 ?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.
?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.Abstract
Considering the fact that iRGD (tumor-homing peptide) demonstrates tumor-targeting and tumor-penetrating activity, and that B16-F10 (murine melanoma) cells overexpress both αv integrin receptor and neuropilin-1 (NRP-1), the purpose of this study was to prepare a novel doxorubicin (DOX)-loaded, iRGD-modified, sterically-stabilized liposome (SSL) (iRGD-SSL-DOX) in order to evaluate its antitumor activity on B16-F10 melanoma cells in vitro and in vivo. The iRGD-SSL-DOX was prepared using a thin-film hydration method. The characteristics of iRGD-SSL-DOX were evaluated. The in vitro leakage of DOX from iRGD-SSL-DOX was tested. The in vitro tumor-targeting and tumor-penetrating characteristics of iRGD-modified liposomes on B16-F10 cells were investigated. The in vivo tumor-targeting and tumor-penetrating activities of iRGD-modified liposomes were performed in B16-F10 tumor-bearing nude mice. The antitumor effect of iRGD-SSL-DOX was evaluated in B16-F10 tumor-bearing C57BL/6 mice in vivo. The average particle size of the iRGD-SSL-DOX was found to be 91 nm with a polydispersity index (PDI) of 0.16. The entrapment efficiency of iRGD-SSL-DOX was 98.36%. The leakage of DOX from iRGD-SSL-DOX at the 24-hour time point was only 7.5%. The results obtained from the in vitro flow cytometry and confocal microscopy, as well as in vivo biodistribution and confocal immunofluorescence microscopy experiments, indicate that the tumor-targeting and tumor-penetrating activity of the iRGD-modified SSL was higher than that of unmodified SSL. In vivo antitumor activity results showed that the antitumor effect of iRGD-SSL-DOX against melanoma tumors was higher than that of SSL-DOX in B16-F10 tumor-bearing mice. In conclusion, the iRGD-SSL-DOX is a tumor-targeting and tumor-penetrating peptide modified liposome which has significant antitumor activity against melanoma tumors.
Introduction
Tumor-targeted drug delivery systems containing antitumor agents are believed to have opened a new era in traditional chemotherapy.Citation1–Citation3 Ligand-modified targeting drug delivery systems can bind with specific tumor surfaces’ overexpressing receptors to increase the drug concentration at the tumor site and significantly improve therapeutic efficacy.Citation4,Citation5 For example, Arg-Gly-Asp (RGD) – or Asn-Gly-Arg (NGR)-modified targeting drug delivery systems have been extensively investigated.Citation6–Citation10
Recently, a tumor-targeting and tumor-penetrating cyclic peptide, tumor-homing peptide iRGD (peptide with Cys-Arg-Gly-Asp-Lys/Arg-Gly-Pro-Asp/Glu-Cys [CRGDK/RGPD/EC]), has been reported to enhance vascularity and tissue permeability in a tumor-specific and neuropilin-1 (NRP-1)-dependent manner.Citation11 The mechanism for how iRGD homes to tumor sites has been explained in detail.Citation11,Citation12 Briefly, iRGD targets tumors by first binding to αv integrins and then being proteolytically cleaved in the tumor to produce CRGDK/R, which has an affinity for NRP-1, in order to trigger tissue penetration. Thus, when nanoparticles, nanogels, or anticancer agents are modified with iRGD, their tissue-penetrating and tumor-targeting properties can be improved.Citation11,Citation13–Citation17 Additionally, when iRGD was coadministrated with drugs or a systemic injection drug delivery system, antitumor activities were also improved.Citation12
In the past, liposomes have been used as drug carriers to improve the pharmacokinetics of drugs, resulting in reduced toxicities and enhanced therapeutic efficacies.Citation18 However, conventional liposomes have reported rapid uptake and accumulation by phagocytic cells of the mononuclear phagocyte system (also know as the reticuloendothelial system [RES]), after systemic administration.Citation19 PEG polyethylene glycol (PEGylation) modification prevents the recognition of conventional liposomes by opsonins and therefore reduces their clearance by cells of the RES.Citation20 Therefore, PEGylated liposomes are often referred to as sterically stabilized liposomes (SSL). Using PEGylated phospholipids, the apparent terminal half-life of a liposome can be extended.Citation21 In addition, SSL could spontaneously accumulate in solid tumors via the enhanced permeability and retention (EPR) effect, presenting passive targeting activity.Citation22,Citation23
Considering the fact that iRGD has a tumor-targeting and tumor-penetrating effect as well as the fact that some tumor cells overexpress both the αv intergrin receptor and NRP-1, in the present study we selected iRGD as a targeting ligand, SSL as a delivery system, and doxorubicin hydrochloride (DOX) as a model drug to prepare a DOX-loaded, iRGD-modified SSL (iRGD-SSL-DOX). Due to overexpression of αv integrin receptor and NRP-1,Citation24,Citation25 the B16-F10 cells were selected as the tumor cell model. The antitumor activity of iRGD-SSL-DOX on B16-F10 cells was investigated in vitro and in vivo.
Materials and methods
Materials
1,2-Distearoyl-sn-Glycero-3-Phosphoethanolamine-N-[Maleimide(polyethylene-Glycol)-2000] (DSPE-PEG-MAL) and 1,2-distearoyl-sn-glycero-3-Phosphoethanolamine-N [methoxy(polyethylene glycol)-2000] (DSPE-PEG) were purchased from Avanti Polar Lipids (Alabaster, AL, USA). Coumarin-6 and cholesterol were obtained from Sigma-Aldrich (St Louis, MO, USA). iRGD peptide (CRGDKGPDC) was synthesized by GL Bio-Chem Co, Ltd (Shanghai, People’s Republic of China). DOX was supplied by Zhejiang Hisun Pharmaceutical Co, Ltd (Taizhou Zhejiang, People’s Republic of China). Near infrared lipophilic carbocyanine dye 1,1′-dioctadecyltetramethyl indotricarbocyanine iodide (DiR) and the fluorescent probe, 1,1′-dioctadecyl-3,3,3′,3′-tetramethylindocarbocyanine perchlorate (DiI) were obtained from Biotium Inc (Hayward CA, USA). Hoechst 33258 and Fluorescein Isothiocyanate (FITC) goat anti-rabbit secondary antibodies (1:1000) were obtained from Molecular Probes Inc (Eugene, OR, USA). Rabbit polyclonal to CD31 (10 μg/mL) and rabbit polyclonal to fibrinogen (10 μg/mL) were purchased from Abcam Inc (Cambridge, Massachusetts, USA). Cell culture media, DMEM (Dulbecco’s Modified Eagle Medium, high glucose), RPMI 1640 (Roswell Park Memorial Institute 1640), penicillin-streptomycin, fetal bovine serum (FBS), and L-glutamine, were obtained from GIBCO Invitrogen Corp (Life Technologies, Carlsbad CA, USA). All other reagents were of analytical grade.
