Abstract
Background
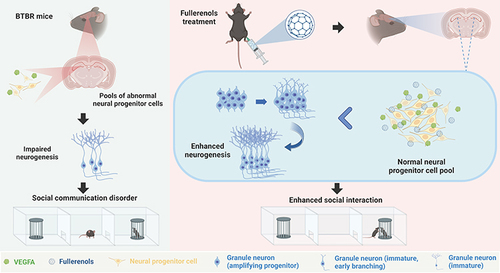
Autism Spectrum Disorder (ASD) is a neurodevelopmental condition that affects social interaction and communication and can cause stereotypic behavior. Fullerenols, a type of carbon nanomaterial known for its neuroprotective properties, have not yet been studied for their potential in treating ASD. We aimed to investigate its role in improving autistic behaviors in BTBR T+Itpr3tf/J (BTBR) mice and its underlying mechanism, which could provide reliable clues for future ASD treatments.
Methods
Our research involved treating C57BL/6J (C57) and BTBR mice with either 0.9% NaCl or fullerenols (10 mg/kg) daily for one week at seven weeks of age. We then conducted ASD-related behavioral tests in the eighth week and used RNA-seq to screen for vital pathways in the mouse hippocampus. Additionally, we used real-time quantitative PCR (RT-qPCR) to verify related pathway genes and evaluated the number of stem cells in the hippocampal dentate gyrus (DG) by Immunofluorescence staining.
Results
Our findings revealed that fullerenols treatment significantly improved the related ASD-like behaviors of BTBR mice, manifested by enhanced social ability and improved cognitive deficits. Immunofluorescence results showed that fullerenols treatment increased the number of DCX+ and SOX2+/GFAP+ cells in the DG region of BTBR mice, indicating an expanded neural progenitor cell (NPC) pool of BTBR mice. RNA-seq analysis of the mouse hippocampus showed that VEGFA was involved in the rescued hippocampal neurogenesis by fullerenols treatment.
Conclusion
In conclusion, our findings suggest that fullerenols treatment improves ASD-like behavior in BTBR mice by upregulating VEGFA, making nanoparticle- fullerenols a promising drug for ASD treatment.
Introduction
Autism Spectrum Disorder (ASD) is a complex and diverse neurodevelopmental condition strongly influenced by genetics and with a significant heritable component.Citation1,Citation2 It is characterized by persistent difficulties in social interaction and communication, as well as repetitive behaviors or restricted interests.Citation3–5 These challenges can vary significantly in severity and presentation among individuals with ASD, making it a complex condition that poses significant challenges for medical professionals and society at large. Although behavioral therapies have been commonly employed to address symptoms and enhance adaptive skills, there is an immediate demand for interventions that specifically address the fundamental features of ASD.
The hippocampus plays a crucial role in several cognitive functions, such as social interaction, learning, memory, and spatial reasoning.Citation6,Citation7 Studies indicate that individuals with ASD exhibit structural and functional differences in their hippocampus when compared to typically developing individuals, including alterations in volume, connectivity, and activation patterns.Citation8–11 These variations may impact social behavior, a key characteristic of ASD, as decreased hippocampal neurogenesis has been linked to deficits in social interaction, cognition, and recognition of social cues. Additionally, cognitive inflexibility, a common trait in individuals with ASD, has been associated with hippocampal neurogenesis.Citation12,Citation13 Our previous studies have shown that enhancing hippocampal neurogenesis can improve social behavior and decrease ASD-like behaviors in autistic animal models.Citation14 It indicates a causal relationship between neurogenesis and social deficits.
Fullerenols are a type of carbon nanomaterial that is derived from fullerene.Citation15,Citation16 They are produced through a hydroxylation reaction that introduces over 20 hydroxyl groups into the carbon cage.Citation17,Citation18 This process has several benefits, such as enhanced water solubility and biocompatibility.Citation19,Citation20 As a nanocomposite with promising applications, fullerenols are less harmful to living organisms and can cross the blood-brain barrier.Citation21,Citation22 Additionally, fullerenols have a range of abilities, including anti-human immunodeficiency virus, antioxidant, anti-inflammatory, and neuroprotective properties.Citation23–30 Recent studies have revealed that fullerenols can restore normal dopaminergic neuron levels in the brain and reduce α-synuclein aggregation, which suggests that it could be neuroprotective agent for Parkinson’s disease.Citation31 Fullerenols have been shown to block glutamate (Glu)-induced neurotoxicity and reduce intracellular free calcium levels in cases of Glu-induced neuronal damage.Citation32 Our studies have demonstrated that fullerenols can improve depression-like behavior induced by lipopolysaccharides injection in mice by restoring weakened neurogenesis in the hippocampus dentate gyrus (DG) region, suggesting its potential significance in promoting neurogenesis.Citation33 It is worth noting that the molecular mechanisms of fullerenols’s role in autism have not yet been fully understood.
BTBR T+Itpr3tf/J (BTBR) mice are commonly used as an animal model for researching idiopathic ASD. These mice exhibit characteristics that resemble some of the core features of ASD in humans, such as impaired social skills, increased repetitive behaviors, and cognitive deficits.Citation34,Citation35 Due to these features, BTBR mice have been widely utilized in preclinical research to study the underlying neurobiological mechanisms of autism and to evaluate potential therapeutic interventions. Previous studies have shown impaired hippocampus neurogenesis in BTBR mice.Citation36–38 Here, we demonstrated that fullerenols treatment can significantly alleviate ASD-like behaviors in BTBR mice. Our results also revealed that fullerenols treatment prevented the exhaustion of the neural progenitor cell (NPC) pool, effectively boosting hippocampus neurogenesis in BTBR mice. These findings suggest that fullerenols treatment may represent a promising pharmacological agent for ASD treatment.
Materials and Methods
Synthesis and Characterization of Fullerenols
Following our previous report, fullerenols were synthesized and subsequently characterized.Citation39 C60 underwent pretreatment via 30-min manual grinding or 1-hour ball milling in an agate mill, with or without NaOH at C60: NaOH ratios of 10:1, 5:1, or 2:1. Then, it was milled with or without 30% H2O2 at molar ratios of 0.139:1 to 0.139:4, matching the pretreatment conditions. Lastly, water was added for hydrolysis, and residual H2O2 was removed using a platinum wire. A dynamic light scattering (DLS) particle size analyzer (Omni, Brookhaven) was utilized to investigate size distribution and dispersion stability. DLS test conditions were 25°C, pure water as the solvent, PH 7, and fullerenols concentration of 20 μg/mL. The molecular structure was also described using an X-ray photoelectron spectrometer (XPS, ESCALAB 250xi, ThermoFisher) and a Fourier transform infrared (FT-IR) spectrometer (iN10-iZ10, ThermoFisher). XPS test conditions: the X-ray source was Al Kα (1486.60eV), and the vacuum of the analysis chamber was 1×10−8 Pa. FT-IR test conditions: 1–2 mg sample and 200 mg pure KBr were ground evenly, placed in a mold, and pressed into a transparent sheet, which could be used for determination. The sample and KBr should be dried and ground to a particle size of less than 2 microns to avoid being affected by scattered light.
Animals
The mice used in this study were housed at the Army Medical University animal facility, including the BTBR from Jackson Laboratories (Bar Harbor, ME, USA) and the C57BL/6J (C57) provided by the Army Medical University. Only male mice that were seven weeks old and weighed between 20–25 g were used for this study, and they were provided with sufficient food and water while being raised on a 12–12 hours light-dark cycle. The laboratory maintained an ambient temperature of 21–23°C and a relative humidity of 50%-70%. All laboratory procedures adhered to the Army Medical University Animal Committee guidelines and followed the principles of laboratory animal care. Every effort was made to minimize the number of Animals used and their suffering. The approval number of the Laboratory Animal Ethics Committee for animal experiments is AMUWEC20230421.
Materials
The fullerenols used in this experiment were synthesized through the methods outlined by the Institute of High Energy Physics and the National Center for Nanoscience and Technology in Beijing, China.Citation39,Citation40 The Materials used, including 0.9% NaCl, Radioimmunoprecipitation assay buffer (RIPA), Bicinchoninic Acid (BCA) Kit, Bovine serum albumin (BSA), Skimmed milk powder, and mouse anti-β-actin were obtained from Shanghai Beyotime Biotechnology. Paraformaldehyde (PFA) was provided by Shanghai Sinopsin Chemical Reagents. TBS buffer powder was purchased from Wuhan Bode Biological Engineering. We utilized Noldus Software Ethovision 11.0, obtained from Beijing Noldus Information Technology. The behavioral equipment is purchased from Shenzhen RWD Life Science. TRIzol Reagent, RevertAid MM kit, and SYBR Green kit were provided by Thermo Fisher Scientific in the United States. Ethylene glycol and 4′,6-diamino-2-phenylindole (DAPI) fluorescent staining reagent were purchased from Sigma Corporation in the United States. Phosphate-buffered saline (PBS), Triton-X-100 solution, and Tween-20 were provided by Sangon Biotech (Shanghai). Immobilon Western HRP Substrate was purchased from Millipore in the United States. Rabbit anti-VEGFA was provided by Wuhan ABclonal Technology. Mouse anti-SOX2 was obtained from Abcam, UK. Rabbit anti-Doublecortin (DCX) was provided by Cell Signaling Technology, Inc., Danvers, MA, and Rabbit anti-GFAP was purchased in Dako, Japan. Cy3 or 488 was provided by Jackson ImmunoResearch, USA.
