 ?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.
?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.Abstract
Atomic force microscope (AFM) has been used incrementally over the last decade in cell biology. Beyond its usefulness in high resolution imaging, AFM also has unique capabilities for probing the viscoelastic properties of living cells in culture and, even more, mapping the spatial distribution of cell mechanical properties, providing thus an indirect indicator of the structure and function of the underlying cytoskeleton and cell organelles. AFM measurements have boosted our understanding of cell mechanics in normal and diseased states and provide future potential in the study of disease pathophysiology and in the establishment of novel diagnostic and treatment options.
Introduction
Functional properties of the various types of cells, the basic building block of all living organisms, have been studied extensively over the last decades and provided the driving force for the exceptional progress of cell biology and physiology. However, no analogous research on the cell mechanical properties in states of health and disease and their pathogenetic significance took place until recently. The mechanical properties of cells include stiffness, nonlinearity, anisotropy, and heterogeneity, as well as several functional aspects, including their relation with individual components of cytoskeleton and cell organelles, cell responsiveness to external mechanical stimulation as well as their remodeling and effects on extracellular matrix.Citation1–Citation4 Indeed, cell mechanical properties have been found to affect substantially several important factors of cell function, such as the shape, deformability, motility, division, and adhesion. Several methods, including magnetic twisting cytometry, laser-tracking microrheology, magnetic tweezers, the optical stretcher, and various cell indenters, have been used for the study of cell mechanical properties in adherent cells. Following its invention in 1986 as a high-resolution imaging tool, atomic force microscope (AFM) has rapidly become a popular method for studying ligand–receptor and cell–cell interactions, typically with the use of AFM tip functionalization with proteins and receptors as well as the mechanical properties of living cells in culture.Citation5–Citation8 Alterations of cell mechanical properties have been reported recently in different diseases such as cancer, arthritis, and cardiovascular disease.Citation9–Citation12 Furthermore, it is tempting to assume that pharmaceutical or genetic treatments might affect the mechanical properties of target cells in vitro. The purpose of this article is to provide a brief introduction to cell biomechanics and its relation to disease; to describe the AFM experiment, including principles of operation and methods of data analysis; to review recent findings in the area of cell mechanics with AFM; and to identify the current limits of the technology and future developments that would enhance transfer to the basic and clinical sciences to aid in the identification of novel cell biomechanical markers that might lead to improved detection, diagnosis, and treatment of disease.
Fundamentals of atomic force microscopy
AFM is a method with unique advantages for the study of cell mechanics, as long as it provides high sensitivity (subnanonewton), high spatial resolution (submicrometer), and the ability to be used for real-time measurements in a physiologic aqueous cell culture environment. Moreover, along with high-resolution scanning, AFM provides the ability to use nanoindentation of living cells, which allows direct correlation of local mechanical properties with underlying cytoskeletal structures,Citation13,Citation14 which has been further potentiated lately by the incorporation of AFM onto standard or confocal inverted fluorescence optical microscope.Citation13,Citation15,Citation16 Unlike most other cell-imaging techniques, atomic force microscopy is based on a direct mechanical interaction between the probe and the sample. In this sense, AFM is inherently an elastography instrument.Citation17 Moreover, AFM can get information about surfaces in situ and in vitro, if not in vivo, in air, in water, buffers, and other ambient media: it can scan surfaces with up to nanometer (molecular) resolution and up to 0.01 nm vertical resolution; it provides true three-dimensional (3D) surface topographical information; it can scan with different forces, starting from virtually zero to large destructive forces, and detect up to single-molecule forces; and it allows measurement of various biophysical properties of materials, such as elasticity, adhesion, hardness, friction, etc. Minimum preparation of the samples is required before and during the experiment. Finally, another advantage of the method is that it is easily accessible and commercially available to most investigators.