Cells
Murine B16-F10 cell line was obtained from Chinese Academy of Sciences Cells Bank (Shanghai, People’s Republic of China). The cells were grown in DMEM medium (high glucose) supplemented with 10% FBS and 1% glutamine penicillin-streptomycin at 37°C and 5% CO2. The human breast adenocarcinoma MCF-7 cell line was supplied by the Institute of Basic Medical Sciences (Chinese Academy of Medical Sciences, Beijing, People’s Republic of China) and cultivated according to the recommended instructions.
Animals
Female BALB (Bagg Albino)/c nude mice weighing 20–25 g (5–6 weeks old) and female C57BL/6 mice weighing 20–24 g (5–6 weeks old) were supplied by the Experimental Animal Center of Peking University Health Science Center (Beijing, People’s Republic of China). The temperature and relative humidity were maintained at 25°C and 45%–55%, respectively. All care and handling of the animals was performed in accordance with the requirements of the Institutional Authority for Laboratory Animal Care of Peking University.
Synthesis of iRGD-PEG-DSPE
iRGD-PEG-DSPE was synthesized from iRGD and DSPE-PEG-maleimide in a single step that coupled iRGD to DSPE-PEG using a method previously reported by our laboratory.Citation9,Citation10,Citation26 In brief, DSPE-PEG-maleimide mixed with iRGD at a 1:1 molar ratio (iRGD:DSPE-PEG-maleimide = 1:1) in Hepes (pH = 6.5). This reaction mixture was gently stirred at room temperature for 48 hours under nitrogen gas. After that, the resulting reaction mixture was placed in a dialysis bag (molecular weight cutoff = 3500 Da) and dialyzed in deionized water for 48 hours to remove the free iRGD. The final solution in the dialysis bag was lyophilized and stored at −20°C until used.
Preparation of iRGD-SSL-DOX
The iRGD-SSL-DOX were prepared by a thin-film hydration method. Briefly, egg phosphatidylcholine, cholesterol, DSPE-PEG, iRGD-PEG-DSPE (65:30:4:1 molar ratio) were dissolved in chloroform. The solvent was evaporated using an RE52 rotary evaporator (Shanghai Yarong Biochemistry Instrument Company, Shanghai, People’s Republic of China) in a round-bottomed flask at 45°C for about 40 minutes to form a solid film. Next, this film was flushed with nitrogen gas for 30 minutes and stored overnight in a desiccator to remove any traces of chloroform. A volume of 2 mL 300 mM citric acid buffer (pH = 4.0) was added in the flask and sonicated in a bath sonicator for 30 minutes to form liposomes. The resulting liposomes were then extruded eleven times through 100 nm polycarbonate membrane filters using a mini-extruder (Avanti Polar Lipids Inc, Alabaster, AL, USA). The obtained liposomes were then passed through a Sephadex G-50 (Sigma-Aldrich, St Louis, MO, USA) column to remove the external citric acid of the liposomes with a 20 mM Hepes buffer solution (HBS) containing 150 mM NaCl (HBS, pH = 7.4). DOX solution was added to the liposomes at a DOX/lipid (weight/weight) ratio of 1:20 to load into these blank liposomes using a pH-gradient method. The mixed solution was incubated for 1 hour at 37°C and then passed through a Sephadex G-50 column to remove the unentrapped DOX with HBS.
The unmodified SSL containing DOX (SSL-DOX) were prepared according to the above procedure except that the equivalent molar amount of iRGD-PEG-DSPE was replaced by DSPE-PEG.
The coumarin-6, DiR or DiI loaded SSL (iRGD-SSL-coumarin-6, and SSL-coumarin-6, iRGD-SSL-DiR and SSL-DiR iRGD-SSL-DiI and SSL-DiI) were also prepared by the thin-film hydration method.
Characterization of iRGD-SSL-DOX
For particle size analysis, the final concentration of DOX in SSL-DOX or iRGD-SSL-DOX was about 0.3 mg/mL. The particle size and zeta potential of iRGD-SSL-DOX were measured by using a Malvern Zetasizer Nano-ZS (Malvern Instruments Ltd, Malvern, Worcestershire, UK) at 25ºC. The liposomal encapsulation efficiency was determined as described below. Briefly, the final liposomes were passed through a Sephadex G-50 column to remove free DOX, followed by disruption with 10% Triton X-100 (volume to volume [v/v]); the DOX in the liposomes was then measured in a spectrofluorometer (RF-5301PC; Shimadzu Corp, Nakagyo-ku, Kyoto, Japan). In addition, the same amount of liposomes was treated as above, except they were passed through a Sephadex G-50 column to obtain the total concentration of DOX. The encapsulation efficiency was calculated by the following formula:
In vitro leakage of DOX from iRGD-SSL-DOX
The leakage of DOX from iRGD-SSL-DOX was tested using a dialysis method. In brief, iRGD-SSL-DOX (1.0 mL, 0.15 mg/mL) was placed in a dialysis bag (MWCO [Molecular Weight Cut-Off] 8,000–14,000). The dialysis bag was then immersed in 20 mL release medium (phosphate buffered saline [PBS] [pH = 7.4]) and incubated in an orbital shaker for 120 hours at 37°C. A collection of 1.0 mL samples was then taken out from the release medium at the predetermined time intervals, and a similar volume of fresh medium was added. The concentration of DOX was determined using a spectrofluorometer (RF-5301PC). The excitation and emission wavelengths were set at 494 nm and 591 nm, respectively. DOX release was measured for 120 hours. After the last sample in the medium was taken out at the 120-hour time point, solution in dialysis bag was mixed with the release medium; a volume of 10 μL of 10% (v/v) Triton X-100 was then added in the released medium. This mixture was determined as a positive control. The percentage DOX release was calculated as (It − I0)/(I100 − I0) × 100%, in which It is the fluorescence at time point t, I0 is the fluorescence at the time the dialysis bag immersed in the released medium, and I100 is the fluorescence of the sample after the addition of Triton X-100 at the 120-hour time point.