Drug Treatment
Fullerenols (FUL) were dissolved in 0.9% NaCl (SAL) and given to mice intraperitoneally at 10 mg/kg.Citation33 The mice were divided randomly into four groups, including C57-SAL, C57-FUL, BTBR-SAL, and BTBR-FUL, with 9 mice in each group. At the age of seven weeks, the mice were injected with either fullerenols or saline solution once a day for seven consecutive days.
Behavioral Assays
All behavioral tests were conducted at the same time of day. Mice were acclimatized in the laboratory for at least 30 minutes before the start of the experiment, with a 24-hour interval between experiments. All behavioral tests were recorded with a camera and analyzed using Etho Vision XT (Noldus Beijing Information Technology Co. Ltd) software.
Three-Chambered Social Approach
The study utilized established protocols to conduct a social approach test using a three-chamber system.Citation41 The mice were placed in transparent polycarbonate devices measuring 60 cm × 40 cm × 22 cm, with doors on two partition walls allowing free movement between the three chambers. The test consisted of two phases: an acclimatization phase and a socialization phase. During the acclimatization phase, the experimental mice were allowed to explore the three side chambers for 10 minutes after being placed in the central chamber. During the socialization phase, a novel mouse (S) and object (O) were introduced on each side. The experimental mouse was placed in the center chamber and allowed to explore all three chambers for 10 minutes. Ethovision 11.0 was used to track the mouse’s total chamber and sniffing time during each phase. To calculate the preference index, the difference in side chamber or sniffing time between the novel mouse and object was divided by the total time spent in both side chambers or sniffing (S-O/total time). After the experiment, the device was cleaned with 75% alcohol and water.
Male-Female Social Reciprocal Interactions
The experiment was conducted in a controlled environment, specifically a soundproof chamber measuring 25 cm × 25 cm × 35 cm, with dim lighting. Experimental mice were introduced to a C57 female mouse in or before estrus, who had not been previously subjected to behavioral testing. The mice were allowed to interact freely for five minutes, with the entire session being recorded via a video system. The C57 female mice were the same age as the experimental mice. The recorded videos were analyzed using Ethovision 11.0 software. We primarily compared changes in social time, which contained nose-to-nose, nose-to-body, nose-to-anogenital, and total social time. Following the experiment, the device was cleaned with 75% alcohol and water.
Self-Grooming Test
The self-grooming test can reflect the stereotyped behavior of mice. We mainly recorded the time to compare the whole grooming time. The mice were placed in separate clean cages of standard size and observed for 20 minutes. The first 10 minutes were considered an acclimatization period, while the subsequent 10 minutes were designated as the testing phase. Following the acclimatization phase, an experienced observer manually reviewed the video recordings of the testing phase without knowledge of the treatment status of each group of mice and calculated the average cumulative time taken to groom all body regions. After each experiment, the equipment was carefully washed with 75% alcohol and water.
Marble Burying Test
The marble burying test can reflect the stereotyped behavior of mice to a certain extent. The more buried glass marbles, the more pronounced the stereotyped behavior. The standard experimental cage, measuring 27 cm × 16.5 cm × 12.5 cm, was filled with corncob bedding material to a 5–6 cm depth. The squirrel cage was then gently moved to create a level surface. Next, twenty black glass marbles, each with a diameter of 1.5 cm, were carefully placed onto the cushion, forming a uniform 4×5 grid pattern. The mice were placed in a laboratory cage under dim light conditions of around 15 lx and allowed to explore freely for 30 minutes. After this time, any marbles with over 75% of their volume buried were recorded. Finally, the device was cleaned using 75% alcohol and water.
Open Field Test
An open-field test was conducted using a standard grey plexiglass device measuring 40 cm × 40 cm × 30 cm to evaluate mice’s motor capacity and anxiety levels. The mice were placed in the central area of the field, and their movements were recorded for 30 minutes with a video camera. Ethovision 11.0 software was utilized to analyze the center time and the total distance traveled by the mice during the 30 minutes. We mainly recorded the movement distance and center time. The movement distance mainly reflected the path movement ability, and the less center time reflected the more anxiety of the mice. Following the experiment, the device was thoroughly cleaned with 75% alcohol and water.
Novel Object Recognition Test
The novel object recognition test is a method used to evaluate the short-term memory of mice. The test was carried out in the standard equipment (40 cm × 40 cm × 30 cm) made of grey plexiglass as described above. The test consisted of two phases: an exploratory phase and a testing phase. During the exploration phase, two identical objects (A) of the same shape, color, size, and texture were placed on opposite sides of the diagonal at the bottom of the device. The mice were allowed to explore the arena for 10 minutes while their activity was recorded. After 2 hours, the second phase began to test the short-term memory ability of the mice. One of the identical objects (A) was replaced with a novel object (B), which was similar in size but differed in shape, color, and texture. The mice were then allowed to explore the arena freely for 10 minutes while their behavior was recorded. To calculate the discriminating index (DI), the formula DI = (tB/(tA + tB)) × 100% was used. After the experiment, the device was thoroughly cleaned with 75% alcohol and water.
Y-Maze Test
The Y-maze test is used to measure spatial working memory ability. The maze has three arms and a central area of 40 cm × 9 cm × 16 cm. To begin the experiment, a mouse was placed in the center area and allowed to explore the maze freely for 8 minutes while being tracked by Ethovision 11.0. The mouse was considered to have entered an arm when its front and rear paws were in the same arm simultaneously. Correct spontaneous alternation performance (SAPs) was a triplet alternation with three overlapping components. The total number of entries and SAPs were analyzed and calculated by Ethovision 11.0 software, and the alternation rate was calculated as [SAPs/(N-2)] × 100%. After the experiment, the device was cleaned with 75% alcohol and water.
Real-Time Quantitative PCR (RT-qPCR)
Mice were anesthetized using 1% sodium pentobarbital solution, whole brains were removed, and hippocampal tissue was isolated for subsequent experiments. RNA was extracted from mouse hippocampal samples using the Trizol method. The samples were lysed in TRIzol Reagent (Invitrogen et al 92008, USA) and ground at 4°C. They were then supplemented with 100 μL chloroform and shaken vigorously for 15 seconds. The mixture was centrifuged at 4°C (13,900 rcf, 15 minutes), the solution was taken, and an equal volume of isopropyl alcohol was added. Centrifuge at 4°C and 13,900 rcf for 10 minutes, discard the supernatant to retain the precipitation, and add 1 mL of 75% ethanol to the resulting rainfall to wash the pellet. After centrifugation at 13,900 rcf for 10 minutes at 4°C, the supernatant was discarded, and 30 μL of RNase-free water was added to the resulting precipitation to complete the RNA extraction. RNA concentration was quantified by NanoDrop spectrophotometer. Total RNA was extracted by reverse transcription using a RevertAid MM kit (Thermoscientific, M1631, USA) according to the manufacturer’s instructions. The SYBR Green kit (Applied Biosystems, A57156) was used for RT-qPCR, and all primer sequences are shown in Supplementary Table 1. The relative mRNA content in mouse hippocampal tissues was calculated by 2− ΔΔCt.
Western Blotting
Hippocampal samples were isolated and homogenized using RIPA (Beyotime, Shanghai, China), and protein concentration was determined using the BCA Kit (Beyotime Biotechnology, Shanghai, China) after centrifugation at 4°C at 13,900 rcf for 30 minutes. The protein samples were subjected to electrophoresis on a 10% SDS-PAGE for 30 minutes at 80 V, followed by 90 minutes at 120 V. The proteins were transferred to a polyvinylidene fluoride membrane at a constant flow of 220 mA for 120 minutes. The membrane was then blocked with 5% skim milk powder at room temperature for 2 hours before being incubated with primary antibodies against rabbit anti-VEGFA (1:1000, ABclonal, Cat.NO: A5708), DCX (1:1000, CST, Cat.4604S), and mouse anti-β-actin (1:10,000, Beyotime, AF0003) at 4°C overnight. After being washed with tris-buffered saline with tween-20, the membrane was incubated with a secondary antibody conjugated to horseradish peroxidase for 2 hours at room temperature. Specific protein bands were visualized on the membrane using the enhanced chemiluminescence method, and the grayscale values of the immune response bands were calculated with Image J (National Institutes of Health, USA), following the manufacturer’s instructions.
