Abstract
The platinum (Pt)-group elements (PGEs) represent a new kind of environmental pollutant and a new hazard for human health. Since their introduction as vehicle-exhaust catalysts, their emissions into the environment have grown considerably compared with their low natural concentration in the earth crust. PGE emissions from vehicle catalysts can be also in the form of nanometer-sized particles (Pt nanoparticles [PtNPs]). These elements, both in their metallic form or as ions solubilized in biological media, are now recognized as potent allergens and sensitizers. Human skin is always exposed to toxic particles; therefore, in the present study we addressed the question of whether polyvinylpyrrolidone-coated PtNPs may have any negative effects on skin cells, including predominantly epidermal keratinocytes. In this study, PtNPs of two sizes were used: 5.8 nm and 57 nm, in concentrations of 6.25, 12.5, and 25 μg/mL. Both types of NPs were protected with polyvinylpyrrolidone. Primary keratinocytes were treated for 24 and 48 hours, then cytotoxicity, genotoxicity, morphology, metabolic activity, and changes in the activation of signaling pathways were investigated in PtNP-treated cells. We found that PtNPs trigger toxic effects on primary keratinocytes, decreasing cell metabolism, but these changes have no effects on cell viability or migration. Moreover, smaller NPs exhibited more deleterious effect on DNA stability than the big ones. Analyzing activation of caspases, we found changes in activity of caspase 9 and caspase 3/7 triggered mainly by smaller NPs. Changes were not so significant in the case of larger nanoparticles. Importantly, we found that PtNPs have antibacterial properties, as is the case with silver NPs (AgNPs). In comparison to our previous study regarding the effects of AgNPs on cell biology, we found that PtNPs do not exhibit such deleterious effects on primary keratinocytes as AgNPs and that they also can be used as potential antibacterial agents, especially in the treatment of Escherichia coli, representing a group of Gram-negative species.
Introduction
The platinum (Pt)-group elements (PGEs) represent a new kind of environmental pollutant and a new hazard for human health. Since their introduction as vehicle-exhaust catalysts, their emissions into the environment have grown considerably compared with their low natural concentration in the earth crust.Citation1 PGE contamination initially occurs in airborne particulate matter, roadside dust, soil, sludge, and water, and afterwards results in bioaccumulation in living organisms through different pathways.Citation2 Traditionally, these elements are considered to be nontoxic for human health, but the massive use of Pt, palladium (Pd), and rhodium (Rh) as nanoparticles (NPs) and automotive catalytic converters, causing their release into the environment, has determined a new risk factor. In particular, PGE emissions from vehicle catalysts could also be in the form of nanometer-sized particles.Citation3 These elements, both in their metallic form or as ions solubilized in biological media, are now recognized as potent allergens and sensitizers.Citation1,Citation4 They have been also associated with asthma, dermatitis, and other serious health problems in humans, resulting from chronic exposure at low concentration levels.Citation5
The effects of PtNPs on different types of cells are not fully clarified. It is generally accepted that PtNPs are not as toxic as silver NPs and exhibit remarkable antioxidant activities, reducing reactive oxygen species (ROS) production.Citation6–Citation8 Because of this strong antioxidative capacity, PtNPs have been effectively used in the cosmetic sector in antiaging formulations. Human skin is constantly exposed to solar ultraviolet photons. Therefore, the formation of ROS, as mediators of photooxidative stress, occurs on a daily basis, especially in countries with enhanced sun exposure. The sun’s ultraviolet ray-driven ROS production in skin cells results in changes in skin structural integrity, signal transduction, gene expression, and even tumorigenic initiation and progression.Citation9,Citation10 The use of antioxidative agents in skin cosmetics could provide positive effects on skin condition. Thus, there is a potential probability that PtNPs will be commonly used in cosmetics in the future.
As there have not been any data concerning the impact of PtNPs on primary keratinocytes so far, in the present study we addressed the question of whether polyvinylpyrrolidone (PVP)-coated PtNPs may have any negative effects on this type of cell.
Materials and methods
Characterization of PtNPs
In this study, PtNPs of two sizes were used: 5.8 nm and 57 nm. Both types of NP were coated with PVP and dissolved in water. The smaller NPs were synthesized by reduction of K2PtCl6, adapting the method described for palladium NPs by Choo et al.Citation11 Transmission electron microscopy (TEM) measurements revealed that PtNPs were as small as 5.8 ± 0.9 nm (number of measured NPs: 100). Total Pt concentration was measured by inductively coupled plasma atomic emission spectroscopy: the determined concentration of PtNPs in the initial NP suspension was 2.0 g/L. A limulus amebocyte lysate endotoxin assay was used to confirm that the PtNP solution was free of bacterial endotoxins and endotoxin-like substances. The bigger NPs were ordered from Particular (Hanover, Germany). The company provided detailed characterization of the NPs. To test whether the NPs were contaminated with bacteria, a limulus amebocyte lysate endotoxin assay was conducted by Clongen Labs (Germantown, MD, USA).
Cell culture and PtNP treatment
Proliferating normal human epidermal keratinocytes (NHEKs) from adult donors derived from three individuals (Lonza, Basel, Switzerland) were cultured in 75 cm2 cell-culture flasks at 37°C in 5% CO2 atmosphere in keratinocyte growth medium (keratinocyte cell basal medium supplemented with KGM-Gold™ SingleQuots™ [Lonza]): bovine pituitary extract, human endothelial growth factor, insulin (bovine), hydrocortisone, gentamicin–amphotericin B (GA-1000), epinephrine, and transferrin.
In several articles, the matter of units used for the NP dose used (mass/number/concentration) has been raised.Citation12,Citation13 We decided to use μg/mL concentrations, as this unit of measurement has been proposed as most appropriate for in vitro toxicity testing of NPs.Citation14 Different concentrations of PtNPs (6.25, 12.5, and 25 μg/mL) were added to the cultured cells 1 day after their seeding on multiwell plates, and these cells were incubated for appropriate time periods (24 and 48 hours).