Flow cytometry
B16-F10 cells were seeded at a density of 3 × 105 cells/well in 6-well plates and incubated at 37°C for 24 hours. After 24 hours incubation, the medium was replaced with SSL-coumarin-6, iRGD-SSL-coumarin-6 or coumarin-6 solution (the final concentration of coumarin-6 was 150 ng/mL). The plates were divided in two groups (with or without trypsin treatment). For group 1 (without trypsin treatment group), after 2 hours incubation at 37°C, the cells were washed three times with PBS solution. For group 2 (with trypsin treatment group), SSL-coumarin-6, iRGD-SSL-coumarin-6 or coumarin-6 solution (the final concentration of coumarin-6 was 150 ng/mL) received 50 μL trypsin solution (250 μg/mL) and was incubated for 5 minutes at 37°C; the soybean inhibitor (50 μL, 30 mg/mL) was then added to stop the reaction. The above solutions were added and incubated with cells for 2 hours at 37°C. The cells were then washed three times with PBS. All cells in both groups were then harvested by trypsinization and centrifuged at 1,000 rpm for 5 minutes and resuspended in 500 μL PBS medium and tested using a FACScan (Becton, Dickinson and Company, Franklin Lakes, NJ, USA). The coumarin-6 in the cells was excited with an argon laser (467 nm) and fluorescence was detected at 502 nm.
For anther flow cytometry experiment, the MCF-7 cells were seeded at a density of 2 × 105 cells/well in 6-well plates and performed as described above in the B16-F10 cell line.
Confocal microscopy studies
Following incubation of B16-F10 cells (3 × 105 cells) on glass-bottomed dishes containing culture medium at 37°C for 24 hours, cell culture media containing SSL-coumarin-6, iRGD-SSL-coumarin-6 or coumarin-6 solution (final drug concentration 150 ng/mL) were added to each dish and incubated for another 2 hours at 37°C. After the medium was removed, the cells were washed with ice-cold PBS followed by fixing with 4% paraformaldehyde in PBS; the cells were then processed in a Hoechst 33258 stain for 20 minutes. The fluorescent images of the cells were inspected using a TCS SP2 confocal microscope (Leica Microsystems, Wetzlar, Germany).
In vivo biodistribution imaging
A volume of 0.1 ml of B16-F10 cells (1 × 106) suspension was administrated via subcutaneous injection into the right armpits of nude female BALB/c nude mice to prepare the B16-F10 tumor-bearing nude mice model. Once the tumor masses in the xenografts reached 200 mm3, a volume of 200 μL physiological saline, SSL-DiR or iRGD-SSL-DiR was intravenously (IV) administrated via the tail vein of the tumor-bearing nude mice at a dose of 1,200 ng/mouse.Citation10 At the predetermined time points, mice were anaesthetized by isoflurane (1.5%) and scanned using a Kodak In-Vivo Imaging System FX PRO (Carestream Health, Inc, Rochester, NY, USA) with an excitation bandpass filter at 730 nm and an emission at 790 nm. The observed time was set at 1 hour, 3 hours, 6 hours, 12 hours, and 24 hours after administration. The exposure time was set at 60 seconds per image. The fluorescent signal intensities in the tumor-bearing nude mice were analyzed using Carestream MI SE software (Carestream Health, Inc). For each near-infrared spectroscopy image, a corresponding X-ray image was taken to identify the anatomical location of the tumor.
Immunohistochemistry
The tumor-bearing nude mice, prepared as described above, received an IV administration of iRGD-SSL-DiI or SSL-DiI at a dose of 200 μg/kg via the tail vein when the tumor masses reached about 200 mm3 in volume; then, after 3 hours administration, the mice were sacrificed, and the tumors were harvested and frozen in optimal cutting temperature compound embedding medium. The tumor sections (6 μm) were incubated with 1% bovine serum albumin for 3 hours at room temperature, followed by incubation with the primary antibody (rabbit polyclonal to CD31 or rabbit polyclonal to fibrinogen) overnight at 4°C; the primary antibodies were then detected with FITC goat anti-rabbit secondary antibodies. Nuclei were counterstained with Hoechst 33258 (5 μg/mL). The sections were put in Gel Mount™ mounting medium (Corporation, Foster City, CA, USA) and observed under a confocal microscope (Leica Microsystems).
In vivo anti-tumor activity of iRGD-SSL-DOX
To prepare the tumor-bearing C57BL/6 mice model, female C57BL/6 mice were inoculated via subcutaneous injection with 0.1 mL B16-F10 cell suspension (1 × 106) in the right armpits.Citation27,Citation28 When the tumor volume reached about 150–200 mm3, the tumor-bearing C57BL/6 mice were randomly assigned to three groups (each group contained six animals): group 1 was IV administrated physiological saline as a control, group 2 was IV administrated SSL-DOX (3 mg/kg, IV, q3d [every 3 days for three doses]), and groups 3 was IV administrated iRGD-SSL-DOX (3 mg/kg, IV, q3d). Throughout the study, mice were weighed and tumors were measured with a caliper twice per week. Tumor volumes were calculated using the following formula: V = length (cm) × width (cm2) × 0.5236. The survival time was calculated from the day of B16-F10 cell inoculation (day 0) to the day of death. Kaplan–Meier survival curves were plotted for each group.
Statistical analysis
All data are shown as the mean ± standard deviation (SD). One-way analysis of variance (ANOVA) was used to determine significance among groups, after which posthoc tests with the Bonferroni correction were used to compare between individual groups. Statistical significance was set at P < 0.05.