Immunofluorescence
According to our previous report, mice were subjected to deep anesthesia with a dose of isoflurane and then transcardiacly infused with 0.01 M PBS (pH 7.4) and 4% PFA in 0.1 M PBS for 15–20 minutes.Citation42 Whole brains were stripped and fixed with 4% PFA for 48 hours at 4°C, followed by dehydration with 4% PFA 30% sucrose solution. Brains were serially cut on an RWD FS800 cryomicrotome to collect coronal brain slices (20 μm) and stored in a cryoprotectants solution (30% ethylene glycol, 30% sucrose dissolved in 0.01 M PBS) at −20°C. The slices were rinsed in 0.01 M PBS to remove the cryoprotectant solution from the sections. It was then incubated in 0.3% Triton X-100 for 30 minutes at 37°C, followed by a blocking buffer (3% BSA and 0.3% Triton X-100 in PBS) for 1 hour at 37°C. Sections were incubated with rabbit anti-DCX (1:500, Cell Signaling Technology, MA, USA), murine anti-SOX2 (1:500, Abcam, USA), and rabbit anti-GFAP (1:500, DAkO, Japan) for 2 hours at 37°C and then transferred to 4°C overnight. After washing with PBS, secondary antibodies conjugated to Cy3 or 488 (1:500, Jackson ImmunoResearch, USA) were incubated at 37°C for 2 hours under low-light conditions. After rinsing with PBS, the sections were immersed in DAPI (Sigma) for counterstaining and, finally, observed and photographed under a Zeiss confocal microscope (LSM880, Oberkochen, Germany).
RNA-Seq Analyses
RNA was extracted from the mouse hippocampus using Trizol Reagent (Invitrogen Life Technologies). After assessing the RNA quality, a sequencing library was created and sequenced on the NovaSeq 6000 platform (Illumina) by Shanghai Personal Biotechnology Cp. Ltd. Using the DESeq2 software package in R, we analyzed the differentially expressed genes (DEGs) between the four groups of C57-SAL, C57-FUL, BTBR-SAL, and BTBR-FUL. Significance was determined with P < 0.05. We utilized the R language Pheatmap software package to conduct a bi-directional clustering analysis of all the different genes of the samples. This provided us with a heatmap based on the expression level of the same gene in various models and the expression patterns of other genes in the same piece. The Euclidean method was used to calculate the distance, and the Complete Linkage method was used to cluster. The R language clusterProfiler software package enriched the gene ontology (GO) pathways with a P value < 0.05. The gene list of the chosen GO pathway was then analyzed using the STRING tool to map the protein-protein interaction (PPI) networks. Finally, we screened the HUB genes of the selected pathways with the cytoHubba plug-in of Cytoscape software. All the raw sequences were deposited in the NCBI Sequence Read Archive (SRA) under the accession number PRJNA1063445.
Statistical Analysis
The sample sizes used are similar to those typically used in the field. For the three-chamber social approach, the paired t-test was used for the chamber and sniffing time comparison. Two-way ANOVA and Tukey post-hoc analysis comparisons were employed to compare multiple groups. All data were analyzed using GraphPad Prism (version 9.5.0, USA) software and presented as mean ± standard error of the mean (SEM). P < 0.05 was considered statistically significant.
Results
Characterization of Fullerenols
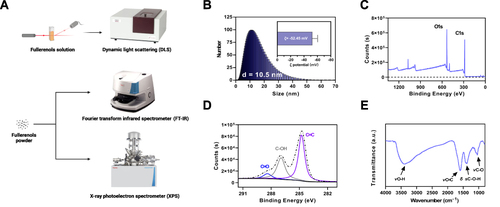
In our previous work, we successfully synthesized fullerenols using a straightforward method involving mechanical chemistry and a catalyst assistant. Our study analyzed the size distribution and dispersion stability of the prepared fullerenols (). Based on our hydrodynamic size distribution analysis, the fullerenols’ diameter was mainly 10.5 nm, and the ζ potential (−52.45 mV) revealed that the fullerenols were stably dispersed (). Further characterization of the molecular structure of the fullerenols was carried out using an XPS and FT-IR. Our XPS spectrum Results indicated that the fullerenols primarily contain carbon and oxygen elements (). The C1s spectra and curve-fitting results corresponded to non-oxidized carbons (C=C, 284.76 eV), monovalent oxidized carbons (C-OH, 286.96 eV), and a few divalent oxidized carbons (C=O, 288.39 eV) (). Meanwhile, the FT-IR spectrum of the as-prepared fullerenols revealed four distinct absorption peaks, including broad O-H stretching vibration (νO-H, 3550–3350 cm−1), C=C stretching vibration (νC=C, 1590 cm−1), O-H in-plane deformation vibration (δ sC-O-H, 1390 cm−1), and C-O stretching vibration (νC-O, 1050 cm−1) (). Our XPS and FT-IR results confirmed the successful Introduction of hydroxyl groups into carbon cages to form hydroxylated fullerenols. Our characterization results indicate that fullerenols possess a delocalized π-conjugated structure, making them highly efficient and effective at scavenging free radicals across a broad spectrum.
Figure 1 Characterization of fullerenols. (A) Schematic diagram of the fullerenols characterization and detection process. (B) Hydrodynamic size distribution and ζ potential of fullerenols. (C) XPS is scanning the spectrum of fullerenols. (D) XPS C1s are scanning the spectrum of fullerenols. (E) FT-IR spectrum of fullerenols.
Fullerenols Treatment Ameliorates Social Deficits in Adult BTBR Mice
Utilizing fullerenols proved effective in ameliorating the social deficits observed in BTBR mice during the three-chambered social approach test, although it did not impact repetitive behavior. This test was conducted to evaluate the social functioning in mice (), and it was discovered that C57 mice displayed a favorable reaction to novel mice, spending more time in their chamber (; Paired t-test: t(8) = 3.669, P = 0.0063) and sniffing them (; Paired t-test: t(8) = 3.526, P = 0.0078). In contrast, BTBR mice exhibited an inherent impairment in social capacity, spending less time in the chamber with novel mice (; Paired t-test: t(8) = 2.265, P = 0.0533) and sniffing them less (; Paired t-test: t(8) = 1.580, P = 0.1528). It is worth noting that the administration of fullerenols did not affect the social ability of C57 mice ( and ; B: Paired t-test: t(8) = 2.878, P = 0.0206; C: Paired t-test: t(8) = 2.995, P = 0.0172). Nevertheless, it significantly increased the chamber (; Paired t-test: t(8) = 2.765, P = 0.0245) and sniffing time (; Paired t-test: t(8) = 2.808, P = 0.0229) of BTBR mice.
Figure 2 Fullerenols treatment reversed social deficits in BTBR mice but did not improve repetitive behavior. (A) Representative heat maps showing the total time and position of C57 and BTBR mice in a 10-minute three-chamber social test. The redder the hue, the longer the mouse spends exploring it. “S” and “O” represent the novel mouse and object, respectively. (B) In the three-chamber test, fullerenols or SAL did not affect the sociability of C57 mice, which stayed longer in the side chamber of the novel mice. BTBR mice showed low interest in the lateral chambers of novel mice, while fullerenols treatment significantly improved the social deficits of BTBR mice. (C) In the three-chamber test, SAL and fullerenols-treated C57 mice spent more time sniffing novel mice, while the time spent sniffing novel mice of BTBR mice showed a decreasing trend. Notably, fullerenols treatment increased the time for BTBR mice to sniff novel mice. (D and E) The preference index from chamber time (D) and sniffing time (E) of BTBR mice was significantly restored to the C57 mice after fullerenols treatment. (F and G) The time of self-grooming (F) and the number of buried marbles (G) in BTBR mice were more than those in C57 mice, and fullerenols treatment did not change the self-grooming time and the number of marbles buried in BTBR mice. Data are presented as mean ± SEM. N = 9. *P < 0.05, **P < 0.01, ***P < 0.001, ****P< 0.0001.
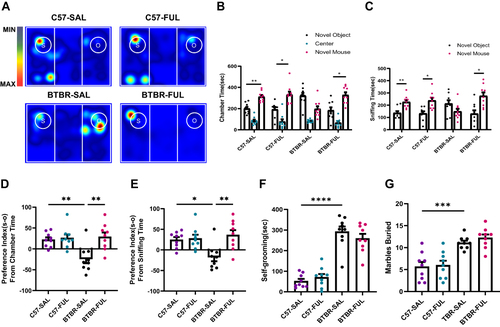
To assess social competence more accurately, we utilized two more precise indicators including the preference index from chamber time and the preference index from sniffing time. Our analysis using a two-way ANOVA revealed significant genotype and drug effects on the preference index from chamber time (; F (1, 32) = 5.700, P = 0.0230 and F (1, 32) = 10.04, P = 0.0034, respectively) as well as genotype × drug interaction effect (; F (1, 32) = 7.278, P = 0.0110). Additionally, we found a significant drug effect in the preference index analysis based on sniffing time (; F (1, 32) = 8.911, P = 0.0054) and genotype × drug interaction effect (; F (1, 32) = 6.991, P = 0.0126), but no significant genotype effect (; F (1, 32) = 3.102, P = 0.0878). Notably, during the adaptation phase of the test, the mice did not exhibit a clear preference for either chamber. These findings suggest that BTBR mice with social deficits can benefit from fullerenols treatment.