In principle, AFM is a relatively simple instrument with demonstrated resolution of fractions of a nanometer, more than 1000 times better than the optical diffraction limit. Its function is based on laser tracking of the deflection of a microscopic-sized cantilever probe as its tip scans, indents, or otherwise interacts with the sample. AFM consists of a microscale rectangular or “V”-shaped cantilever, typically made of silicon or silicon nitride, with a sharp tip (probe) at its end, with a tip radius of curvature on the order of 50–100 nm. It is this tip that actually comes in contact with the cell, while the cantilever serves as a soft spring to measure the contact force. The tip dimension determines the spatial resolution of the instrument. Therefore, sharpened pyramids, etched silicon cones, carbon nanotubes, and other high-aspect ratio tips have been developed to scan samples with ultra-high resolution.Citation18,Citation19 However, such tips have been shown to penetrate the cell membrane and cause damage to living cells, whereas the standard pyramid tip apparently does not penetrate the cell membrane.Citation20 Modified AFM probes with glass or polystyrene microsphere tips also have been used for some cellular applications to yield a more easily characterized tip geometry, though at the expense of decreased spatial resolution.Citation21,Citation22 The physical and geometric properties of the cantilever determine its spring constant, k, which typically ranges from 0.01 N/m to 1.0 N/m for cell mechanics applications and which is used to convert the measured cantilever deflection, h, into a contact force, F = k × h. When the tip is brought into proximity of a sample surface, forces between the tip and the sample lead to a deflection of the cantilever. Typically, the deflection is measured using a laser spot reflected from the top surface of the cantilever into an array of photodiodes. AFM probes often are coated with a thin layer of gold to increase reflectivity, especially for cell mechanics applications in which the laser intensity may be attenuated by the phenol red present in standard cell culture medium. As long as the parameters used to construct the topographical image of the sample are the z-position of the probe at each x–y pixel location, accuracy in the z-direction as well as in the x–y plane are critical in the optimal function of AFM, and this is achieved at a subnanometer level by ultrasensitive piezoelectric positioners used in the movement of the AFM probe. Although piezoelectric materials inherently are nonlinear and hysteretic, these effects can be overcome by software compensation (open-loop design) or direct strain-gauge monitoring (closed-loop design) to yield very precise positioning of the AFM tip. In contrast to the standard AFM configuration, where the sample is positioned relative to a stationary probe, for cell biology applications the entire AFM is placed on the stage of an inverted light microscope to allow simultaneous visualization, including fluorescence microscopy, of the cells,Citation21 in a configuration where the AFM probe is moved relative to a stationary sample.
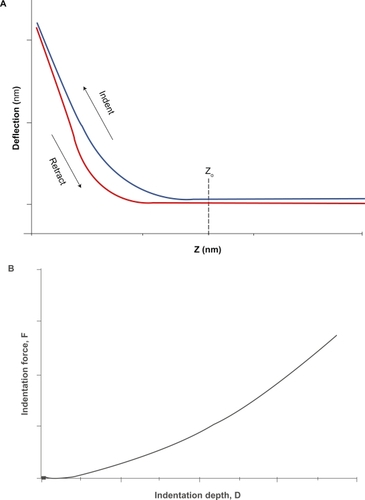
Although both primary forms of AFM imaging, ie, tapping and contact mode, have been used in cell elastography, contact mode is easier and more convenient to use than tapping mode, as long as it is more conducive to switching back and forth between imaging and “force mode,” in which nanoindentation is used to obtain quantitative stiffness measurements, while it can also give high-resolution images with cell viability sustained for several hours.Citation23 The indentation response depends on the spring constant of the probe, the geometry of the tip, and the mechanical properties of the sample. One also can vary the rate of indentation to study viscoelastic properties. Thus, by monitoring the z-position and deflection of the probe (the so-called “force curve”) (), one can obtain an indentation curve of indentation force versus depth () that can be analyzed to extract the elastic material properties of the sample as discussed below. Force mapping is a hybrid combination of imaging and force probing that involves making a series of in dentations in an array covering a region of interest on the sample and reconstructing an isoforce image from the z-position at which the probe reaches a preset constant deflection (ie, contact force).Citation24 In such images, larger z-values are interpreted as softer regions of the sample because a greater motion of the probe would have been required to achieve the preset force. However, in samples such as living cells, such images are complicated by the highly variable topography of the cell, which also influences the z-position at which a given contact force is achieved. Therefore, it is more accurate to analyze the indentation data and create an image that directly represents the elastic properties obtained at each pixel location. This is the method of AFM elastography. The retraction curve also contains useful information. Differences between the indentation and retraction curves reflect viscoelastic hysteresis of the sample. Upon retraction of the probe, the AFM tip may adhere to the sample and cause negative deflections of the probe. Such retraction events are the focus of experiments on protein unfolding,Citation25 receptor–ligand binding,Citation26 and cell–cell adhesion.Citation27
Figure 1 The AFM cell indentation experiment. A) The force curve obtained by measurements of cantilever deflection versus z-position during advancement and retraction of the probe. This curve provides information about the viscoelastic properties of the cell. Once the raw force curve is obtained and the contact point (Z0) identified, cell mechanical properties are obtained from the analysis of the curve of indentation force (F = k × h) versus depth (D = (Z − Z0) − h) B).