Results
Preparation of iRGD-SSL-DOX
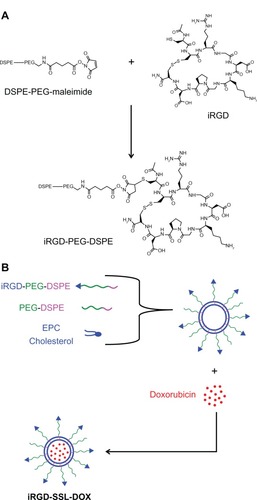
As shown in , iRGD was connected to the terminal of PEG through a reaction between the maleimide group of PEG and the cysteine sulfhydryl group of the iRGD peptides, through a nucleophilic addition reaction. The MALDI (matrix-assisted laser desorption/ionization) – TOF (time-of-fight mass spectrometer) – MS (mass spectrometry) results indicated that iRGD was successfully conjugated with DSPE-PEG-maleimide (data not shown). The procedure of iRGD-SSL-DOX preparation is illustrated in .
Figure 1 The preparation of iRGD-SSL-DOX.
Note: (A)The synthesis of iRGD-PEG-DSPE and (B) the preparation of iRGD-SSL-DOX.
Abbreviations: iRGD-SSL-DOX, doxorubicin-loaded iRGD-modified sterically-stabilized liposome; iRGD, tumor-homing peptide; SSL, sterically-stabilized liposome; DOX, doxorubicin; DSPE-PEG-MAL, 1,2-Distearoyl-sn-Glycero-3-Phosphoethanolamine-N-[Maleimide(polyethylene-Glycol)-2000; PEG, polyethylene glycol; DSPE, 1,2-Distearoyl-sn-glycero-3-phosphoethanolamine; EPC, egg phosphatidylcholine.
Characterization of iRGD-SSL-DOX
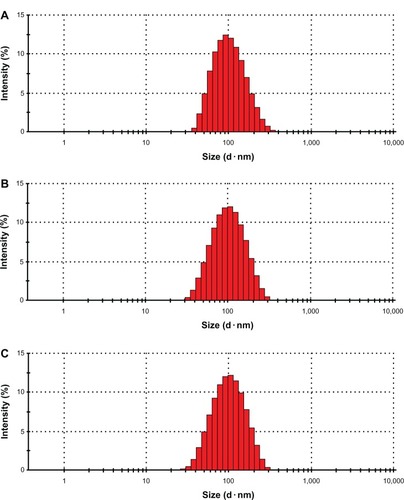
As shown in , the average particle size of iRGD-SSL-DOX was about 91 nm ± 0.8 nm, with a polydispersity index (PDI) of 0.16 ± 0.003. The zeta potential of iRGD-SSL-DOX was slightly negative. The entrapment efficiency of iRGD-SSL-DOX indicated that DOX was almost entrapped within the liposomes. shows the typical particle size and distribution of iRGD-SSL-DOX.
Table 1 The characteristics of iRGD-SSL-DOX (n = 3)
Figure 2 The typical particle size and distribution of SSL, SSL-DOX, and iRGD-SSL-DOX.
Notes: The typical particle size and distribution of (A) SSL, (B) SSL-DOX, and (C) iRGD-SSL-DOX.
Abbreviations: SSL, sterically-stabilized liposome; SSL-DOX, sterically-stabilized liposome containing DOX; DOX, doxorubicin; iRGD-SSL-DOX, DOX-loaded iRGD-modified SSL; iRGD, tumor-homing peptide.
In vitro leakage of DOX from iRGD-SSL-DOX
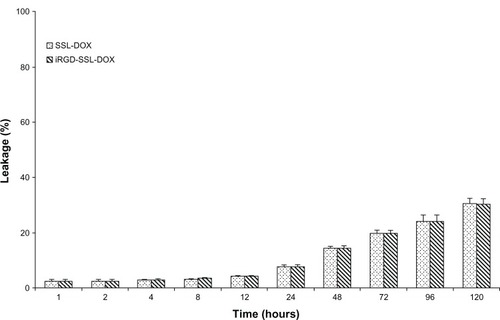
The in vitro leakage of DOX from iRGD-SSL-DOX is shown in . For iRGD-SSL-DOX groups, the leakage of DOX at the 24-hour time point was only 7.5%, showing the stability of the iRGD-SSL-DOX; even at 120 hours, the leakage of DOX form iRGD-SSL-DOX was only about 30%. In addition, the leakage behavior of DOX from SSL-DOX was similar to that of iRGD-SSL-DOX.
Figure 3 The leakage of DOX from iRGD-SSL-DOX.
Notes: The medium was PBS (pH = 7.4). The temperature was controlled at 37°C. Each datum exhibits the mean ± SD (n = 3).
Abbreviations: DOX, doxorubicin; iRGD-SSL-DOX, DOX-loaded iRGD-modified SSL; SSL, sterically-stabilized liposome; PBS, phosphate buffered saline; SD, standard deviation.
Flow cytometry analysis
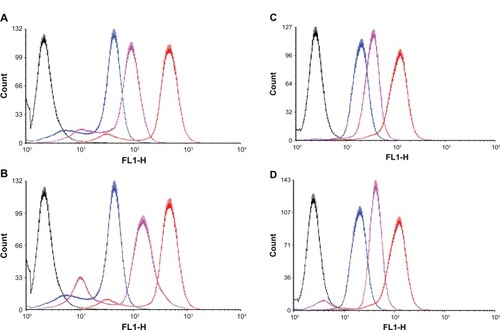
The total coumarin-6 uptake by B16-F10 or MCF-7 cells for coumarin-6 formulations was quantified by flow cytometry determination. Without trypsin treatment groups, the cellular coumarin-6 fluorescence intensity for iRGD-SSL-coumarin-6 in B16-F10 cells was about 2.5-fold higher than that of SSL-coumarin-6 (). For trypsin treatment groups, as shown in , the cellular coumarin-6 fluorescence intensity for iRGD-SSL-coumarin-6 in B16-F10 cells was about threefold higher than that of SSL-coumarin-6 (). In addition, the cellular coumarin-6 level for iRGD-SSL-coumarin-6 with or without trypsin treatment in MCF-7 cells was about 1.8- or 2.3-fold higher than that for SSL-coumarin-6, respectively ().