Stereotypes and repetitive behaviors are also reflected in the BTBR of autistic model mice as one of the core symptoms of ASD. Self-grooming and marble-burying tests examined repetitive and stereotyped behavior in mice. Through two-way ANOVA, we found that only the differences between the genotype were detected in the self-grooming and marble burying tests ( and ; F: F (1, 32) =120.9, P < 0.0001; G: F (1, 32) = 45.70; P < 0.0001), without the drug effect ( and ; F: F (1, 32) = 0.1456, P = 0.7053; G: F (1, 32) = 0.7005; P = 0.4088) and genotype × drug interaction effect ( and ; F: F (1, 32) = 1.761, P = 0.1939; G: F (1, 32) = 0.2031; P = 0.6553). Consistent with previous studies, the self-grooming time and the number of buried marbles in BTBR mice were significantly higher than in C57 mice.Citation43 In these trials, no adverse effects were found on C57 mice after treatment with fullerenols. Overall, the above data showed that fullerenols treatment significantly increased the social interaction ability of BTBR mice without altering repetitive and stereotyped behavior.
Fullerenols Treatment Increased the Ability of Male-Female Reciprocal Social in BTBR Mice
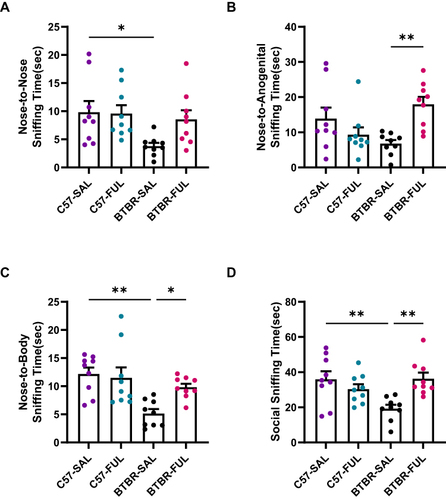
Our study utilized the male-female reciprocal social method to evaluate direct sniffing behavior. The results of our two-way ANOVA analysis revealed a significant genotype effect on nose-to-nose sniffing time (; F (1, 32) = 5.578, P = 0.0244) but no significant drug effect (; F (1, 32) = 2.285, P = 0.1404) or genotype × drug interaction effect (; F (1, 32) = 2.801, P = 0.1039). We also observed a significant genotype × drug interaction effect in nose-to-anogenital sniffing time (; F (1, 32) = 12.86, P = 0.0011), but no significant genotype effect (; F (1, 32) = 0.1187, P = 0.7327) or drug effect (; F (1, 32) = 2.211, P = 0.1468). Our analysis of nose-to-body and total social sniffing time revealed a significant genotype effect on nose-to-body sniffing time (; F (1, 32) = 13.20, P = 0.0010) and genotype × drug interaction effect (; F (1, 32) = 4.966, P = 0.0330), while the drug effect was not significant (; F (1, 32) = 2.766, P = 0.1060). Similarly, we found a significant genotype × drug interaction effect in total social sniffing time (; F (1, 32) = 11.52, P = 0.0019) but no significant genotype effect (; F (1, 32) = 2.595, P = 0.1170) or drug effect (; F (1, 32) = 2.995, P = 0.0932). Additionally, post-hoc tests showed that C57 mice exhibited significantly higher levels of nose-to-nose (; P < 0.05), nose-to-body (; P < 0.01), and total social sniffing time (; P < 0.01). At the same time, compared with the BTBR group, the BTBR mice treated with fullerenols had improved nose-to-anogenital (; P < 0.01), nose-to-body (; P < 0.05), and total social sniffing time (; P < 0.01). Notably, the male-female reciprocal sociability of fullerenols-treated C57 mice was not affected.
Figure 3 Fullerenols can ameliorate the deficits of male-female reciprocal social interaction in BTBR mice. (A–D) The nose-to-nose (A), nose-to-body (C), and social sniffing time (D) of BTBR mice were significantly lower than those of C57 mice but did not include nose-to-anogenital time (B). After fullerenols treatment, the nose-to-nose sniffing time of BTBR mice showed an increasing trend (A). In contrast, the nose-to-anogenital (B), nose-to-body (C), and social sniffing time (D) were markedly increased. Data are presented as mean ± SEM. N = 9. *P < 0.05, **P < 0.01.
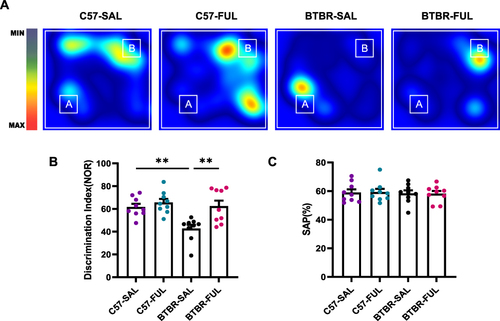
Fullerenols Treatment Rescued Short-Term Memory Deficits in BTBR Mice, but Did Not Alter the Performance of the Y-Maze Test
A novel object recognition task was employed to evaluate the short-term memory abilities of mice (). The results of the two-way ANOVA revealed a significant genotype effect among the four groups of mice (; F (1, 32) = 9.057, P = 0.0051), drug effect (; F (1, 32) = 10.26, P = 0.0031), and genotype × drug interaction effect (; F (1, 32) = 4.556, P = 0.0406). Further analysis demonstrated that the BTBR mice had a significantly lower discrimination index than the C57 mice (; P < 0.01). However, treatment with fullerenols led to a significant increase in the discrimination index of BTBR mice (; P < 0.01), indicating that fullerenols may be an effective treatment for improving impaired short-term memory in BTBR mice. The Y-maze test was also used to assess working memory capacity, and the results demonstrated that neither genotype nor fullerenols treatment had a significant effect (; F (1, 32) = 0.1673, P = 0.6853; F (1, 32) = 0.001015, P = 0.9748, respectively), and had no genotype × drug interaction effect (; F (1, 32) = 0.01590, P = 0.9005). This suggests that genotype or fullerenols treatment did not impact working memory abilities.
Figure 4 Fullerenols improved short-term memory deficits in BTBR mice without affecting working memory. (A) Representative heat maps show the time C57 and BTBR mice spent exploring objects in a novel object recognition task. “A” and “B” represent familiar and novel objects. (B) The recognition index of BTBR mice was lower than that of C57 mice, which was reversed by fullerenols treatment. (C) There was no significant difference in SAPs in the Y-maze test among the four groups. Data are presented as mean ± SEM. N = 9. **P < 0.01.
Fullerenols Treatment Did Not Affect Locomotor or Anxiety Behavior in BTBR Mice
We conducted an open-field test on mice to avoid confusion regarding sociability and repetitive tests (). Notably, we observed a significant decrease in the distance traveled by the mice per 5-minute segment over time (; F (5, 160) = 41.098, P < 0.001). This suggests that the mice gradually became more familiar with the open arena. Interestingly, the drug effect (; F (1, 32) = 0.5464, P = 0.4652) and genotype × drug interaction effect (; F (1, 32) = 0.1615, P = 0.6904) did not have a significant impact on the total travel distance in 30 minutes, according to the two-way ANOVA. However, we did find that genotype played a crucial role in the full length traveled at 30 minutes (; F (1, 32) = 5.525, P = 0.0251) and the 0–5 minutes period (; F (1, 32) = 60.86, P < 0.0001). Specifically, BTBR mice traveled longer distances in 0–5 minutes (; P < 0.0001) compared to C57 mice, indicating that BTBR mice exhibited enhanced locomotor activity within the first 5 minutes before the start of the open-air experiment, which is consistent with previous reports.Citation44
Figure 5 Fullerenols treatment did not alter the exploratory locomotion and anxiety level of C57 and BTBR mice in the open-field test. (A) Representative trajectory diagrams of four groups of mice in the open-field test within 30 minutes. (B) Total distance traveled in 30 minutes. (C) The distance traveled by the segment per 5 minutes. (D) BTBR mice had less center time in the open-field, and fullerenols treatment did not affect the total 30 minutes of center time spent by C57 and BTBR mice. (E) The 30-minute central time was divided into six 5-minute segments. Data are expressed as mean ± SEM. N = 9. *P < 0.05, **P < 0.01, ****P < 0.0001.
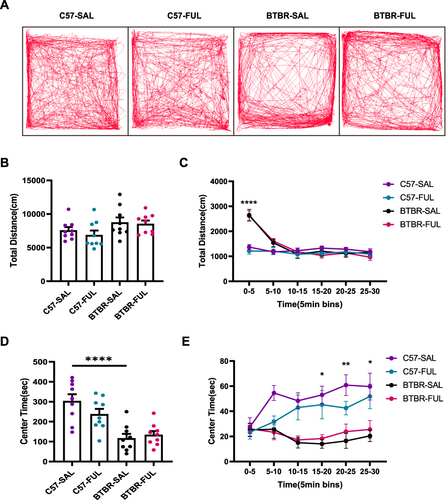
We measured the time spent in the central region to assess anxiety in the mice. After acclimatization, we found that the higher the anxiety level of the mice, the more likely they were to prefer the peripheral area of the open-field arena rather than the central area. Two-way ANOVA showed no drug effect (; F (1, 32) = 0.8984, P = 0.3503) and genotype × drug interaction effect (; F (1, 32) = 2.635, P = 0.1143), but genotype had a significant effect on the center spent time within 30 minutes (; F (1, 32) = 31.68, P < 0.0001), and 15–20 minutes (; F (1, 32) = 22.36, P < 0.0001), 20–25 minutes (; F (1, 32) = 23.68, P < 0.0001) and 25–30 minutes (; F (1, 32) = 15.42, P = 0.0004). Compared with the C57 mice, BTBR mice were observed to be more effective at a total of 30 minutes (; P < 0.0001) and in 15–20 minutes (; P < 0.01), 20–25 minutes (; P < 0.001), and 25–30 minutes (; P < 0.05) showed a decrease in spent time in the central region, indicating that the anxiety level of BTBR mice increased in the last 15 minutes. After fullerenols treatment, the anxiety level of BTBR mice remained unchanged ( and ). This suggests that fullerenols-induced changes in social behavior in BTBR mice are independent of general changes in motor function or anxiety behavior.