Transmission electron microscopy
Keratinocytes cultured in multiwell plates were fixed in 5% and 2.5% glutaraldehyde in 0.1 M cacodylate buffer, pH 7.3, at 10°C (Polysciences, Warrington, PA, USA) for 24 hours in each concentration. Next, they were washed several times in the buffer, removed from the wells, and transferred to Eppendorf tubes. They were postfixed in 1.5% osmium tetroxide in cacodylate buffer for 1 hour. After fixation, cells were washed in cacodylate buffer again and dehydrated in 50%, 70%, 96%, and 100% ethanol. Finally, they were treated with propylene oxide twice for 5 minutes and embedded in a Poly/Bed 812 (Polysciences). After polymerization, 70 nm-thick sections were cut on a Reichert (Depew, NY, USA) Ultracut ultramicrotome, collected on copper Formvar-coated single-slot grids, and contrasted with uranyl acetate and lead citrate. Sections were viewed and photographed using a JEOL (Tokyo, Japan) 100 SX TEM.
Viability and cytotoxicity tests
To measure cell viability and cytotoxicity, MTT (3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide) and adenosine triphosphate (ATP) assays were carried out. For the MTT test, NHEKs were seeded on 96-well plates (4 × 103 cells/well). Following 24- and 48-hour stimulation with PtNPs (6.25, 12.5, and 25 μg/mL), thiazolyl blue tetrazolium bromide (Sigma-Aldrich, St Louis, MO, USA) was added for an additional 3.5 hours to a final concentration of 500 ng/mL. The next steps of the assay were done as described before.Citation15 Absorbance was measured in an Infinite M200 microplate reader (Tecan, Männedorf, Switzerland) at 570 nm with a reference wavelength of 650 nm. Absorbance of PtNPs in cell-culture medium measured in the absence of cells (NP background) was subsequently subtracted from the total absorbance of the NP-treated cells. Four independent experiments were done, each performed in triplicate. The mean absorbance value for each PtNP concentration was divided by the mean value for control cells and thus presented as a percentage of control (control treated as 100%).
Cell viability was analyzed by quantification of intracellular ATP content, as described before.Citation16 Cells seeded on 96-well plates (4 × 103 cells/well) were treated for 24 and 48 hours with PtNPs. The ATPlite™ luminescence assay system (PerkinElmer, Waltham, MA, USA) was used for ATP determination. Luminescence was measured using the Infinite M200 microplate reader. The mean luminescence value for each PtNP concentration was divided by the mean value for control cells, and presented as percentage of control (control treated as 100%).
Caspase 3/7 and 9 activity
The activity of caspases 3 and 7 was measured using Caspase-Glo 3/7 Assay (Promega, Fitchburg, WI, USA), as described before.Citation15 NHEKs (8 × 103 per well) were seeded on twelve-well plates and exposed to 12.5 and 25 ug/mL PtNPs for 24 and 48 hours. Protein extracts (3 μg) were mixed with 40 μL of Caspase-Glo® 3/7 Reagent on white 96-well plates. After 120 minutes of incubation, luminescence was measured with the Infinite M200 microplate reader. The experiment was repeated three times, and each was performed in duplicate. Mean luminescence value for each PtNP concentration was divided by the mean value for control cells and thus presented as percentage of control (control treated as 100%).
For determination of caspase 9 activity, NHEKs were seeded on twelve-well plates at a density of 8 × 103 cells per well and stimulated the next day with 12.5 or 25 μg/mL of PtNPs for 24 or 48 hours. Unstimulated cells served as a control. Cells were lysed in 50 μL of radioimmunopre-cipitation assay buffer containing protease and phosphatase inhibitors (10 μM; Roche, Basel, Switzerland). Supernatants were collected by spinning of samples at 14,000 rpm for 15 minutes at 4°C. Three micrograms of isolated protein was mixed with 30 μL of Caspase-Glo 9 substrate on a white 96-well plate. The plate was incubated in the darkness for 2 hours, and luminescence was measured with the Infinite M200 microplate reader. Three independent experiments were carried out, each performed in duplicate. The mean luminescence value for each PtNP concentration was divided by the mean value for control cells and thus presented as a percentage of control (control treated as 100%).
Comet assay
The level of DNA damage was tested by electrophoresis of single cells in agarose gel, with the experiment being carried out exactly as described before,Citation16 and two concentrations of PtNPs (12.5 and 25 μg/mL) were used at two time points (24 or 48 hours). The data were collected for 50 randomly selected comets from each slide. Three independent experiments were done, with two replicates each (number of cells per condition = 300).
Cell migration
NHEKs were seeded on six-well plates (4 × 104 per well). At 24 and 48 hours after PtNP addition, cell migration was recorded for 90 minutes with 1.5-minute time lapse using the Leica DMI6000 B microscope (Leica Microsystems, Wetzlar, Germany) equipped with Leica Application Suite Advanced Fluorescence software. The tracks of individual cells were determined from series of changes in the cell-centroid positions, pooled and analyzed to determine the velocity of cell movement. All analyses were done using Hiro software version 1.0.0.4.Citation16 The experiment was performed three times, and the data were collected each time from 50 cells. Final data are presented as mean values for 150 cells.