Cell mechanics
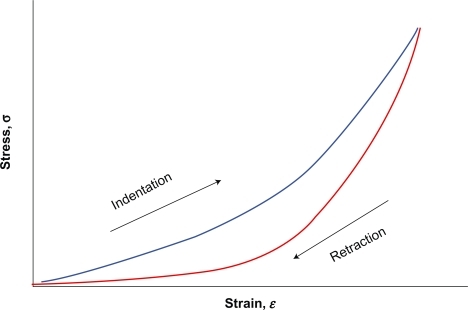
Knowledge of the relation of cell deformation (ie, strain) to internal forces and externally applied loads (ie, stress) acting on the cell is a sine qua non for the study of cell mechanics. As long as stiffness, defined as the slope of the force (F) -deformation (ΔL) curve, depends on geometric characteristics, which vary at each sample and testing device used, it is far better to study the related normalized quantities stress (σ = F/A) and strain ɛ = (ΔL/Lo), which are independent of size or geometry and rather reflect the underlying properties of the cell. The standard constitutive relation for solid materials is Hooke’s law, which states that stress is proportional to strain (σ = Eɛ), where E is the constant of proportionality called the Young’s modulus. Materials that follow Hooke’s law (eg, rubber, steel, bone) are called linear elastic. On the other hand, Newtonian fluids (eg, water, blood plasma) follow another similar constitutive relation for fluid materials which states that stress is proportional to the rate of strain (σ = μdɛ/dt), where the constant of proportionality, μ, is called the viscosity. However, being viscoelastic materials and characterized by heterogeneity, anisotropy, a nonlinear stress–strain relationship and hysteresis between the loading and unloading portions of the stress–strain curve, most soft biological tissues as well as individual cells are more complex than these simple idealized materials ().Citation28 Within cells the aqueous gel nature of the cytoplasm,Citation29,Citation30 heterogeneously distributed actin filaments, intermediate filaments, and microtubules,Citation31 cell adhesiveness,Citation32 or the presence of nucleus and other organellesCitation33,Citation34 are important factors that affect the mechanical properties of the cells. It is clear, therefore, that the mechanical behavior of such tissues and cells is not defined adequately by Young’s modulus. Constitutive equations that combine elastic and viscous properties are required to mathematically model their stress–strain behavior. Consequently, reported measurements of the Young’s modulus of cells must be interpreted with caution.
Figure 2 A nonlinear stress–strain relation (solid line) characterizes most biological soft tissues, with a viscoelastic hysteresis between loading and unloading segments of the curve, as opposed to the linear stress–strain curve of an idealized elastic material which is characterized by the Young’s modulus obtained from the slope of the line.