Fullerenols Treatment Enhanced DG Neurogenesis in BTBR Mice
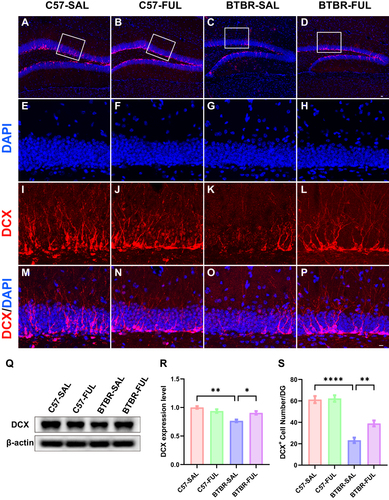
It has been previously established that defects in hippocampal neurogenesis correlate with autism-like behaviors.Citation13,Citation45,Citation46 In this study, we utilized a specific marker, DCX, associated with immature neurons to evaluate the impact of fullerenols on neurogenesis in the DG region (). DCX is a microtubule-associated protein that regulates development and is expressed in young migrating and differentiating neurons.Citation47,Citation48 It is commonly used as a marker for neurogenesis in the adult brain. Our study revealed that the number of DCX+ cells in the DG region was significantly affected by genotype factors (; F (1, 16) = 96.30, P < 0.0001) and fullerenols (; F (1, 16) = 10.04, P = 0.0060), with genotype × drug interaction effect being observed as well (; F (1, 16) = 5.376, P = 0.0340). DCX protein expression was confirmed through western blotting, and the analysis revealed significant genotype effects on DCX protein levels among the four groups (; F (1, 8) = 23.19, P = 0.0013) and genotype × drug interaction effect (; F (1, 8) = 13.36, P = 0.0065). Although the fullerenols effect was not significant (; F (1, 8) = 1.928, P = 0.2025), the analysis found the number of DCX+ cells in the DG region of BTBR mice (; P < 0.0001) and protein expression levels ( and ; P < 0.01) were significantly lower compared to C57 mice. However, this trend was reversed after fullerenols treatment (, P < 0.01; and , P < 0.05).
Figure 6 Fullerenols treatment increased the DCX+ cells in the DG and DCX protein levels in the hippocampus of BTBR mice. (A-D) Representative images of DCX-positive cells in the DG of four groups of mice. (E-P) Immunolabeled DCX (red), DAPI (blue), and DCX+/DAPI+ images at high magnification. (Q) Representative Western blotting of DCX in mouse hippocampus in each group. (R) Quantification of DCX protein level in the hippocampus. (S) Quantification of the number of DCX+ cells in the DG. The number of DCX+ cells in the DG and protein expression in the hippocampus of BTBR mice were significantly lower than those in C57 mice, which were rescued by fullerenols intervention. Data are presented as mean ± SEM. N = 3–5. The scale bar = 20 μm is shown in Figure D and applied to A-D. The scale bar = 10 μm is shown in Figure P and applied to E-P. *P < 0.05, **P < 0.01, ****P < 0.0001.
Fullerenols Treatment Rescues the Pool of NPC in the DG of BTBR Mice
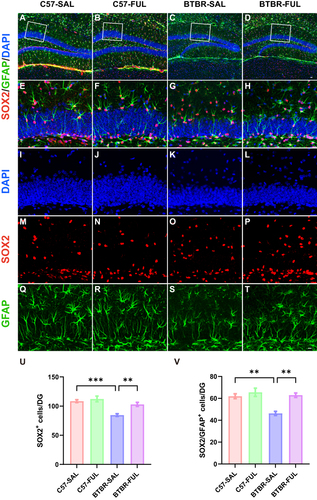
In a previous study, it was discovered that there was a significant decrease in DG neurogenesis in BTBR mice when compared to C57 mice. This decrease could be attributed to a reduced pool of NPC in the DG of BTBR mice. Therefore, we then evaluated the changes in the NPC pool in the DG region of BTBR mice (). NPCs are persistent in the hippocampus of the central nervous system and continue into adulthood.Citation49 It has shown that the transcription factor SOX2 is vital for maintaining stem cell proliferation and determining the fate of differentiation.Citation49–52 A deficiency in SOX2 can lead to neurodegeneration and negatively impact neurogenesis.Citation53 Through conducting a two-way ANOVA, the effects of genotype (; F (1, 16) = 24.31, P = 0.0002), drug (; F (1, 16) = 10.84, P = 0.0046), and genotype × drug interaction effect (; F (1, 16) = 4.742, P = 0.0447) on the number of SOX2+ cells were observed. The results showed that the number of SOX2+ cells in the DG of BTBR mice was notably lower than that of C57 mice (; P < 0.001), which aligns with the NPC pool in BTBR mice and previous reports (; P < 0.01).Citation14 However, there was positive news as SOX2+ (; P < 0.01) and SOX2+/GFAP+ radial glia cells (; P < 0.01) exhibited a significant increase in BTBR mice DG treated with fullerenols compared to the control group. This suggests that fullerenols treatment could effectively reverse the NPC pool capacity in BTBR mice DG.
Figure 7 Fullerenols treatment restored the NPC pool in the DG of the BTBR mouse. (A–D) Representative images of SOX2+/GFAP+ double-positive NPCs in the DG of four groups of mice. (E–H) High-magnification of the selected area. (I–T) Immunostaining of DAPI (blue), SOX2 (red), and GFAP (green). (U) Quantification of the number of SOX2+ cells in the DG. (V) Quantification of SOX2+/GFAP+ cells in the DG. Data are presented as mean ± SEM. N = 5. The scale bar = 20 μm is shown in Figure D and applied to (A–D). The scale bar = 10 μm is shown in Figure T and applied to (E–T). **P < 0.01, ***P < 0.001.
The Therapeutic Effects of Fullerenols via VEGFA Have Been Linked to Neurogenesis
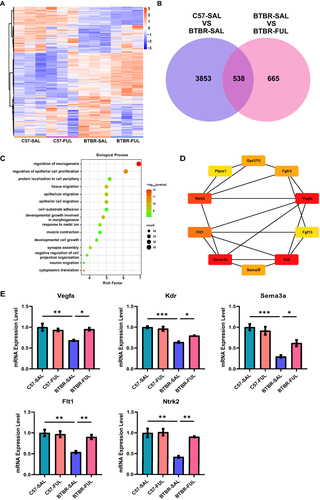
We thoroughly examined the genes and pathways involved to understand better how fullerenols improve social interaction in BTBR mice. Using high-throughput RNA sequencing, we could identify differences in mRNA expression in the hippocampus of four different groups of mice. Our analysis produced heat maps of gene expression profiles (), and we found 3853 DEGs in C57-SAL vs BTBR-SAL and 665 regulated DEGs in BTBR-SAL vs BTBR-FUL (). Of these, 538 DEGs overlapped (). We then conducted GO pathway enrichment on these genes. We found they were mainly related to neurogenesis, such as neurogenesis regulation, epithelial cell proliferation, and neuronal migration (). We found that regulating the neurogenesis pathway was particularly important, and we identified the top ten HUB genes (). We validated the top five HUB genes using RT-qPCR and found they were down-regulated in BTBR mice but restored after fullerenols supplementation ().
Figure 8 Fullerenols altered the hippocampus transcriptome in four groups of mice. (A) Hierarchical clustering gene expression heatmap of the four groups of mice. (B) Venn diagram for the treatment of co-regulated DEGs by fullerenols. (C) The top 15 GO pathways in BP enrichment. (D) HUB genes in neurogenesis regulatory pathways. (E) RT-qPCR verification of HUB genes in the regulatory pathways of neurogenesis (Vegfa: genotype effect: F (1, 8) = 8.740, P = 0.0182; drug effect: F (1, 8) = 4.033, P = 0.0795; genotype × drug interaction effect: F (1, 8) = 11.23, P = 0.0101. Sema3a: genotype effect: F (1, 12) = 44.98, P < 0.0001; drug effect: F (1, 12) = 2.494, P = 0.1403; genotype × drug interaction effect: F (1, 12) = 7.387, P = 0.0187. Kdr: genotype effect: F (1, 8) = 70.41, P < 0.0001; drug effect: F (1, 8) = 3.682, P = 0.0913; genotype × drug interaction effect: F (1, 8) = 9.470, P = 0.0152. Flt1: genotype effect: F (1, 12) = 17.61, P = 0.0012; drug effect: F (1, 12) = 7.021, P = 0.0212; genotype × drug interaction effect: F (1, 12) = 9.987, P = 0.0082. Ntrk2: genotype effect: F (1, 8) = 24.72, P = 0.0011; drug effect: F (1, 8) = 13.56, P = 0.0062; genotype × drug interaction effect: F (1, 8) = 11.56, P = 0.0094), the data are presented as mean ± SEM, N = 3–4, *P < 0.05, **P < 0.01, ***P < 0.001.