Western blot
Activation of mitogen-activated protein kinases (MAPKs) and protein kinase B (Akt) was investigated by Western blot using procedures previously described.Citation16 The following antibodies and dilutions were used: Akt (1:1,000; Cell Signaling Technology, Danvers, MA, USA), Phospho-Akt (1:2,000; Cell Signaling Technology), stress-activated protein kinase (SAPK)/c-Jun N-terminal kinase (JNK) (1:500; Cell Signaling Technology), Phospho-SAPK/JNK (1:500; Cell Signaling Technology), p38 MAPK (1:1,000; Cell Signaling Technology), Phospho-p38 MAPK (1:500; Cell Signaling Technology), phosphorylated extracellular signal-regulated kinase (p-ERK)-1/2 MAPK (1:2,000; Cell Signaling Technology), Phospho-ERK1/2 MAPK (1:1,000; Cell Signaling Technology). Tubulin (1:2,000, Calbiochem; Merck Millipore, Billerica, MA, USA) was used as a loading control. The following secondary antibodies were used: peroxidase-conjugated antirabbit (1:3,000–1:10,000; Cell Signaling Technology) and peroxidase-conjugated antimouse (1:20,000, Pharmingen; BD Biosciences, San Jose, CA, USA).
Colony-reduction assay
The assay was performed as described previously by Sieprawska-Lupa et al.Citation17 Briefly, Escherichia coli (33694; American Type Culture Collection [ATCC], Manassas, VA, USA) and Staphylococcus aureus (ATCC 29213) were grown to the midexponential phase in Luria-Bertani broth or tryptic soy broth, respectively. Then, bacteria were washed four times in phosphate-buffered saline and diluted to a final concentration of 2 × 105 colony-forming units/mL in Roswell Park Memorial Institute 1640 supplemented with 10% Hank’s balanced salt solution. PtNPs were diluted in water to a designated concentration. Nine parts bacteria were incubated with one part PtNPs for 2 hours at 37°C. As a control, bacteria were incubated in the same solution, but without the PtNPs. Each assay point was repeated at least three times. Each sample was diluted 1:100, 1:200, and 1:400 and plated in triplicate on tryptic soy broth or Luria-Bertani agar plates. After 24 hours’ incubation at 37°C, colonies were counted. Data are shown as the percentage of bacteria survival. The control sample was arbitrarily set as 100% survival.
Cell-cycle analysis by flow cytometry
The impact of different concentrations of two sizes of PtNPs (0, 12.5, and 25 μg/mL) on cell-cycle and DNA accumulation was examined in keratinocytes following 24- and 48-hour incubation by flow cytometry. Control untreated cells (1 × 106) as well as the cells following 12.5 μg/mL and 25 μg/mL treatment with PtNPs at two time points were fixed in suspension employing fridge-cold 70% ethanol. The cells were further washed in phosphate-buffered saline without Ca2+ and Mg2+ (Invitrogen; Life Technologies, Carlsbad, CA, USA), treated with DNase-free ribonuclease (100 μg/mL, Sigma-Aldrich) to remove the RNA fraction, and subsequently stained with propidium iodide (50 μg/mL, Sigma-Aldrich) to visualize DNA content. Samples were evaluated by flow cytometry (LSR II; BD Biosciences).
Statistical analysis
All results are the means of at least three independent experiments ± standard deviation. The data were analyzed using Student’s t-test, and only in case of comet-assay data were calculated by Tukey’s rank-invariant resampling test. Statistical significance was accepted at the level of P ≤ 0.05.
Results
Ultrastructure of PtNP-treated keratinocytes
To verify the interaction between PtNPs and normal human keratinocytes, we treated the cells with 5.8 nm and 57 nm PtNPs and performed TEM analysis. When compared with the control (–), keratinocytes treated with PtNPs in 12.5 μg/mL concentration for 24 hours showed cytoplasmic endosomes filled with electron-dense material, probably aggregates of PtNPs (). When the concentration of NPs was increased to 25 μg/mL, there were more endosomes/lysosome-like organelles visible in the cytoplasm (). Longer exposure of keratinocytes to PtNPs increased the number and size of lysosomes in the cytoplasm filled with NPs and cell-engulfed material (). There were no obvious ultrastructural changes noticed in the nucleus except changes in its shape (). In some cells, the nucleolus was observed on the nucleus territory ().
Figure 1 (A–I) Ultrastructure of keratinocytes treated with 5.8 nm platinum nanoparticles (PtNPs). (A–C) Keratinocytes from the control fixed after 24 hours (A) and 48 hours (B and C) of in vitro culturing. (D and E) Endosome/lysosome-like organelles in a keratinocyte treated for 24 hours with 12.5 μg/mL PtNPs. Electron-dense material of an NP aggregate is observed inside an endosome/lysosome. (F and G) Part of a keratinocyte exposed for 48 hours to 12.5 μg/mL and (G) 25 μg/mL PtNPs. (H) Endosomes/lysosomes in a keratinocyte treated with 25 μg/mL PtNPs and fixed after 24 hours. (I) Keratinocytes treated for 24 hours with 5.8 nm PtNPs or 57 nm PtNPs at the indicated concentrations. Arrows indicate groups of 57 nm PtNPs localized inside treated keratinocytes.
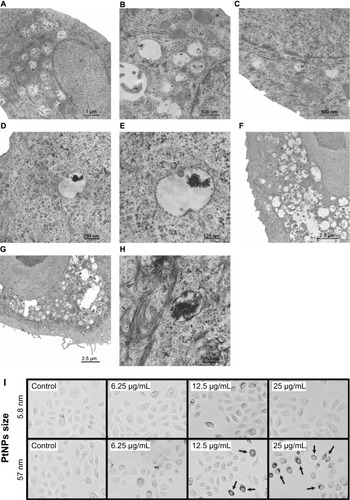
Besides weak changes in the nucleus, no clear changes in cell morphology were observed in keratinocytes treated for 24 hours with 5.8 nm PtNPs (). While the cells treated with 57 nm PtNPs looked generally fine, strong accumulation of NPs was observed when higher doses were used (12.5 and 25 μg/mL, ). The material accumulated around cell nuclei. This suggests that 57 nm PtNPs undergo endocytosis, similarly to 5.8 nm PtNPs, but for small NPs TEM is necessary for the visualization of NP endocytosis.