Since the earliest AFM studies of soft biological samples,Citation35,Citation36 the prevalent method of analyzing AFM indentation data has been application of the so-called “Hertz model” of contact between two elastic bodies.Citation37 In particular, the equations relating force and depth for indentation with a cone and a sphere, respectively, are given by:
and
where α is the semi-included angle of the cone tip, R is the sphere radius, and ν is the Poisson’s ratio that determines the amount of lateral expansion that accompanies axial compression (note that ν = 0.5 for water and other incompressible materials, and this value often is assumed for cells). As analysis based on the Hertz theory has been traditionally applied on thin films of gelatin, polyacrylamide, and similar materials, which actually satisfy several key assumptions of the theory (eg, thick films with homogeneous, isotropic, linear elastic material properties, axis symmetry; infinitesimal deformations; infinite sample thickness and dimensions; and a smooth sample surface), it is important to keep in mind that caution must be exercised when such theoretical solutions are applied to the more complex AFM–cell indentation problem, as long as it is not self-evident that these assumptions can be extrapolated to living cells.Citation38–Citation41 Fidelity of the calculated elastic properties also requires accurate identification of the contact point,Citation42 accurate calibration of the probe spring constant,Citation43 and accurate representation of the detailed tip geometry,Citation39,Citation40 each of which can be challenging procedures. To address some of the practical and theoretical limitations of the Hertz theory, various alternative approaches for analyzing AFM indentation data have been developed.Citation38–Citation40,Citation44,Citation45 A detailed analysis of these approaches is beyond the scope of the present paper. The multitude of the analysis theories used in cell indentation reveals the fact that a satisfying and more exact data analysis theory is still required, but this does not reduce the value of ongoing AFM indentation tests for the study of cell mechanical properties.
AFM elastography studies
As emphasized above, beyond imaging the surface topography, AFM is capable of mapping the elastic properties of living cells, which has yielded interesting insights into a number of physiologic cell processes. Several studies have used AFM over the last few years in the measurement of various mechanical propertiesCitation46,Citation47 of various cell types, such as fibroblasts,Citation44 endothelial,Citation48,Citation49 vertebrate cells,Citation50 etc, even between different regions within the same cell,Citation51–Citation53 and under various conditions. To elucidate how cell mechanical properties are related to the structure and function of the underlying cytoskeleton, a number of studies have examined the effects of chemical treatments or genetic mutations that target specific cytoskeletal constituents. In general, apart from the fact that internal cell organelles contribute to the overall cell mechanics, the actin cytoskeleton has a dominant effect on cell stiffness measured with AFM.Citation54–Citation56 Correlation of regional cell mechanics with underlying cytoskeletal components by combining AFM and fluorescent microscopy with immunolabeling showed that actin and intermediate filaments make a major contribution to elastic properties, whereas microtubules make a negligible contribution to cell elastic properties. Clearly, cell mechanics is an important indicator of cytoskeletal structure and function.Citation57 In particular, actin stress fibers are prominent linear structures comprised of actin and myosinCitation58 that provide a contractile apparatus in many cultured nonmuscle cell types, as well as in vascular endothelial cells in some physiologic conditions.Citation59,Citation60 AFM force mapping studies show that these structures are very stiff compared to any other cellular component.Citation4 Taken together with the growing data relating cell mechanical properties to cytoskeletal structure and substrate adhesion, these studies underscore the tremendous potential for AFM elastography of living cells to provide novel biomechanical markers that will enhance the detection, diagnosis, and treatment of disease.
Of particular interest are the growing number of studies that demonstrate a close association between cell mechanical properties and various disease conditions. For example, cultured myotubes from a dystrophin-deficient rat model of Duchenne muscular dystrophy were only one-fourth as stiff as normal cells,Citation61 and recent evidence suggests that some muscle types are protected from dystrophin deficiency by upregulating specialized accessory proteins that act to preserve cell stiffness.Citation62 In osteoarthritis, cartilage chondrocytes exhibit elevated viscoelastic moduli compared to cells from normal tissue,Citation34 which may underlie the dissimilar responses of these cells to external mechanical stimulation.Citation63 Differences in mechanical properties between normal hepatocytes and hepatocellular carcinoma cells were restricted to the elastic moduli, while the viscous modulus was unaltered.Citation64 On the other hand, pressure-overload ventricular hypertrophy specifically increases viscous damping (without affecting elastic stiffness) in passive cardiac myocytes.Citation11 Therefore, methods of elastographic mapping must be developed that are sensitive