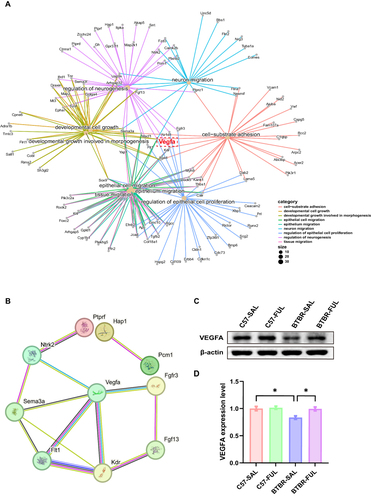
We presented our findings through a Gene-Concept Network, mapping the connections between genes and biological processes. Our analysis revealed that Vegfa is a crucial bridge gene that links neurogenesis-related biological processes (). By studying the PPI network of genes involved in neurogenesis, we identified that Vegfa interacts with seven genes, including Ntrk2, Sema3a, Flt1, Kdr, Fgf13, and Fgfr3 (). These results suggest that Vegfa could play a significant role in regulating neurogenesis. To further investigate, we conducted a protein expression level analysis of VEGFA in the hippocampus of four groups of mice (). Interestingly, our results showed genotype and drug effects among the four groups of mice (; F (1, 8) = 7.853, P = 0.0231 and F (1, 8) = 7.138, P = 0.0283, respectively). Although the genotype × drug interaction effect was not significant (; F (1, 8) = 4.776, P = 0.0603), we observed that BTBR mice had significantly lower VEGFA expression levels compared to the control group (; P < 0.05). However, after administration, VEGFA expression levels increased dramatically (; P < 0.05). Our RNA sequencing findings suggest that Vegfa plays a vital role in neurogenesis and the therapeutic effects of fullerenols on BTBR in a mouse model of autism.
Figure 9 The effects of fullerenols on bridge gene Vegfa in the hippocampus. (A) Gene-Concept Network of related biological processes of GO terms. (B) A PPI network was constructed using the STRING online tool to regulate neurogenesis terms. (C) Representative western blotting of VEGFA in DG region of mice in each group. (D) The density of critical protein VEGFA was quantified, N = 3, and the data are presented as mean ± SEM, *P < 0.05.
Discussion
Our study revealed that administering fullerenols can effectively improve ASD-like behavior in BTBR mice, such as enhancing social skills and cognitive abilities. Our research has also shown that fullerenols treatment promotes neurogenesis in the DG and expands the NPC pool in BTBR mice. Additionally, our RNA sequence analysis indicates that the Vegfa gene, which plays a crucial role in neurogenesis, significantly increases DG neurogenesis of BTBR mice following fullerenols supplementation. These results strongly support the therapeutic potential of fullerenols and present a novel approach to addressing ASD, where VEGFA-regulated neurogenesis may serve as a promising treatment option.
The findings indicate that administering fullerenols can offer potential benefits in addressing the behavioral symptoms of ASD in BTBR mice. These mice exhibit social impairment, communication disorders, and repetitive, stereotyped behavior with narrow interests, which align with the primary symptoms of ASD.Citation54–56 Fullerenols have been shown to improve cognitive impairment in BTBR mice, as evidenced by a strong preference for the novel object in the novel object test. Additionally, fullerenols have been shown to alleviate impaired sociability and male-female reciprocal social function. However, our study did not find any significant impact on stereotypical behavior in BTBR mice as measured by self-grooming and marble-burying tests. Additionally, we evaluated the effects of fullerenols treatment on exploration in BTBR mice using an open-field test, which showed no effect on locomotor activity. We observed no adverse effects in the C57 mice due to fullerenols supplementation.
Neurogenesis is a vital process that occurs during prenatal development and early postnatal life, and disruptions in this process can have significant and long-lasting effects on brain development and function.Citation57–61 Research has shown that individuals with ASD have abnormalities in neurogenesis, including changes in neural stem cell (NSC) proliferation, migration, and differentiation.Citation45,Citation62,Citation63 Dysmorphic manifestations of hippocampal neurogenesis are often observed in mouse models of ASD.Citation38,Citation64,Citation65 Our study examined neonatal neurons and NPC pool in BTBR mice with an autism model and confirmed a deficit in neurogenesis. However, we found that treatment with fullerenols restored impaired hippocampus neurogenesis in BTBR mice, as evidenced by increased DCX-positive neurons in the hippocampal DG. Additionally, fullerenols treatment expanded NPC populations in the DG of BTBR mice, as demonstrated by an increased number of SOX2+/GFAP+ cells. Our findings that suggest improving hippocampal neurogenesis through fullerenol treatment can improve autism-like behaviors.
According to our RNA-seq data analysis of the hippocampal region, fullerenols impact various biological processes. Our GO biological process analysis showed that the treatment of fullerenols affected the regulation of neurogenesis, epithelial cell proliferation, and neuronal migration. To illustrate this further, we constructed a gene concept network highlighting BP’s vital enrichment components and their associated genes. Our research revealed that VEGFA was the critical gene that bridged these essential integrals. After conducting a PPI analysis, we discovered that VEGFA interacted with seven genes, including Ntrk2, Sema3a, Flt1, Kdr, Fgf13, and Fgfr3. These findings suggest that VEGFA could be crucial in the neurogenesis regulatory pathway.
It has been reported that VEGFA is essential for neurogenesis through multiple channels. Firstly, VEGFA can directly boost the proliferation and survival of NSCs responsible for producing new neurons.Citation66 By binding to receptors on NSCs, VEGFA can promote proliferation and improve their survival rates, leading to a pool of NSC available for neurogenesis.Citation67–71 VEGFA can also influence the differentiation of NSCs into specific neuronal lineages.Citation67,Citation72 Studies have shown that VEGFA can promote the differentiation of NSCs into neurons, particularly in areas of the brain associated with learning and memory, such as the hippocampus.Citation73,Citation74 Moreover, VEGFA can modulate the migration of newly generated neurons. By promoting the growth and remodeling of blood vessels in the brain, VEGFA can create a supportive environment for neuron migration to their appropriate locations during brain development.Citation75–77 Western blotting confirmed decreased VEGFA levels in BTBR mice were restored after fullerenols supplementation. Interestingly, in a study on motor-mediated hippocampal neurogenesis, the authors found that blocking VEGF eliminated motor-induced neurogenesis.Citation78 It suggests that VEGFA may play a role in the regulation of neurogenesis and could be a potential target for fullerenols therapy.
Conclusion
According to the study, administering fullerenols to adult BTBR mice resulted in significant rescue in their social impairments and cognitive dysfunction. Additionally, fullerenols treatment was found to promote neurogenesis in the hippocampus of BTBR mice. Transcriptomic analysis of the treatment revealed increased NPCs and premature neurons through the up-regulation of Vegfa. These exciting new findings suggest that fullerenols may have potential applications in treating ASD and pave the way for further research in this area.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Disclosure
The authors report no potential conflicts of interest in this article.
Acknowledgments
Thanks to BioRender, our graphical abstract was created by BioRender.com, with permission.
Additional information
Funding
References
- Lord C, Elsabbagh M, Baird G, Veenstra-Vanderweele J. Autism spectrum disorder. Lancet. 2018;392(10146):508–520. doi:10.1016/s0140-6736(18)31129-2
- Arberas C, Ruggieri V. Autismo. Aspectos genéticos y biológicos [Autism. Genetic and biological aspects]. Medicina. 2019;79(Suppl 1):16–21. Spanish.