Figure 4 The flow cytometric measurement of coumarin-6 uptake from SSL-coumarin-6 or iRGD-SSL-coumarin-6 by B16-F10 cells and MCF-7 cells, with or without trypsin treatment, at the 2 hours incubation time point.
Notes: (A) B16-F10 cells, without trypsin treatment groups. (B) B16-F10 cells, with trypsin treatment groups. (C) MCF-7 cells, without trypsin treatment groups. (D) MCF-7 cells, with trypsin treatment groups. Black exhibits control, blue exhibits incubation with SSL-coumarin-6, pink exhibits incubation with iRGD-SSL-coumarin-6, and red exhibits coumarin-6.
Abbreviations: SSL, sterically-stabilized liposome; iRGD, tumor-homing peptide; MCF-7, breast cancer cell line.
Confocal microscopy studies
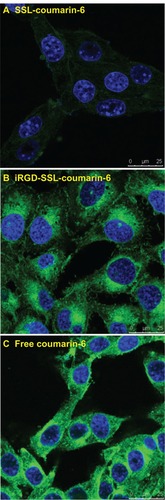
The confocal microscopic images of B16-F10 cells after 2 hours incubation with coumarin-6 formulations are shown in . For the iRGD-SSL-coumarin-6 treatment group, the images show a more intense fluorescence of coumarin-6 () compared with the fluorescence found in the SSL-coumarin-6 treatment group (). As shown in , free coumarin-6 readily partitioned into the lipid membranes and then diffused into the three types of cells, leading to a greater cellular accumulation due to its highly hydrophobic nature. Therefore, free coumarin-6 was selected as the positive control group.
Figure 5 The confocal microscopy images of B16-F10 cells incubated with (A) SSL-coumarin-6, (B) iRGD-SSL-coumarin-6 or (C) free coumarin-6, for 2 hours at 37°C. Fluorescence of coumarin-6 exhibits green; fluorescence of Hoechst 33258 exhibits blue.
Abbreviations: iRGD, tumor-homing peptide; SSL, sterically-stabilized liposome.
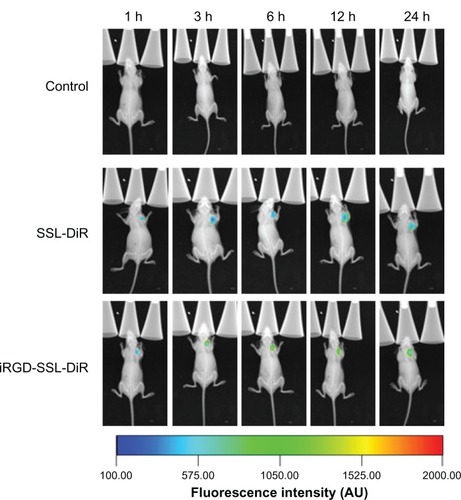
Biodistribution of DiR-loaded liposomes in tumor-bearing nude mice
The distribution and tumor accumulation of fluorescent DiR in B16-F10 tumor-bearing nude mice is shown in . Compared with control group, the DiR fluorescence signal in tumor site was observed in SSL-DiR treatment and iRGD-SSL-DiR treatment groups from the 1-hour to the 24-hour time point. The fluorescence signal of DiR in the tumor site was stronger in the iRGD-SSL-DiR treatment group than that found in the SSL-DiR treatment group, at all observed time points.
Figure 6 In vivo image of biodistribution of iRGD-SSL-DiR in B16-F10 tumor-bearing nude mice.
Note: In vivo whole body imaging of B16-F10 tumor-bearing nude mice after IV administered physiological saline (as control), SSL-DiR, and iRGD-SSL-DiR, respectively.
Abbreviations: SSL, sterically-stabilized liposome; DiR, carbocyanine dye 1,1′-dioctadecyltetramethyl indotricarbocyanine iodide; iRGD, tumor-homing peptide; IV, intravenous.
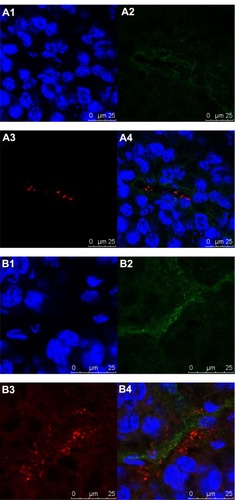
Immunohistochemistry study
In the immunohistochemistry study, the nuclei stained with Hoechst 33258 are represented by a blue fluorescence (), the blood vessels stained with CD31 are represented by a green fluorescence (), while DiI is represented by a red fluorescence (). At the 3-hour time point after administration, iRGD-SSL-DiI mostly penetrated from the tumor blood vessels and gradually accumulated to tumor cells (), while SSL-DiI still existed in the tumor blood vessels (). In addition, it could also be observed that the red fluorescence intensity of DiI in the iRGD-SSL-DiI treatment group was stronger than that of the SSL-DiI treatment group.
Figure 7 The localization of iRGD-SSL-DiI in tumor section.
Notes: B16-F10 tumor-bearing nude mice were IV administered with SSL-DiI or iRGD-SSL-DiI at a dose of 1200 ng/animal. At the 3-hour time point, the mice were sacrificed, and the tumors were harvested. The tumor sections were immune-stained and examined for fluorescence. Images in cluster represent (A) SSL-DiI treatment group, and (B) iRGD-SSL-DiI treatment group. Nuclei were counterstained with Hoechst 33258 (blue; A1 and B1). Tumor blood vessels represented a green fluorescence (antiCD31, green; A2 and B2). DiI represented a red fluorescence (red; A3 and B3). Merge (A4 and B4).
Abbreviations: iRGD, tumor-homing peptide; SSL, sterically-stabilized liposome; DiI, fluorescent probe, 1,1′-dioctadecyl-3,3,3′,3′-tetramethylindocarbocyanine perchlorate; IV, intravenous.