Effects of PtNPs on cell viability and metabolism
To estimate the cytotoxic effects of PtNPs on keratinocytes, we carried out a viability assay (MTT) and metabolic activity test (ATP content). Cells were treated with PtNPs of two sizes (5.8 and 57 nm) in three concentrations: 6.25, 12.5 and 25 μg/mL. The experiment was conducted at two time points: 24 and 48 hours. Untreated cells served as a control.
In the MTT assay, we observed that both the size and the concentrations of NPs altered their influence on cell viability. However, the observed changes were not statistically significant ().
Figure 2 (A and B) Biological activity of cells treated with platinum nanoparticles (PtNPs). (A) An MTT (3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide) assay was carried out to measure cell viability. Normal human epidermal keratinocytes (NHEKs; plated at a density of 4 × 103 per well of 96-well plates) were treated with PtNPs of two sizes (5.8 and 57 nm) in concentrations of 6.25, 12.5, and 25 μg/mL at indicated time points. Untreated cells served as a control. For statistics, Student’s t-test was performed. The graphs (left panel and right panel) represent the mean ± standard deviation (SD) of five independent experiments, each performed in quintuplicate (number of wells = 25). (B) An adenosine triphosphate assay was performed to determine metabolic activity of cells. NHEKs (plated as in (A)) were treated with PtNPs as described in (A). Untreated cells served as a control. For statistics, Student’s t-test was performed (*P < 0.05; **P < 0.01; ***P < 0.001). The graphs represent the mean ± SD of three independent experiments, each performed in triplicate (number of wells = 9).
Abbreviation: ATP, adenosine triphosphate.
![Figure 2 (A and B) Biological activity of cells treated with platinum nanoparticles (PtNPs). (A) An MTT (3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide) assay was carried out to measure cell viability. Normal human epidermal keratinocytes (NHEKs; plated at a density of 4 × 103 per well of 96-well plates) were treated with PtNPs of two sizes (5.8 and 57 nm) in concentrations of 6.25, 12.5, and 25 μg/mL at indicated time points. Untreated cells served as a control. For statistics, Student’s t-test was performed. The graphs (left panel and right panel) represent the mean ± standard deviation (SD) of five independent experiments, each performed in quintuplicate (number of wells = 25). (B) An adenosine triphosphate assay was performed to determine metabolic activity of cells. NHEKs (plated as in (A)) were treated with PtNPs as described in (A). Untreated cells served as a control. For statistics, Student’s t-test was performed (*P < 0.05; **P < 0.01; ***P < 0.001). The graphs represent the mean ± SD of three independent experiments, each performed in triplicate (number of wells = 9).Abbreviation: ATP, adenosine triphosphate.](/cms/asset/fc146c71-3542-417f-b7d9-a6b76c5630f9/dijn_a_49612_f0002_b.jpg)
In the case of metabolic activity, we noticed dose- and time-dependent decrease of ATP content in cells treated with PtNPs. Both NP fractions decreased cell metabolism, but this effect was stronger for the cells treated with the smaller NPs than with the bigger ones, and in most cases it was more evident after 24 hours than after 48 hours following PtNP administration. Generally, for the highest concentration of 5.8 nm PtNPs (25 μg/mL), we observed a decrease in cell metabolism by 29% after 24 hours and by 24% after 48 hours of exposure. For the lowest concentration, the changes were 18% and 20% decrease after treatment of cells for 24 and 48 hours, respectively (, left panel). In case of cells treated with the bigger NPs, ATP content was decreased by 16% and 27%, respectively, after administration of the highest concentration of NPs (25 μg/mL) for 24 and 48 hours and 12% after 24 hours for the lowest concentration (6.25 μg/mL). There was no statistically significant change after 48 hours (, right panel).
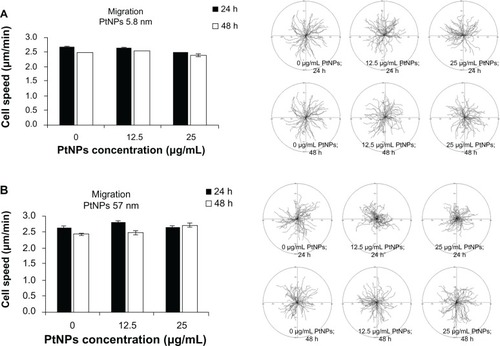
Cell migration
Time-lapse monitoring of cell movement was performed to estimate the influence of PtNPs on the migratory potential of keratinocytes. As in the other experiments, 5.8 and 57 nm PtNPs were incubated with keratinocytes for 24 and 48 hours, and untreated cells served as a control. We did not observe any significant inhibition of cell migration for used concentrations of PtNPs used in the experiment (). Thus, the treatment of cells with PtNPs at indicated concentrations and time points has no effect on the migratory activity of keratinocytes.
Figure 3 (A and B) Cell migration. Normal human epidermal keratinocytes seeded on six-well plates (4 × 104 per well) were exposed to 5.8 (A) and 57 (B) nm platinum nanoparticles (PtNPs) in concentrations of 12.5 and 25 μg/mL for 24 hours and 48 hours. Cell migration was recorded for 90 minutes with 1.5-minute time lapse using a Leica DMI6000B microscope equipped with Leica Application Suite Advanced Fluorescence software. The tracks of individual cells were determined from the series of changes in the cell centroid positions, pooled, and analyzed to determine the total length of the cell trajectory (left panels). All analyses were performed using Hiro software version 1.0.0.4. Presented data are means ± standard deviation from cell-speed values for 150 cells, measured in three independent experiments. Student’s t-test was carried out for statistics (right panels).