- Lord C, Brugha TS, Charman T, et al. Autism spectrum disorder. Nat Rev Dis Primers. 2020;6(1):5. doi:10.1038/s41572-019-0138-4
- Hannan AJ. Repeat DNA expands our understanding of autism spectrum disorder. Nature. 2021;589(7841):200–202. doi:10.1038/d41586-020-03658-7
- Lai MC, Lombardo MV, Baron-Cohen S. Autism. Lancet. 2014;383(9920):896–910. doi:10.1016/s0140-6736(13)61539-1
- Nicolson R, DeVito TJ, Vidal CN, et al. Detection and mapping of hippocampal abnormalities in autism. Psychiatry Res. 2006;148(1):11–21. doi:10.1016/j.pscychresns.2006.02.005
- DeLong GR. Autism, amnesia, hippocampus, and learning. Neurosci Biobehav Rev. 1992;16(1):63–70. doi:10.1016/s0149-7634(05)80052-1
- Banker SM, Gu X, Schiller D, Foss-Feig JH. Hippocampal contributions to social and cognitive deficits in autism spectrum disorder. Trends Neurosci. 2021;44(10):793–807. doi:10.1016/j.tins.2021.08.005
- Maier S, Tebartz van Elst L, Beier D, et al. Increased hippocampal volumes in adults with high functioning autism spectrum disorder and an IQ>100: a manual morphometric study. Psychiatry Res. 2015;234(1):152–155. doi:10.1016/j.pscychresns.2015.08.002
- Jacob Y, Morris LS, Verma G, Rutter SB, Balchandani P, Murrough JW. Altered hippocampus and amygdala subregion connectome hierarchy in major depressive disorder. Transl Psychiatr. 2022;12(1):209. doi:10.1038/s41398-022-01976-0
- Liu J, Okada NJ, Cummings KK, et al. Emerging atypicalities in functional connectivity of language-related networks in young infants at high familial risk for ASD. Dev Cogn Neurosci. 2020;45:100814. doi:10.1016/j.dcn.2020.100814
- Walgrave H, Balusu S, Snoeck S, et al. Restoring miR-132 expression rescues adult hippocampal neurogenesis and memory deficits in Alzheimer’s disease. Cell Stem Cell. 2021;28(10):1805–1821.e8. doi:10.1016/j.stem.2021.05.001
- Liu C, Liu J, Gong H, Liu T, Li X, Fan X. Implication of Hippocampal Neurogenesis in Autism Spectrum Disorder: pathogenesis and Therapeutic Implications. Curr Neuropharmacol. 2023;21(11):2266–2282. doi:10.2174/1570159x21666221220155455
- Zhong H, Xiao R, Ruan R, et al. Neonatal curcumin treatment restores hippocampal neurogenesis and improves autism-related behaviors in a mouse model of autism. Psychopharmacology. 2020;237(12):3539–3552. doi:10.1007/s00213-020-05634-5
- Kovel ES, Sachkova AS, Vnukova NG, Churilov GN, Knyazeva EM, Kudryasheva NS. Antioxidant activity and toxicity of fullerenols via bioluminescence signaling: role of oxygen substituents. Int J Mol Sci. 2019;20(9). doi:10.3390/ijms20092324
- Sachkova AS, Kovel ES, Churilov GN, et al. On mechanism of antioxidant effect of fullerenols. Biochem Biophys Rep. 2017;9:1–8. doi:10.1016/j.bbrep.2016.10.011
- Injac R, Prijatelj M, Strukelj B. Fullerenol nanoparticles: toxicity and antioxidant activity. Methods Mol Biol. 2013;1028:75–100. doi:10.1007/978-1-62703-475-3_5
- Liu Z, Zou Y, Zhang Q, Chen P, Liu Y, Qian Z. Distinct binding dynamics, sites and interactions of fullerene and fullerenols with amyloid-β peptides revealed by molecular dynamics simulations. Int J Mol Sci. 2019;20(8). doi:10.3390/ijms20082048
- Grebowski J, Kazmierska P, Krokosz A. Fullerenols as a new therapeutic approach in nanomedicine. Biomed Res Int. 2013;2013:751913. doi:10.1155/2013/751913
- Sun Y, Kakinen A, Zhang C, et al. Amphiphilic surface chemistry of fullerenols is necessary for inhibiting the amyloid aggregation of alpha-synuclein NACore. Nanoscale. 2019;11(24):11933–11945. doi:10.1039/c9nr02407g
- Zha Y, Jin Y, Wang X, Chen L, Zhang X, Wang M. Long-term maintenance of synaptic plasticity by Fullerenol Ameliorates lead-induced-impaired learning and memory in vivo. J Nanobiotechnol. 2022;20(1):348. doi:10.1186/s12951-022-01550-2
- Shi Q, Fang C, Zhang Z, Yan C, Zhang X. Visualization of the tissue distribution of fullerenols in zebrafish (Danio rerio) using imaging mass spectrometry. Anal Bioanal Chem. 2020;412(27):7649–7658. doi:10.1007/s00216-020-02902-3
- Xu L, Liu Y, Chen Z, et al. Morphologically virus-like fullerenol nanoparticles act as the dual-functional nanoadjuvant for HIV-1 vaccine. Adv Mater. 2013;25(41):5928–5936. doi:10.1002/adma.201300583
- Yin JJ, Lao F, Fu PP, et al. The scavenging of reactive oxygen species and the potential for cell protection by functionalized fullerene materials. Biomaterials. 2009;30(4):611–621. doi:10.1016/j.biomaterials.2008.09.061
- Tsai MC, Chen YH, Chiang LY. Polyhydroxylated C60, fullerenol, a novel free-radical trapper, prevented hydrogen peroxide- and cumene hydroperoxide-elicited changes in rat hippocampus in-vitro. J Pharm Pharmacol. 1997;49(4):438–445. doi:10.1111/j.2042-7158.1997.tb06821.x
- Zhou Y, Zhen M, Ma H, Li J, Shu C, Wang C. Inhalable gadofullerenol/[70] fullerenol as high-efficiency ROS scavengers for pulmonary fibrosis therapy. Nanomedicine. 2018;14(4):1361–1369. doi:10.1016/j.nano.2018.03.008
- Cao H, Zhang L, Qu Z, et al. The protective effect of hydroxylated fullerene pretreatment on pilocarpine-induced status epilepticus. Brain Res. 2021;1764:147468. doi:10.1016/j.brainres.2021.147468
- Grębowski J, Kaźmierska P, Krokosz A. Fulerenol - właściwości i zastosowanie w naukach biomedycznych [Fullerenol - properties and applications in biomedical sciences]. Postepy Hig Med Dosw. 2013;67:859–872. Polish. doi:10.5604/17322693.1063743
- Zhao Y, Shen X, Ma R, Hou Y, Qian Y, Fan C. Biological and biocompatible characteristics of fullerenols nanomaterials for tissue engineering. Histol Histopathol. 2021;36(7):725–731. doi:10.14670/hh-18-316
- Zha YY, Yang B, Tang ML, et al. Concentration-dependent effects of fullerenol on cultured hippocampal neuron viability. Int J Nanomed. 2012;7:3099–3109. doi:10.2147/ijn.S30934
- Golomidov I, Bolshakova O, Komissarov A, et al. The neuroprotective effect of fullerenols on a model of Parkinson’s disease in Drosophila melanogaster. Biochem Biophys Res Commun. 2020;523(2):446–451. doi:10.1016/j.bbrc.2019.12.075
- Jin H, Chen WQ, Tang XW, et al. Polyhydroxylated C(60), fullerenols, as glutamate receptor antagonists and neuroprotective agents. J Neurosci Res. 2000;62(4):600–607. doi:10.1002/1097-4547(20001115)62:4<600::Aid-jnr15>3.0.Co;2-f
- Kong A, Liu T, Deng S, et al. Novel antidepressant-like properties of the fullerenol in an LPS-induced depressive mouse model. Int Immunopharmacol. 2023;116:109792. doi:10.1016/j.intimp.2023.109792
- Ciernia AV, Link VM, Careaga M, LaSalle JM, Ashwood P. Genetic variants drive altered epigenetic regulation of endotoxin response in BTBR macrophages. Brain Behav Immun. 2020;89:20–31. doi:10.1016/j.bbi.2020.05.058
- Schwartzer JJ, Onore CE, Rose D, Ashwood P. C57BL/6J bone marrow transplant increases sociability in BTBR T(+) Itpr3(tf)/J mice. Brain Behav Immun. 2017;59:55–61. doi:10.1016/j.bbi.2016.05.019
- Abookasis D, Lerman D, Roth H, Tfilin M, Turgeman G. Optically derived metabolic and hemodynamic parameters predict hippocampal neurogenesis in the BTBR mouse model of autism. J Biophotonics. 2018;11(3). doi:10.1002/jbio.201600322
- Segal-Gavish H, Karvat G, Barak N, et al. Mesenchymal Stem Cell Transplantation Promotes Neurogenesis And Ameliorates Autism Related Behaviors in BTBR mice. Autism Res. 2016;9(1):17–32. doi:10.1002/aur.1530
- Stephenson DT, O’Neill SM, Narayan S, et al. Histopathologic characterization of the BTBR mouse model of autistic-like behavior reveals selective changes in neurodevelopmental proteins and adult hippocampal neurogenesis. Mol Autism. 2011;2(1):7. doi:10.1186/2040-2392-2-7
- Zhao M, Wang C, Xie J, Ji C, Gu Z. Eco-friendly and scalable synthesis of fullerenols with high free radical scavenging ability for skin radioprotection. Small. 2021;17(37):e2102035. doi:10.1002/smll.202102035