to changes in viscous as well as elastic properties of the cell. Erythrocytes from patients with sickle cell disease are stiffer and more viscous than are normal red blood cells.Citation12,Citation65 These mechanical properties are restored to near-normal values in patients treated with hydroxyurea,Citation12,Citation66 which suggests that measurements of cell mechanics also may be used to monitor the efficacy of therapeutic interventions. An area where AFM has been used rather extensively over the last several years is oncology. The reason why cancer cell mechanics measurement is more tempting compared to other diseases is that oncogenically transformed cells differ from normal cells in terms of cell growth, morphology, cell–cell interaction, organization of cytoskeleton, and interactions with the extracellular matrix.Citation67 Recently with the use of “sonocytology”, a method invented by Dr James K Gimzewski’s group,Citation68 which permits the detection by AFM of the vibrations of the cell membrane and their procession as a regular sound signal amplified up to the level of audible sound, it was discovered that cancerous cells emit a slightly different sound than healthy cells. More recently, in their ex vivo studies, Cross and colleaguesCitation69,Citation70 measured the stiffness of live metastatic cancer cells taken from the body (pleural) fluids of patients with suspected lung, breast and pancreas cancer, reported that cell stiffness of metastatic cancer cells is more than 70% softer than the benign cells that line the body cavity, and concluded that nanomechanical analysis correlates well with immunohistochemical testing currently used for detecting cancer. Because cell mechanical properties are determined largely, though not solely, by the underlying cytoskeleton, any disease process that alters the composition, organization, kinetics, or crosslinking of the cytoskeleton is likely to be detectable using single-cell elastography. Data on the mechanical properties of different cell types are critically needed to establish methodological criteria and guidelines for comparing measured mechanical properties with a normal population, as is being done for clinical hemorheology.Citation71,Citation72 Thus, the development of tools for reliable and rapid characterization of cell mechanical properties is essential. Finally, there are great perspectives in the use of AFM to study morphological and functional properties of various microorganisms, leading to the so-called “nanomicrobiology”.Citation73
Limitations of AFM elastography
All of its advantages notwithstanding, AFM still has a number of limitations. We have identified many limitations when applying the standard Hertz theory, and preliminary finite element models have motivated novel experiments and yielded alternative methods of analysis that promise to increase the information that can be obtained from AFM indentation tests.Citation40 Finite element models also can accommodate challenging aspects of the AFM indentation problem such as nonaxisymmetry of the tip geometry, inclination angle of the cantilever relative to the cell surface, the irregular topography of the cell, and the more complex cell mechanical properties, including nonlinearity, viscoelasticity, anisotropy, heterogeneity, and even multiphasic material composition. Such computational methods also will be critical in evaluating alternative theoretical models of the cell, including discrete structurally based models of the cytoskeleton.Citation74,Citation75 Furthermore, other limitations inherent to AFM technique itself are often impeding. In particular, accuracy of the cantilever spring constant, defined predominantly by variations in the thicknessCitation76 and stoichiometry,Citation77 has been one major limitation of using the AFM for quantitative measurements of mechanical properties, as long as it can result in spring-constant variability of nearly an order of magnitude between batch-produced wafers,Citation78 a disadvantage necessitating individual calibration for applications such as cell elastography, in which accuracy of the contact force is critical. Beyond the fact that eventually all measurements are made on the nonflat surface of the cells, another source of error in estimating mechanical properties from indentation tests is the identification of the exact point of contact between the AFM tip and the sample. In indentation on soft samples the transition from pre-contact to post-contact is smooth and obscured by noise in the data, not allowing thereby the contact point to be detected as a discontinuity in the slope (first derivative) or a spike in the curvature (second derivative) of the raw force curve, as is the case in stiff samples.Citation79 Another limitation of AFM elastography is that typical experiments are time-consuming and copious, so that a limited number of cells can be analyzed in one day. Another major challenge is to position automatically such irregular biological samples under the probe tip for consistent and rapid sequential testing and to avoid as much as possible the lateral drag of the cell by the tip. As AFM elastography evolves, the mechanical tests become increasingly sophisticated, and data sets become increasingly large and complex, computational methods and imaging techniques will play a critical role in the analysis and visualization of cell mechanics data, but also for simulation purposes to better understand how forces at the cantilever tip are manifest as the raw force curve data.