Cell cycle
The impact of PtNPs on cell-cycle and DNA accumulation in keratinocytes was evaluated by employing flow cytometry. The data showed no significant difference in content of quiescent (in the G0/G1 phase) or proliferating cells represented in both S and G2/M phases. Moreover, we did not observe any considerable accumulation of polyploidy following treatment with PtNPs of two sizes (). The effect was similar for cells treated with the agent for 24 and 48 hours. Similarly, the ratio between quiescent and proliferating cells (computed as ratio of cells in the G0/G1 phase to the sum of cells in both the S and G2/M phases) confirmed that there was no major impact of PtNPs on cell proliferation and accumulation of cells with an abnormal, polyploid number of chromosomes ().
Figure 4 (A and B) Cell-cycle phases and DNA content in keratinocytes by flow cytometry following treatment with platinum nanoparticles (PtNPs). (A) Percentage content of cells in each cell-cycle phase (G0/G1, S, G2/M) as well as polyploid cells (>4 n) following 24- and 48-hour treatment with different doses of 5.8 and 57 nm PtNPs. The bars represent average data (means ± standard deviation [SD]). (B) Representative histograms showing DNA content of cells following treatment with PtNPs and stained with propidium iodide. The numbers represent the ratio of nondividing, quiescent, and proliferating cells (Q/P) in each sample, calculated as ratio between cells in G0/G1 phase to sum of S and G2/M phases. The numbers represent mean data ± SD.
Abbreviation: Ctrl, control.
![Figure 4 (A and B) Cell-cycle phases and DNA content in keratinocytes by flow cytometry following treatment with platinum nanoparticles (PtNPs). (A) Percentage content of cells in each cell-cycle phase (G0/G1, S, G2/M) as well as polyploid cells (>4 n) following 24- and 48-hour treatment with different doses of 5.8 and 57 nm PtNPs. The bars represent average data (means ± standard deviation [SD]). (B) Representative histograms showing DNA content of cells following treatment with PtNPs and stained with propidium iodide. The numbers represent the ratio of nondividing, quiescent, and proliferating cells (Q/P) in each sample, calculated as ratio between cells in G0/G1 phase to sum of S and G2/M phases. The numbers represent mean data ± SD.Abbreviation: Ctrl, control.](/cms/asset/88628668-bd2e-42e3-9239-53a8db842e75/dijn_a_49612_f0004_b.jpg)
Comet assay
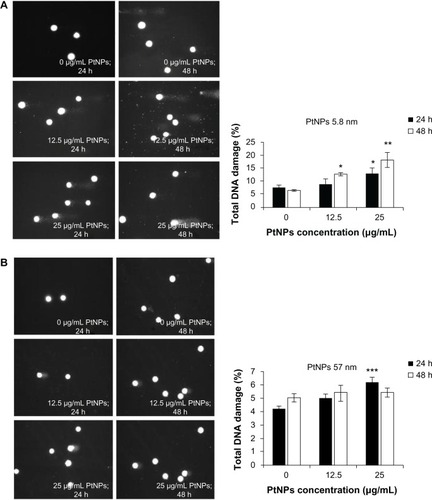
To measure DNA damage in individual cells, single-cell gel electrophoresis (comet assay) was carried out. Genotoxic effects triggered by 5.8 nm PtNPs were time- and dose-dependent. Smaller NPs caused significant changes in the level of damaged DNA, as observed after 48 hours of treatment at a concentration of 12.5 μg/mL. The use of a higher concentration resulted in the augmentation of the DNA comet formation, reaching a level of 13.2% and 18.2% after 24 and 48 hours, respectively (). In the experiment where the bigger NPs were used, the amount of DNA in a comet tail was slightly higher, but statistically significant in comparison to control only for the concentration 25 μg/mL and after 24 hours of incubation. Other time points and lower concentrations were not statistically significant, and the effects were not as visible as in the case of smaller NPs ().
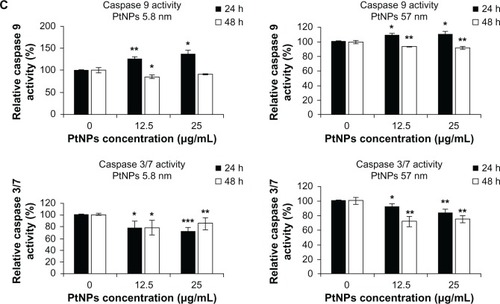
Figure 5 (A–C) Determination of DNA quality and activation of caspases in platinum nanoparticle (PtNP)-treated normal human epidermal keratinocytes (NHEKs). (A and B) DNA damage in NHEKs was investigated by comet assay. Cells were plated at a density of 2.1 × 103 cells/cm2. After 24 hours, NHEKs were treated with 12.5 and 25 μg/mL for 24 and 48 hours. Mean values of the percentage of DNA in the comet tail are presented. Images were made using the computer program Comet Plus (left panels). Statistical analyses were calculated using Tukey’s rank-invariant resampling test (*P < 0.05; **P < 0.01; ***P < 0.001). The data represent the means ± standard deviation (SD) from DNA damage in 100 cells, measured in three independent experiments (right panels). (C) Activation of caspases. The apoptotic effect triggered by PtNPs was measured by the determination of caspase 9 and 3/7 activity. A luminescent assay was performed using 3 μg protein isolated from NHEKs after incubation with 12.5 and 25 μg/mL PtNPs for 24 and 48 hours. The graph represents the means ± SD of three independent experiments. Student’s t-test was carried out for statistics (*P < 0.05; **P < 0.01; ***P < 0.001).