In vivo antitumor activity of iRGD-SSL-DOX
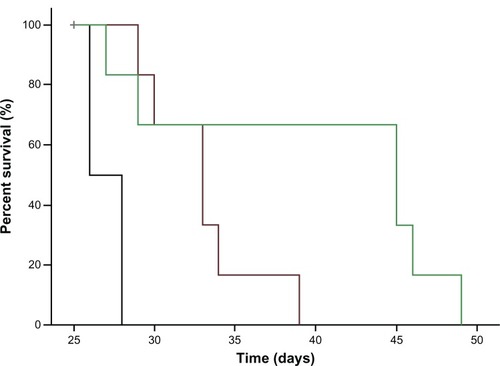
The antitumor effect of iRGD-SSL-DOX was evaluated in B16-F10 tumor-bearing C57BL/6 mice. The tumor growth was significantly inhibited in SSL-DOX and iRGD-SSL-DOX treatment groups compared with the physiological saline treatment group (P < 0.01), as shown in . iRGD-SSL-DOX significantly inhibited the growth of B16-F10 tumors compared with that in the SSL-DOX treatment groups (P < 0.01). The average tumor size at day 22 in the SSL-DOX and iRGD-SSL-DOX group was 3,310 mm3 ± 1,022 mm3 and 1205 mm3 ± 496 mm3, respectively, compared with 6,975 mm3 ± 1,100 mm3 in the physiological saline group (P < 0.01). The corresponding tumor growth inhibition in the SSL-DOX and iRGD-SSL-DOX treated groups was 52.5% and 82.7%, respectively. The Kaplan– Meier survival curve is represented in . After administration three times, the median survival time of mice treated with iRGD-SSL-DOX (43.5 days) was significantly longer than that of mice treated with physiological saline (27 days, P < 0.01) and SSL-DOX (33 days, P < 0.01), respectively.
Figure 8 In vivo antitumor activity of iRGD-SSL-DOX.
Notes: C57BL/6 mice were inoculated SC with B16-F10 cells and treated with physiological saline, SSL-DOX (3 mg/kg, IV, q3d), iRGD-SSL-DOX (3 mg/kg, IV, q3d). The formulations were given via the tail vein for all administrations. The tumors were measured with a caliper twice per week throughout the study. **P < 0.01 versus physiological saline as control; ††P < 0.01 versus SSL-DOX treatment group. ↑, administration.
Abbreviations: iRGD-SSL-DOX, doxorubicin-loaded iRGD-modified sterically-stabilized liposome; iRGD, tumor-homing peptide; SSL, sterically-stabilized liposome; DOX, doxorubicin; SC, subcutaneous; IV, intravenous; q3d, every 3 days for three doses.
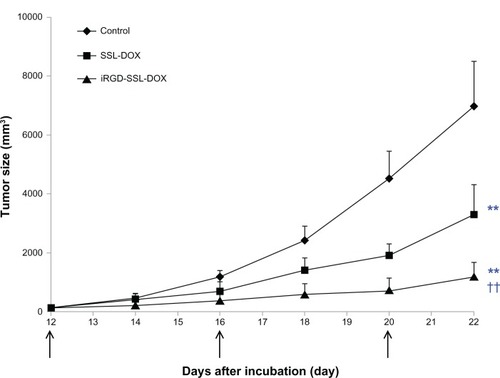
Figure 9 Kaplan–Meier survival curves of B16-F10 tumor-bearing C57BL/6 mice treated with iRGD-SSL-DOX.
Notes: Kaplan–Meier survival curves of B16-F10 tumor-bearing C57BL/6 mice treated with physiological saline (black), SSL-DOX (3 mg/kg, IV, q3d) (brown), iRGD-SSL-DOX (3 mg/kg, IV, q3d) (green). Results indicated that the iRGD-SSL-DOX (43.5 days) significantly improved the median survival time of mice as compared with that treated with SSL-DOX (33 days, P < 0.01) and physiological saline (27 days, P < 0.01), respectively.
Abbreviations: iRGD-SSL-DOX, doxorubicin-loaded iRGD-modified sterically-stabilized liposome; iRGD, tumor-homing peptide; SSL, sterically-stabilized liposome; DOX, doxorubicin; IV, intravenous; q3d, every 3 days for three doses.
Discussion
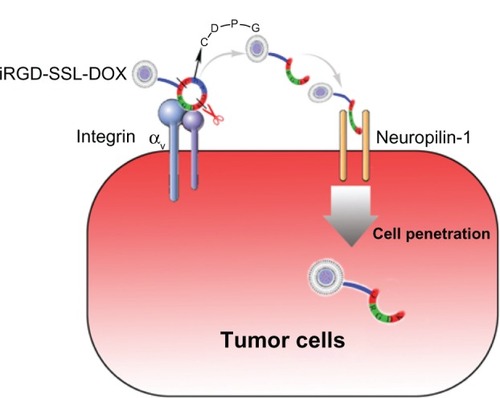
iRGD is a tumor-targeting and tumor-penetrating peptide which can enhance the permeability of tumor cells mediated by integrins and NRP-1 which upregulate tumor cells.Citation11,Citation12,Citation29 Considering the mechanism of iRGD, in the present study, we designed and prepared a iRGD-SSL-DOX and suggested that the iRGD in the iRGD-SSL-DOX bound firstly with αν integrins and then with NRP-1, which can mediate the delivery system penetrating into the tumor cells. The possible role of iRGD-SSL-DOX in tumor-targeting and tumor-penetrating activities is illustrated in .
Figure 10 The detailed scheme of tumor-targeting and tumor-penetrating effect of iRGD-SSL-DOX.
Note: The RGD motif in iRGD-SSL-DOX mediates binding to αν integrins firstly on tumor cells followed by a proteolytic cleavage, exposing a binding motif of CRGDK sequences for NRP-1, which can then mediate penetration into tumor cells.
Abbreviations: iRGD-SSL-DOX, doxorubicin-loaded iRGD-modified sterically-stabilized liposome; iRGD, tumor-homing peptide; SSL, sterically-stabilized liposome; DOX, doxorubicin; CRGDK, Cys-Arg-Gly-Asp-Lys.