- Chen X, Yang J, Li M, et al. Fullerenol protects cornea from ultraviolet B exposure. Redox Biol. 2022;54:102360. doi:10.1016/j.redox.2022.102360
- Yang M, Silverman JL, Crawley JN. Automated three-chambered social approach task for mice. Curr Protoc Neurosci. 2011;56(1):8–26. doi:10.1002/0471142301.ns0826s56
- Zhang Q, Yang C, Liu T, et al. Citalopram restores short-term memory deficit and non-cognitive behaviors in APP/PS1 mice while halting the advance of Alzheimer’s disease-like pathology. Neuropharmacology. 2018;131:475–486. doi:10.1016/j.neuropharm.2017.12.021
- Liu J, Liu C, Gao Z, et al. GW4064 alters gut microbiota composition and counteracts autism-associated behaviors in BTBR T+tf/J mice. Front Cell Infect Microbiol. 2022;12:911259. doi:10.3389/fcimb.2022.911259
- Luo Y, Lv K, Du Z, et al. Minocycline improves autism-related behaviors by modulating microglia polarization in a mouse model of autism. Int Immunopharmacol. 2023;122:110594. doi:10.1016/j.intimp.2023.110594
- Wegiel J, Kuchna I, Nowicki K, et al. The neuropathology of autism: defects of neurogenesis and neuronal migration, and dysplastic changes. Acta Neuropathol. 2010;119(6):755–770. doi:10.1007/s00401-010-0655-4
- Zhang R, Cai Y, Xiao R, et al. Human amniotic epithelial cell transplantation promotes neurogenesis and ameliorates social deficits in BTBR mice. Stem Cell Res Ther. 2019;10(1):153. doi:10.1186/s13287-019-1267-0
- Francis F, Koulakoff A, Boucher D, et al. Doublecortin is a developmentally regulated, microtubule-associated protein expressed in migrating and differentiating neurons. Neuron. 1999;23(2):247–256. doi:10.1016/s0896-6273(00)80777-1
- Murata Y, Oka A, Iseki A, et al. Prolonged sleep deprivation decreases cell proliferation and immature newborn neurons in both dorsal and ventral hippocampus of male rats. Neurosci Res. 2018;131:45–51. doi:10.1016/j.neures.2017.08.008
- Graham V, Khudyakov J, Ellis P, Pevny L. SOX2 functions to maintain neural progenitor identity. Neuron. 2003;39(5):749–765. doi:10.1016/s0896-6273(03)00497-5
- Favaro R, Valotta M, Ferri AL, et al. Hippocampal development and neural stem cell maintenance require Sox2-dependent regulation of Shh. Nat Neurosci. 2009;12(10):1248–1256. doi:10.1038/nn.2397
- Toda T, Hsu JY, Linker SB, et al. Nup153 interacts with Sox2 to enable bimodal gene regulation and maintenance of neural progenitor cells. Cell Stem Cell. 2017;21(5):618–634.e7. doi:10.1016/j.stem.2017.08.012
- Avilion AA, Nicolis SK, Pevny LH, Perez L, Vivian N, Lovell-Badge R. Multipotent cell lineages in early mouse development depend on SOX2 function. Genes Dev. 2003;17(1):126–140. doi:10.1101/gad.224503
- Ferri AL, Cavallaro M, Braida D, et al. Sox2 deficiency causes neurodegeneration and impaired neurogenesis in the adult mouse brain. Development. 2004;131(15):3805–3819. doi:10.1242/dev.01204
- Vijaya Shankara J, Horsley KG, Cheng N, Rho JM, Antle MC. Circadian responses to light in the BTBR mouse. J Biol Rhythms. 2022;37(5):498–515. doi:10.1177/07487304221102279
- Mutovina A, Ayriyants K, Mezhlumyan E, et al. Unique features of the immune response in BTBR mice. Int J Mol Sci. 2022;23(24). doi:10.3390/ijms232415577
- Uddin MN, Yao Y, Manley K, Lawrence DA. Development, phenotypes of immune cells in BTBR T(+)Itpr3(tf)/J mice. Cell Immunol. 2020;358:104223. doi:10.1016/j.cellimm.2020.104223
- Gage FH. Mammalian neural stem cells. Science. 2000;287(5457):1433–1438. doi:10.1126/science.287.5457.1433
- Kriegstein A, Alvarez-Buylla A. The glial nature of embryonic and adult neural stem cells. Annu Rev Neurosci. 2009;32:149–184. doi:10.1146/annurev.neuro.051508.135600
- Villalba A, Götz M, Borrell V. The regulation of cortical neurogenesis. Curr Top Dev Biol. 2021;142:1–66. doi:10.1016/bs.ctdb.2020.10.003
- Wang C, Qin J, Jiao J, Ji F. Ccdc25 regulates neurogenesis during the brain development. Dev Neurobiol. 2023;83(3–4):91–103. doi:10.1002/dneu.22911
- Pombero A, Garcia-Lopez R, Estirado A, Martinez S. Vascular pattern of the dentate gyrus is regulated by neural progenitors. Brain Struct Funct. 2018;223(4):1971–1987. doi:10.1007/s00429-017-1603-z
- Courchesne E, Gazestani VH, Lewis NE. Prenatal origins of ASD: the when, what, and how of ASD development. Trends Neurosci. 2020;43(5):326–342. doi:10.1016/j.tins.2020.03.005
- Packer A. Neocortical neurogenesis and the etiology of autism spectrum disorder. Neurosci Biobehav Rev. 2016;64:185–195. doi:10.1016/j.neubiorev.2016.03.002
- Luhach K, Kulkarni GT, Singh VP, Sharma B. Vinpocetine amended prenatal valproic acid induced features of ASD possibly by altering markers of neuronal function, inflammation, and oxidative stress. Autism Res. 2021;14(11):2270–2286. doi:10.1002/aur.2597
- Cai Y, Zhong H, Li X, Xiao R, Wang L, Fan X. The liver x receptor agonist TO901317 ameliorates behavioral deficits in two mouse models of autism. Front Cell Neurosci. 2019;13:213. doi:10.3389/fncel.2019.00213
- Sun J, Zhou W, Ma D, Yang Y. Endothelial cells promote neural stem cell proliferation and differentiation associated with VEGF activated Notch and Pten signaling. Dev Dyn. 2010;239(9):2345–2353. doi:10.1002/dvdy.22377
- Sun S, Xu Y, Yu N, et al. Catalpol Alleviates Ischemic Stroke Through Promoting Angiogenesis and Facilitating Proliferation and Differentiation of Neural Stem Cells via the VEGF-A/KDR Pathway. Mol Neurobiol. 2023;60(11):6227–6247. doi:10.1007/s12035-023-03459-9
- Yang Y, Wei H, Zhou X, Zhang F, Wang C. Hyperbaric oxygen promotes neural stem cell proliferation by activating vascular endothelial growth factor/extracellular signal-regulated kinase signaling after traumatic brain injury. Neuroreport. 2017;28(18):1232–1238. doi:10.1097/wnr.0000000000000901
- Schänzer A, Wachs FP, Wilhelm D, et al. Direct stimulation of adult neural stem cells in vitro and neurogenesis in vivo by vascular endothelial growth factor. Brain Pathol. 2004;14(3):237–248. doi:10.1111/j.1750-3639.2004.tb00060.x
- Maurer MH, Tripps WK, Feldmann RE, Kuschinsky W. Expression of vascular endothelial growth factor and its receptors in rat neural stem cells. Neurosci Lett. 2003;344(3):165–168. doi:10.1016/s0304-3940(03)00407-5
- Jin K, Zhu Y, Sun Y, Mao XO, Xie L, Greenberg DA. Vascular endothelial growth factor (VEGF) stimulates neurogenesis in vitro and in vivo. Proc Natl Acad Sci U S A. 2002;99(18):11946–11950. doi:10.1073/pnas.182296499
- Sun J, Sha B, Zhou W, Yang Y. VEGF-mediated angiogenesis stimulates neural stem cell proliferation and differentiation in the premature brain. Biochem Biophys Res Commun. 2010;394(1):146–152. doi:10.1016/j.bbrc.2010.02.132
- During MJ, Cao L. VEGF, a mediator of the effect of experience on hippocampal neurogenesis. Curr Alzheimer Res. 2006;3(1):29–33. doi:10.2174/156720506775697133
- Licht T, Keshet E. Delineating multiple functions of VEGF-A in the adult brain. Cell Mol Life Sci. 2013;70(10):1727–1737. doi:10.1007/s00018-013-1280-x
- Barber M, Andrews WD, Memi F, et al. Vascular-derived vegfa promotes cortical interneuron migration and proximity to the vasculature in the developing forebrain. Cereb Cortex. 2018;28(7):2577–2593. doi:10.1093/cercor/bhy082
- Rosenstein JM, Krum JM, Ruhrberg C. VEGF in the nervous system. Organogenesis. 2010;6(2):107–114. doi:10.4161/org.6.2.11687
- Ruiz de Almodovar C, Coulon C, Salin PA, et al. Matrix-binding vascular endothelial growth factor (VEGF) isoforms guide granule cell migration in the cerebellum via VEGF receptor Flk1. J Neurosci. 2010;30(45):15052–15066. doi:10.1523/jneurosci.0477-10.2010
- Fabel K, Fabel K, Tam B, et al. VEGF is necessary for exercise-induced adult hippocampal neurogenesis. Eur J Neurosci. 2003;18(10):2803–2812. doi:10.1111/j.1460-9568.2003.03041.x