Conclusions
Over the last decade, AFM rapidly has become one of the most widely used and versatile tools for studying living cells. In particular, AFM elastography, which uniquely incorporates the capability for mechanical measurements and imaging of cell topography, holds great promise in the field of cell biology. A growing body of evidence relating cell mechanical properties to cytoskeletal structure and substrate adhesion suggests that single-cell elastography may provide sensitive indicators of the presence of disease. However, a number of technical and practical hurdles remain in the way of obtaining accurate and meaningful cell mechanics measurements with sufficient throughput that they will be practical for reliably examining large populations of cells. Nevertheless, as one powerful method of questioning, the future holds tremendous potential for AFM elastography of living cells to provide novel biomechanical markers that will enhance the detection, diagnosis, and treatment of disease.
Disclosure
The authors report no conflicts of interest in this work.
References
- ZhuCBaoGWangNCell mechanics: mechanical response, cell adhesion, and molecular deformationAnnu Rev Biomed Eng2000218922611701511
- ElsonELCellular mechanics as an indicator of cytoskeletal structure and functionAnnu Rev Biophys Chem198817397430
- PouratiJManiotisASpiegelDIs cytoskeletal tension a major determinant of cell deformability in adherent endothelial cellsAm J Physiol1998247C1283C12899612215
- RotschCRadmacherMDrug-induced changes of cytoskeletal structure and mechanics in fibroblsts: an atomic force microscopy studyBiophys J20007852053510620315
- TrickeyWRVailTPGuilakFThe role of the cytoskeleton in the viscoelastic properties of human articular chondrocytesJ Orthop Res20042213113914656671
- HeidelmannSRKaechSBuxbaumREMatusADirect observations of the mechanical behaviours of the cytoskeleton in living fibroblastsJ Cell Biol199914510912210189372
- SatoMNagayamaKKataokaNSasakiMHaneKLocal mechanical properties measured by atomic force microscopy for cultured bovine endothelial cells exposed to shear stressJ Biomech20003312713510609525
- CostaKDLeeEJHolmesJWCreating alignment and anisotropy in engineered heart tissue: role of boundary conditions in a model three-dimensional culture systemTissue Eng2003956757713678436
- LekkaMLaidlerPGilDLekkiJStachuraZHrynkiewiczAZElasticity of normal and cancerous human bladder cells studied by scanning force microscopyEur Biophys J19992831231610394623
- JonesWRTing-BeallHPLeeGMKelleySSHochmuthRMGuilakFAlterations in the Young’s modulus and volumetric properties of chondrocytes isolated from normal and osteoarthritic human cartilageJ Biomech19993211912710052916
- ZileMRRichardsonKCowlesMKConstitutive properties of adult mammalian cardiac muscle cellsCirculation1998985675799714115
- BrandãoMMFontesABarjas-CastroMLOptical tweezers for measuring red blood cell elasticity: application to the study of drug response in sickle cell diseaseEur J Hematol200370207211
- RotschCRadmacherMDrug-induced changes of cytoskeletal structure and mechanics in fibroblasts: an atomic force microscopy studyBiophys J20007852053510620315
- HassanEAHeinzWFAntonikMDRelative microelastic mapping of living cells by atomic force microscopyBiophys J199874156415789512052
- HagaHSasakiSKawabataKItoEUshikiTSambongiTElasticity mapping of living fibroblasts by AFM and immunofluorescence observation of the cytoskeletonUltramicroscopy20008225325810741677
- HortonMACharrasGBallestremCLehenkariPIntegration of atomic force and confocal microscopySingle Mols20001135137
- CostaKDSingle-cell elastography: probing for disease with atomic force microscopeDis Markers2003–200419139154
- TortoneseMCantilevers and tips for atomic force microscopyIEEE Eng Med Biol Mag19971628339086369
- DaiHHafnerJHRinzlerAGColbertDTSmalleyRENanotubes as nanoprobes in scanning probe microscopyNature1996384147150
- HaydonPGLartiusRParpuraVMarchese-RagonaSPMembrane deformation of living glial cells using atomic force microscopyJ Microsc19961821141208683560
- LehenkariPPCharrasGTNykanenAHortonMAAdapting atomic force microscopy for cell biologyUltramicroscopy20008228929510741681