Activation of apoptosis
As an indicator of apoptosis-related processes, caspase 3/7 and 9 activity was measured. NPs of both sizes induced caspase 9 after treatment of keratinocytes for 24 hours. In case of smaller NPs, caspase activity increased about 24% for the concentration 12.5 μg/mL and 35% for 25 μg/mL. The use of 57 nm NPs resulted in 8% and 10% activation of caspase 9 for lower and higher concentrations, respectively, in comparison to control cells. However, after 48 hours of keratinocyte incubation with NPs, the activity of caspase 9 was below the control value for both types of PtNPs (, upper panel). Distinct results were obtained for caspase 3/7. Both small as well bigger NPs caused a decrease in activity of these caspases in comparison to basic activity present in control cells (, lower panel). The decline in caspase 3/7 activity was insignificantly stronger in comparison to control cells after 24 hours than after 48 hours in the case of cells incubated with smaller NPs (, lower left panel). In the case of bigger NPs, we observed another tendency: caspase 3/7 activity was slightly below the basic level after 24 hours, and then a more significant decline was observed after 48 hours of treatment (, lower right panel).
Activation of signaling pathways by PtNPs
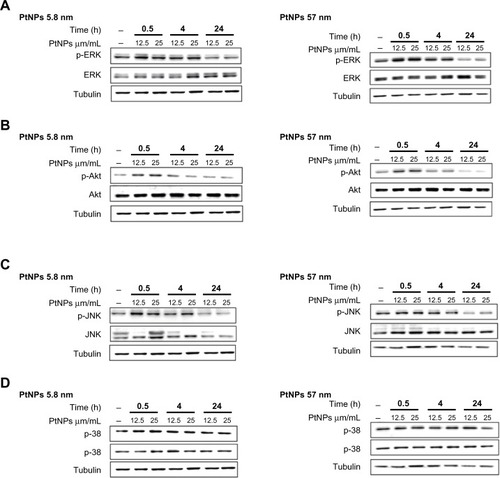
MAPKs (p38, JNK and ERK1/2) and Akt are involved in relaying extracellular stimulations to intracellular responses. The MAPKs coordinately regulate cell proliferation, differentiation, motility, and survival. Akt is a key regulator of cell metabolism, but plays also a crucial role in the regulation of such cellular processes as growth, metabolism, survival, and proliferation. These kinases are quickly activated upon stress conditions to trigger signals towards appropriate transcription factors.
Treating keratinocytes with PtNPs, we observed enhanced phosphorylation of ERK1/2, JNK, and Akt, but not p38 (–). The activation of these kinases was observed at two time points of the experiment (0.5 and 4 hours), and then after 24 hours the phosphorylation was as in the control or even below the signal for control cells (in the case of Akt and ERK1/2 and cells treated with 57 nm PtNPs; ).
Figure 6 (A–D) Activation of ERK, Akt, p38, and JNK by Western blot analysis. Normal human epidermal keratinocyte cells were cultured in 60 mm tissue-culture dishes at a density of 9.5 × 103 cells/cm2. The cells were exposed to 12.5 and 25 μg/mL PtNPs for 0.5, 4, and 24 hours. Untreated cells, harvested together at the 0.5-hour time point, served as a control. At the time of NP addition, the whole medium was exchanged in both treated and untreated cells. Tubulin was used as an internal control to monitor for equal loading. The data are representative of three independent experiments.
Abbreviations: PtNPs, platinum nanoparticles; p-ERK, phosphorylated extracellular signal-regulated kinase; p-Akt, phosphorylated protein kinase B; p-JNK, phosphorylated c-Jun N-terminal kinase; Akt, protein kinase B.
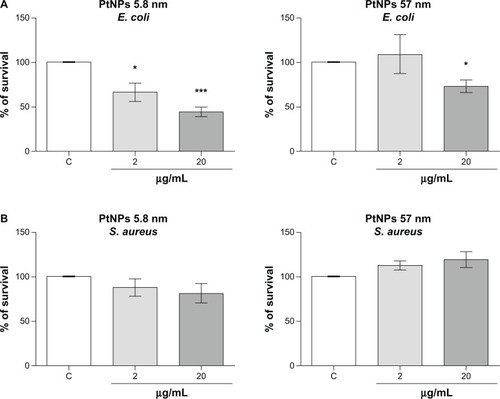
Antimicrobial activity
The antimicrobial activity of silver NPs is well known. However the properties of PtNPs in this field have not been fully investigated.Citation18 Therefore, we determined the antimicrobial properties of PtNPs against Gram-negative (E. coli) and Gram-positive (S. aureus) bacteria. Both pathogens were treated with two concentrations (2 and 20 μg/mL) of PtNPs of two sizes (5.8 nm and 57 nm). Viability inhibition was significant in a concentration-dependent manner only in the case of treatment of E. coli with the smallest NPs. Notably, significant growth inhibition was observed already at the 2 μg/mL concentration of PtNPs, with bacterial growth estimated at about 66.5% ± 10.1% of control cells, the viability of which was set as 100% (). The data obtained for S. aureus were not significant. In the case of bigger PtNPs, the decrease in E. coli survival was observed only at the highest concentration used in our study (20 μg/mL), and was estimated at about 70% of the control cells (). Bigger NPs did not display antimicrobial properties towards S. aureus. Obtained data demonstrated the differential antimicrobial potential of PtNPs.
Figure 7 (A and B) Antibacterial activity of platinum nanoparticles (Pt-NPs). Concentration-dependent antibacterial effect of Pt-NPs on Escherichia coli (A) and Staphylococcus aureus (B). Antimicrobial activity was determined using a colony-reduction assay. Bacteria were incubated alone or with PtNPs at concentrations of 2–200 μg/mL for 2 hours at 37°C, then plated onto agar plates to determine the numbers of colony-forming units. Percentage survival was calculated and compared to bacteria grown without PtNPs (100% survival). Mean values ± standard error of mean of three independent experiments performed in triplicate are shown.
Notes: * P < 0.05; ***P < 0.001.
Abbreviations: E. coli, Escherichia coli; S. aureus, Staphylococcus aureus.
Discussion
NPs have become more and more commonly used in scientific studies and in industry. Although NPs have comprehensive applications, their potential effects on organisms and environment are not well clarified yet; in particular, their long-term effects have not been estimated. There is a growing list of reports indicating that NPs might be medically and environmentally toxic, as their high surface-to-volume ratio makes the particles of some metals very reactive or catalytic.Citation19 There is also evidence that NPs pass through cell membranes and interact with cellular structures, and thus have a direct impact on cell functioning and consequently on cell viability.
