vaccine vector for respiratory syncytial virusNanomedicine2009546347219341819
- TyagiRKSharmaPKVyasSPMehtaAVarious carrier system(s)- mediated genetic vaccination strategies against malaria. Expert Rev Vaccines. 2008;7:499–520Retraction in Expert Rev Vaccines2011101244
- TahaMASinghSRDennisVABiodegradable PLGA85/15 nanoparticles as a delivery vehicle for Chlamydia trachomatis recombinant MOMP-187 peptideNanotechnology20122332510122824940
- FeltOBuriPGurnyRChitosan: a unique polysaccharide for drug deliveryDrug Dev Ind Pharm1998249799939876553
- MacLaughlinFCMumperRJWangJChitosan and depolymerized chitosan oligomers as condensing carriers for in vivo plasmid deliveryJ Controlled Rel199856259272
- MaoSShuaiXUngerFSimonMThe depolymerization of chitosan: effects on physiochemical and biological propertiesInt J Pharm2004281455415288342
- GanQWangTCochraneCMcCarronPModulation of surface charge, particle size and morphological properties of chitosan-TPP nanoparticles intended for gene deliveryColloids Surf B Biointerfaces200544657316024239
- JanesKACalvoPAlonsoMJPolysaccharide colloidal particles as delivery systems for macromoleculesAdv Drug Deliv Rev200147839711251247
- ArturssonPLindmarkTDavisSSIllumLEffect of chitosan on the permeability of monolayers of intestinal epithelial cells (Caco-2)Pharm Res199411135813617816770
- GanQWangTChitosan nanoparticles as protein delivery carrier- Systematic examination of fabrication conditions for efficient loading and releaseColloids Surf B Biointerfaces200759243417555948
- SunYZhangSPengXPreparation, characterization and transfection efficacy of chitosan nanoparticles containing the intestinal trefoil factor geneMol Biol Rep20123994595221573797
- Yu-HongLMin-WenFZhiCQiZHai-RuiYChitosan-DNA microparticles as mucosal delivery system: synthesis, characterization and release in vitroChin Med2005118936941
- PanyamJLabhasetwarVBiodegradable nanoparticles for drug and gene delivery to cells and tissuesAdv Drug Deliv Rev20035532934712628320
- Ravi KumarMNBakowskyULehrCMPreparation and characterization of cationic PLGA nanospheres as DNA carriersBiomaterials2004251771177714738840
- BozkirASakaOMChitosan-DNA nanoparticles: effect on DNA integrity, bacterial transformation and transfection efficiencyJ Drug Target20041228128815512779
- MaoHQRoyKTroung-LeVLJanesKALinKYWangYChitosan-DNA nanoparticles as gene carriers: synthesis, characterization and transfection efficiencyJ Control Release20017039942111182210
- BorchardGChitosans for gene deliveryAdv Drug Deliv Rev20015214515011718938
- ZhaoKShiXZhaoYPreparation and immunological effectiveness of a swine influenza DNA vaccine encapsulated in chitosan nanoparticlesVaccine2011298549855621945253
- HejaziRAmijiMChitosan-based gastrointestinal delivery systemsJ Control Release20038915116512711440
- HuYJiangXDingYGeHYuanYYangCSynthesis and characterization of chitosan-poly(acrylic acid) nanoparticlesBiomaterials2002233193320112102191
- Köping-HöggårdMTubulekasIGuanHEdwardsKNilssonMVårumKMChitosan as a non-viral gene delivery system. Structure-property relationships and characteristics compared with polyethylenimine in vitro and after lung administration in vivoGene Ther200181108112111526458
- Waeckerle-MenYGroettrupMPLGA microspheres for improved antigen delivery to dendritic cells as cellular vaccinesAdv Drug Deliv Rev20055747548215560953
- Waeckerle-MenYAllmenEUGanderBEncapsulation of proteins and peptides into biodegradable poly(d,l-lactide-co-glycolide) microspheres prolongs and enhances antigen presentation by human dendritic cellsVaccine2006241847185716288821
- GuerraGDCerraiPTricoliMMaltiniSdel GuerraRSIn-vitro cytotoxicity testing of chitosan-containing polyelectrolyte complexesJ Mater Sci Mater Med19989737615348910
- MumperRJClaspellJMRollandAPNovel polymeric condensing carriers for gene deliveryProc Natl Acad Sci U S A199522178179
- IllumLJabbal-GillIHinchcliffeMFisherANDavisSSChitosan as a novel nasal delivery system for vaccinesAdv Drug Deliv Rev200151819611516781
- VinogradovSVKabanovAVNanosized cationic hyrogels for drug delivery: preparation, properties and interactions with cellsAdv Drug Deliv Rev20025413514711755709
- HsuSHHoTTTsengTCNanoparticle uptake and gene transfer efficiency for MSCs on chitosan and chitosan-hyaluronan substratesBiomaterials2012333639365022364729
- SakaOMBozkirAFormulation and in vitro characterization of PEGylated chitosan and polyethylene imine polymers with thrombospondin-I gene bearing pDNAJ Biomed Mater Res B Appl Biomater201210098499222279036
- JeanMAlamehMBuschmannMDMerzoukiAEffective and safe gene-based delivery of GLP-1 using chitosan/plasmid-DNA therapeutic nanocomplexes in an animal model of type 2 diabetesGene Ther20111880781621412280