Integrins are central regulators in multicellular biology. Many human pathologies (including inflammation, cancer, fibrosis, and infectious diseases) involve integrin adhesion.Citation30 Integrins play a direct role in tumor progression, specifically in tumor cell survival, tumor angiogenesis, and metastasis.Citation31 αv integrins are highly expressed on endothelial cells and tumor cells, and could bind with the RGD peptide sequence.Citation32 The neuropilin receptors were first discovered as regulators of nervous system development and then identified as receptors for vascular endothelial growth factor.Citation33 It has been reported that NRP-1 is overexpressed on several types of tumor cells, such as prostate cancer, colorectal cancer, lung cancer, breast cancer, and astrocytomas.Citation34 Considering both overexpression of αv integrin receptor and NRP-1 in B16-F10 cells,Citation24,Citation25 in the present study, we selected the B16-F10 cells as a model for tumor cells so as to investigate the antitumor activity of iRGD-SSL-DOX in vitro and in vivo.
The targeting effect of the iRGD-modified SSL on B16-F10 cells was observed in our flow cytometry and confocal experiments ( and ); this effect was due to iRGD recognizing the αv integrin receptors with the RGD motif. The in vivo biodistribution results also demonstrated the tumor-targeting effect of the iRGD-modified SSL (). When iRGD-SSL-coumarin-6 was treated with trypsin, the coumarin-6 fluorescence-intensity ratio of iRGD-SSL-coumarin-6/SSL-coumarin-6 was further increased, compared with the ratio for the treatment groups without trypsin (). We suggest that the iRGD might partly proteolytically cleave to produce the CRGDK fragment, when iRGD-SSL-coumarin-6 was treated with trypsin, which has the affinity for NRP-1, to trigger the penetration. Interestingly, we also observed that the coumarin-6 fluorescence intensity for iRGD-SSL-coumarin-6 (with or without trypsin treatment) was also higher than that for SSL-coumarin-6 () in αv integrin-negative MCF-7 cells.Citation35,Citation36 We suggest that this is due to the expression of NRP-1 in the MCF-7 cells.Citation37 When iRGD-SSL-coumarin-6 or iRGD-SSL-coumarin-6 treated with trypsin was incubated with MCF-7 cells, the iRGD might partly proteolytic cleave to produce the CRGDK fragment which has the necessary affinity for NRP-1 to trigger the penetration. These above results might partly explain the tumor-penetrating action of the iRGD, which proteolytically cleaved to produce the CRGDK fragment mediated with NRP-1. In addition, our immunohistochemistry study indicated that the iRGD-SSL-DiI mostly penetrated from the tumor blood vessels, and gradually accumulated to tumor cells, compared with the results for SSL-DiI (, respectively).
Considering the EPR effect, as well as ligand-modified SSL as an active-targeting delivery system,Citation38–Citation40 we selected iRGD as a ligand and developed a novel iRGD-modified SSL. Our particle size, distribution, and in vitro leakage results indicated that the prepared iRGD-SSL-DOX is desirable.
Because the antitumor activity of the DOX delivery system on melanoma has been demonstrated,Citation27,Citation41–Citation43 in the present study we selected DOX as a model drug to prepare the iRGD-SSL-DOX. The antitumor activity of iRGD-SSL-DOX was evaluated in a B16-F10 bearing animal model. In addition, because the antitumor activity of free DOX in a B16-F10 bearing animal model is limited,Citation43 in the present in vivo antitumor activity study we did select free DOX as a treatment group. Our in vivo antitumor activity results indicated iRGD could be used as a tumor-targeting and tumor-penetrating ligand for tumor targeting drug delivery systems.
Conclusion
In the present study, we prepared a novel iRGD-SSL-DOX with the aim of evaluating its antitumor activity on B16-F10 melanoma cells, in vitro and in vivo. The tumor-targeting and tumor-penetrating activity of the iRGD-modified SSL was demonstrated by in vitro flow cytometry and confocal microscopy, as well as in vivo biodistribution and confocal immunofluorescence microscopy experiments. The antitumor activity of the iRGD-SSL-DOX against melanoma tumors was confirmed in our in vivo B16-F10 tumor-bearing mice. The iRGD-SSL-DOX is a tumor-targeting and tumor-penetrating peptide modified liposome which has significant antitumor activity against melanoma tumors.
Acknowledgments
The authors gratefully acknowledge the financial support from the National Natural Science Foundation of China (No 81172992), the National Basic Research Program of China (973 Program 2009CB930300 and 2013CB932501), and the Innovation Team of the Ministry of Education of China (No BMU20110263).
Disclosure
The authors report no conflicts of interest in this work.