PtNPs might have potential application as scavengers of ROS in different types of cells. ROS are important modulators of key signaling pathways,Citation20 but may also cause oxidative stress when they appear in high concentrations. Such activity may eventually lead to cell damage, caused by DNA damage, lipid peroxidation, and oxidations of amino acids.Citation21 It has been shown that PtNPs influence many processes by modulating the level of ROS production. Kim and coworkersCitation7 showed that PtNPs inhibit formation of osteoclasts by impairing the receptor activator of nuclear factor-κB ligand (RANKL) signaling. Thus, PtNPs could protect against bone loss by modulation of oxidative stress. Similar data were delivered by Nomura et al,Citation8 showing that PtNPs inhibit RANKL-stimulated osteoclast differentiation via their ROS-scavenging property. Moreover, a study by Clark et alCitation22 showed that PtNPs reduce oxidant-mediated apoptosis in target cells, judging by the activation of caspase 3. Nevertheless, there are also reports suggesting that PtNPs might have toxic effects on cells. They cause, for example, DNA-strand breaks in human colon carcinoma cells (HT29) in a concentration-, time- and size-dependent manner. However, such DNA damage was not accompanied by the formation of ROS.Citation23,Citation24
In this study, we used PtNPs coated with PVP, which represent a polymer increasingly widespread in medicine and foodstuffs. First of all, PVP is used as a binder in many pharmaceutical tablets. Also, PVP is used as a stabilizer of different kinds of food (E1201) and also in the wine industry as a fining agent for white wine. This chemical is approved for many uses (inactive ingredients in FDA-approved drugs).
In human primary keratinocytes treated with 5.8 nm PtNPs, we observed at the TEM level that PtNPs cause cell membrane folding around the NPs, producing a sac-like bubble into which the material is incorporated and NPs are local-ized within intracellular vacuoles. The localization of PtNPs in intracellular vacuoles suggests endosomal containment and an uptake mechanism involving endocytosis. Such a mechanism has already been described for silver NPs.Citation25,Citation26 Cells treated with NPs displayed changed morphology after 24 and 48 hours of incubation in comparison to control cells, but only when analyzed by TEM. No morphological changes could be observed for 5.8 nm PtNPs treated for 24 hours, when bright-field microscopy was applied. For the cells treated with larger NPs (57 nm), the accumulation of material was observed using bright-field microscopy already after 24 hours of exposition. Nevertheless, the data from cell-viability assays and migratory tests were not significantly different from those obtained for the untreated cells.
We observed a difference in toxicity level between smaller and bigger PtNPs. Smaller NPs triggered a stronger decrease in the metabolic activity of treated cells and stronger DNA damage, as well as higher activity of signaling pathways. Similarly to what was observed in the case of silver NPs, the size of PtNPs was an important factor in the induction of genotoxic and cytotoxic effects.Citation7,Citation27,Citation28 Western blot data showing activation of signaling pathways important for cell survival and proliferation indicate that PtNPs trigger stressful conditions for keratinocytes during the first few hours of treatment, resulting in enhanced phosphorylation of ERK1/2, JNK, and Akt. However, the activity of these kinases returns to the level observed in control cells after 24 hours, and thus PtNP-related stress does not affect homeostasis in a stable way. Interestingly, the decrease in cell metabolism was accompanied by a slight decrease in activation of caspase 9 after 24-hour treatment of cells with NPs, and then after 48 hours a decline of activity was observed. In the case of caspase 3/7, a significant decline of activity was seen at both time points of experiment. Such a phenomenon, that caspase activity is reduced under toxin treatment, was observed by Tao and coworkers when cells were treated with anticancer drugs (doxorubicin, dactinomycin).Citation29 These authors suggested that this might be due to the effects of caspase leakage, ATP depletion, or cell death.Citation29
We showed here that PtNPs display antimicrobial properties against E. coli and S. aureus, pathogens that can cause serious local and systemic life-threatening infections. We demonstrated that the susceptibility of E. coli to PtNPs is significantly higher in comparison to S. aureus. Our results corroborate the data obtained using silver NPs, since the minimal inhibitory concentration of silver NPs against E. coli was estimated to be ten times lower in comparison to S. aureus.Citation30 Such an effect is probably a consequence of the thick peptidoglycan layer of the S. aureus cell wall, preventing the penetration of NPs inside the cytoplasm.Citation31 Nowadays, in the era of multiresistant strain appearance, the discovery of new, nonantibiotic compounds with antimicrobial potential is a promising tool in the treatment of infectious diseases.
We have recently shown that silver NPs possess quite strong genotoxic and cytotoxic effects on human primary keratinocytes.Citation16 However, they are among the most commercialized NPs due to their antimicrobial potential. We are surrounded by therapeutic agents, household products, textiles, and cosmetics containing silver NPs. Data presented in this study show that an important advantage of PtNP use may be related to their antibacterial properties and lack of significant toxic effects on keratinocyte biology and function, as was shown in the case of silver NPs.Citation16 However, it has to be mentioned that effects observed in vitro may differ from the effects triggered by NPs on human skin. The skin acts as a barrier to potentially dangerous materials, and it is not clear yet whether NPs penetrate the skin’s surface. Further in vivo studies are essential to clarify this issue. In contrast to previous studies, recently obtained data with the use of confocal microscopy allowed Campbell et al to show that fluorescent polystyrene NPs applied in aqueous suspension could infiltrate only the stratum disjunctum, ie, skin layers in the final stages of desquamation.Citation32 Thus, it is possible that the use of NPs on the skin directly does not trigger such toxic effects as observed in vitro. Nevertheless, one could consider whether instead of more toxic silver NPs, PtNPs could be used in various body products.