References
- LammersTKiesslingFHenninkWEStormGDrug targeting to tumors: principles, pitfalls and (pre-) clinical progressJ Control Release2012161217518721945285
- ValenciaPMHanewich-HollatzMHGaoWEffects of ligands with different water solubilities on self-assembly and properties of targeted nanoparticlesBiomaterials201132266226623321658757
- XiaoZLevy-NissenbaumEAlexisFEngineering of targeted nanoparticles for cancer therapy using internalizing aptamers isolated by cell-uptake selectionACS Nano20126169670422214176
- WangWChengDGongFMiaoXShuaiXDesign of multifunctional micelle for tumor-targeted intracellular drug release and fluorescent imagingAdv Mater201224111512022143956
- Eldar-BoockAMillerKSanchisJLupuRVicentMJSatchi-FainaroRIntegrin-assisted drug delivery of nano-scaled polymer therapeutics bearing paclitaxelBiomaterials201132153862387421376390
- YangZLuoXZhangXLiuJJiangQTargeted delivery of 10-hydroxycamptothecin to human breast cancers by cyclic RGD-modified lipid-polymer hybrid nanoparticlesBiomed Mater20138202501223507576
- ChenZDengJZhaoYTaoTCyclic RGD peptide-modified liposomal drug delivery system: enhanced cellular uptake in vitro and improved pharmacokinetics in ratsInt J Nanomedicine201273803381122888235
- CaiLLLiuPLiXRGD peptide-mediated chitosan-based polymeric micelles targeting delivery for integrin-overexpressing tumor cellsInt J Nanomedicine201163499350822282676
- ZhaoBJKeXYHuangYThe antiangiogenic efficacy of NGR-modified PEG-DSPE micelles containing paclitaxel (NGR-M-PTX) for the treatment of glioma in ratsJ Drug Target201119538239020677914
- LuoLMHuangYZhaoBXAnti-tumor and anti-angiogenic effect of metronomic cyclic NGR-modified liposomes containing paclitaxelBiomaterials20133441102111423127332
- SugaharaKNTeesaluTKarmaliPPTissue-penetrating delivery of compounds and nanoparticles into tumorsCancer Cell200916651052019962669
- SugaharaKNTeesaluTKarmaliPPCoadministration of a tumor-penetrating peptide enhances the efficacy of cancer drugsScience201032859811031103520378772
- ZhuZXieCLiuQThe effect of hydrophilic chain length and iRGD on drug delivery from poly(ε-caprolactone)-poly(N-vinylpyrrolidone) nanoparticlesBiomaterials201132359525953521903260
- SongWLiMTangZMethoxypoly(ethylene glycol)-block-poly(L-glutamic acid)-loaded cisplatin and a combination with iRGD for the treatment of non-small-cell lung cancersMacromol Biosci201212111514152323070837
- WangXZhenXWangJZhangJWuWJiangXDoxorubicin delivery to 3D multicellular spheroids and tumors based on boronic acid-rich chitosan nanoparticlesBiomaterials201334194667467923537667
- SuSWangHLiuXWuYNieGiRGD-coupled responsive fluorescent nanogel for targeted drug deliveryBiomaterials201334133523353323410678
- Hai-TaoZHui-ChengLZheng-WuLChang-HongGA tumor-penetrating peptide modification enhances the antitumor activity of endostatin in vivoAnticancer Drugs201122540941521427563
- AllenTMCullisPRLiposomal drug delivery systems: from concept to clinical applicationsAdv Drug Deliv Rev2013651364823036225
- HuwylerJDreweJKrähenbuhlSTumor targeting using liposomal antineoplastic drugsInt J Nanomedicine200831212918488413
- MoghimiSMPatelHMOpsonophagocytosis of liposomes by peritoneal macrophages and bone marrow reticuloendothelial cellsBiochim Biophys Acta1992113532692741623013
- LasicDDDoxorubicin in sterically stabilized liposomesNature199638065745615628606781
- YuanFDellianMFukumuraDVascular permeability in a human tumor xenograft: molecular size dependence and cutoff sizeCancer Res19955517375237567641188
- MaruyamaKIntracellular targeting delivery of liposomal drugs to solid tumors based on EPR effectsAdv Drug Deliv Rev201163316116920869415
- RatheeshAIngleAGudeRPPentoxifylline modulates cell surface integrin expression and integrin mediated adhesion of B16F10 cells to extracellular matrix componentsCancer Biol Ther20076111743175217986856
- MazurekAMOlbrytMThe influence of neuropilin-1 silencing on semaphorin 3A and 3C activity in B16(F10) murine melanoma cellsNeoplasma2012591435122082309
- ZhaoBXZhaoYHuangYThe efficiency of tumor-specific pH-responsive peptide-modified polymeric micelles containing paclitaxelBiomaterials20123382508252022197569
- YangTWangYLiZTargeted delivery of a combination therapy consisting of combretastatin A4 and low-dose doxorubicin against tumor neovasculatureNanomedicine201281819221664295
- WangYYangTWangXMaterializing sequential killing of tumor vasculature and tumor cells via targeted polymeric micelle systemJ Control Release2011149329930621044651
- MarcucciFCortiAHow to improve exposure of tumor cells to drugs: promoter drugs increase tumor uptake and penetration of effector drugsAdv Drug Deliv Rev2012641536821983328
- GoodmanSLPicardMIntegrins as therapeutic targetsTrends Pharmacol Sci201233740541222633092
- NemethJANakadaMTTrikhaMAlpha-v integrins as therapeutic targets in oncologyCancer Invest200725763264618027153
- TuckerGCAlpha v integrin inhibitors and cancer therapyCurr Opin Investig Drugs200346722731
- BagriATessier-LavigneMWattsRJNeuropilins in tumor biologyClin Cancer Res20091561860186419240167
- EllisLMThe role of neuropilins in cancerMol Cancer Ther2006551099110716731741
- CaoJWanSTianJFast clearing RGD-based near-infrared fluorescent probes for in vivo tumor diagnosisContrast Media Mol Imaging20127439040222649045
- BiswasSWangXMoralesARAhnHYBelfieldKDIntegrin-targeting block copolymer probes for two-photon fluorescence bioimagingBiomacromolecules201112244144921190348
- NasarrePConstantinBRouhaudLSemaphorin SEMA3F and VEGF have opposing effects on cell attachment and spreadingNeoplasia200351839212659673
- VailDMAmanteaMAColbernGTMartinFJHilgerRAWorkingPKPegylated liposomal doxorubicin: proof of principle using preclinical animal models and pharmacokinetic studiesSemin Oncol2004316 Suppl 13163515717736
- AcharyaSSahooSKPLGA nanoparticles containing various anticancer agents and tumour delivery by EPR effectAdv Drug Deliv Rev201163317018320965219
- PastorinoFBrignoleCMarimpietriDVascular damage and anti-angiogenic effects of tumor vessel-targeted liposomal chemotherapyCancer Res200363217400740914612539
- Al-JamalWTAl-AhmadyZSKostarelosKPharmacokinetics & tissue distribution of temperature-sensitive liposomal doxorubicin in tumor-bearing mice triggered with mild hyperthermiaBiomaterials201233184608461722459195
- DaiWYangTWangYPeptide PHSCNK as an integrin α5β1 antagonist targets stealth liposomes to integrin-overexpressing melanomaNanomedicine2012871152116122306158
- FanYDuWHeBThe reduction of tumor interstitial fluid pressure by liposomal imatinib and its effect on combination therapy with liposomal doxorubicinBiomaterials20133492277228823290525