Acknowledgments
This study was supported by the European Community grant COST action BM0903 and the Polish Ministry of Science and Higher Education grant 776/N-COST/2010/0, both awarded to Jolanta Jura. Francesca Larese Filon and Jolanta Jura are members of the management committee of COST Action BM0903.
Disclosure
The authors report no conflicts of interest in this work.
References
- RavindraKBencsLVan GriekenRPlatinum group elements in the environment and their health riskSci Total Environ20043181–314314654273
- ContiMEAlimontiABoccaBEnvironmental exposure to platinum group elements released by automotive catalytic converters: the risk for childrenInt J Environ Health200823439462
- EkKHMorrisonGMRauchSEnvironmental routes for platinum group elements to biological materials – a reviewSci Total Environ20043343352138
- BurasteroSEPaolucciCFabbriMAmbient pollutants as adjuvant for allergic sensitization: the emerging role of platinum group elementsJ Biol Regul Homeost Agents200923420721520003759
- WisemanCLZereiniFAirborne particulate matter, platinum group elements and human health: a review of recent evidenceSci Total Environ200940782493250019181366
- AsharaniPVLianwuYGongZValiyaveettilSComparison of the toxicity of silver, gold and platinum nanoparticles in developing zebrafish embryosNanotoxicology201151435421417687
- KimWKKimJCParkHJPlatinum nanoparticles reduce ovariectomy-induced bone loss by decreasing osteoclastogenesisExp Mol Med201244743243922525805
- NomuraMYoshimuraYKikuiriTPlatinum nanoparticles suppress osteoclastogenesis through scavenging of reactive oxygen species produced in RAW264.7 cellsJ Pharmacol Sci2011117424325222083043
- LiebelFKaurSRuvoloEKolliasNSouthallMDIrradiation of skin with visible light induces reactive oxygen species and matrix-degrading enzymesJ Invest Dermatol201213271901190722318388
- SwalwellHLatimerJHaywoodRMBirch-MachinMAInvestigating the role of melanin in UVA/UVB- and hydrogen peroxide-induced cellular and mitochondrial ROS production and mitochondrial DNA damage in human melanoma cellsFree Radic Biol Med201252362663422178978
- ChooHPLiewKYLiuHFFactors affecting the size of polymer stabilized Pd nanoparticlesJ Mater Chem2002124934937
- TeeguardenJGHinderliterPMOrrGThrallBDPoundsJGParticokinetics in vitro: dosimetry considerations for in vitro nanoparticle toxicity assessmentsToxicol Sci200795230031217098817
- AsefaTTaoZBiocompatibility of mesoporous silica nanoparticlesChem Res Toxicol201225112265228422823891
- LisonDThomassenLCRabolliVNominal and effective dosimetry of silica nanoparticles in cytotoxicity assaysToxicol Sci2008104115516218400775
- WegrzynPYarwoodSJFieglerNMimitin – a novel cytokine-regulated mitochondrial proteinBMC Cell Biol2009102319331698
- SzmydRGoralczykAGSkalniakLEffect of silver nanoparticles on human primary keratinocytesBiol Chem2013394111312323091270
- Sieprawska-LupaMMydelPKrawczykKDegradation of human antimicrobial peptide LL-37 by Staphylococcus aureus-derived proteinasesAntimicrob Agents Chemother200448124673467915561843
- ChwalibogASawoszEHotowyAVisualization of interaction between inorganic nanoparticles and bacteria or fungiInt J Nanomedicine201051085109421270959
- OberdorsterGOberdorsterEOberdorsterJNanotoxicology: an emerging discipline evolving from studies of ultrafine particlesEnviron Health Perspect2005113782383916002369
- ThannickalVJFanburgBLReactive oxygen species in cell signalingAm J Physiol Lung Cell Mol Physiol20002796L1005L102811076791
- IchihashiMUedaMBudiyantoAUV-induced skin damageToxicology20031891–2213912821280
- ClarkAZhuASunKPettyHRCerium oxide and platinum nanoparticles protect cells from oxidant-mediated apoptosisJ Nanopart Res201113105547555522039334
- PelkaJGehrkeHEsselenMCellular uptake of platinum nanoparticles in human colon carcinoma cells and their impact on cellular redox systems and DNA integrityChem Res Toxicol200922464965919290672
- GehrkeHPelkaJHartingerCGPlatinum nanoparticles and their cellular uptake and DNA platination at non-cytotoxic concentrationsArch Toxicol201185779981221229235
- AsharaniPVHandeMPValiyaveettilSAnti-proliferative activity of silver nanoparticlesBMC Cell Biol2009106519761582
- GreulichCDiendorfJSimonTEggelerGEppleMKollerMUptake and intracellular distribution of silver nanoparticles in human mesenchymal stem cellsActa Biomater20117134735420709196
- OsborneOJJohnstonBDMogerJEffects of particle size and coating on nanoscale Ag and TiO(2) exposure in zebrafish (Danio rerio) embryosNanotoxicology Epub10292012
- ShiJXuBSunXMaCYuCZhangHLight induced toxicity reduction of silver nanoparticles to Tetrahymena pyriformis: effect of particle sizeAquat Toxicol2013132133536023399445
- TaoZGoodismanJPenefskyHSSouidAKCaspase activation by anticancer drugs: the caspase stormMol Pharm20074458359517439154
- KimJSKukEYuKNAntimicrobial effects of silver nanoparticlesNanomedicine2007319510117379174
- TagliettiADiaz FernandezYAAmatoEAntibacterial activity of glutathione-coated silver nanoparticles against Gram positive and Gram negative bacteriaLangmuir201228218140814822546237
- CampbellCSContreras-RojasLRDelgado-CharroMBGuyRHObjective assessment of nanoparticle disposition in mammalian skin after topical exposureJ Control Release2012162120120722732479