- MahaffyREShihCKMacKintoshFCKasJScanning probe-based frequency-dependent microrheology of polymer gels and biological cellsPhys Rev Lett20008588088310991422
- HohJHSchoenenbergerCASurface morphology and mechanical properties of MDCK monolayers by atomic force microscopyJ Cell Sci1994107110511147929621
- RadmacherMFritzMKacherCMClevelandJPHansmaPKMeasuring the viscoelastic properties of human platelets with the atomic force microscopeBiophys J1996705565678770233
- ReifMGautelMOesterheltFFernandezJMGaubHEReversible unfolding of individual titin immunoglobulin domains by AFMScience1997276110911129148804
- YuanCChenAKolbPMoyVTEnergy landscape of the streptavidinbiotin complexes measured by atomic force microscopyBiochemistry200039102191022310956011
- BenoitMGabrielDGerischGGaubHEDiscrete interactions in cell adhesion measured by single-molecule force spectroscopyNature Cell Biol2000231331710854320
- FungYCBiomechanics: Mechanical properties of living tissues2nd edNew York, NYSpringer-Verlag1993
- BaggeUSkalakRAtteforsRGranulocyte rheology, experimental studies in an in vitro micro-flow systemAdv Microcirc197772948
- EvansEYeungAApparent viscosity and cortical tension of blood granulocytes determined by micropipet aspirationBiophys J1989561511602752085
- WangNMechanical interactions among cytoskeletal filamentsHypertension1998321621659674654
- PouratiJManiotisASpiegelDIs cytoskeletal tension a major determinant of cell deformability in adherent endothelial cells?Am J Physiol1998247C1283C12899612215
- PetersenNOMcConnaugheyWBElsonELDependence of locally measured cellular deformability on position on the cell, temperature, and cytochalasin BProc Nat Acad Sci U S A19827953275331
- GuilakFThe deformation behaviour and viscoelastic properties of chondrocytes in articular cartilageBiorheology200037274410912176
- WeisenhornALKhorsandiMKasasSGotzosVButtHJDeformation and height anomaly of soft surfaces studied with an AFMNanotechnology19934106113
- RadmacherMFritzMHansmaPKImaging soft samples with the atomic force microscope: gelatin in water and propanolBiophys J1995692642707669903
- HertzHÜber die Berührung fester elastischer Körper (On the contact of elastic solids)J Reine Angew Mathematik188192156171
- McElfreshMBaesuEBalhornRBelakJAllenMJRuddRECombining constitutive materials modelling with atomic force microscopy to understand the mechanical properties of living cellsProc Natl Acad Sci U S A2002996493649711983924
- MathurABCollinsworthAMReichertWMKrausWETruskeyGAEndothelial, cardiac muscle and skeletal muscle exhibit different viscous and elastic properties as determined by atomic force microscopyJ Biomech2001341545155311716856
- CostaKDYinFCAnalysis of indentation: implications for measuring mechanical properties with atomic force microscopyJ Biomech Eng199912146247110529912
- CharrasGTHortonMADetermination of cellular strains by combined atomic force microscopy and finite element modellingBiophys J20028385887912124270
- DimitriadisEKHorkayFMarescaJKacharBChadwickRSDetermination of elastic moduli of thin layers of soft material using the atomic force microscopeBiophys J2002822798281011964265
- SendenTJDuckerWAExperimental determination of spring constants in atomic force microscopyLangmuir19941010031004
- RotschCJacobsonKRadmacherMDimensional and mechanical dynamics of active and stable edges in motile fibroblasts investigated by using atomic force microscopyProc Natl Acad Sci U S A1999969219269927669
- RicciDTedescoMGrattarolaMMechanical and morphological properties of living 3T6 cells probed via scanning force microscopyMicrosc Res Tech1997361651719080406
- BraetFde ZangerRSeynaeveCBaekelandMWisseEA comparative atomic force microscopy study on living skin fibroblasts and liver endothelial cellsJ Electron Microsc200150283290
- CollinsworthAMZhangSKrausWETruskeyGAApparent elastic modulus and hysteresis of skeletal muscle cells throughout differentiationAm J Physiol Cell Physiol2002283C1219C122712225985
- KataokaNIwakiKHashimotoKMeasurements of endothelial cell-to-cell and cell-to-substrate gaps and micromechanical properties of endothelial cells during monocyte adhesionProc Natl Acad Sci U S A200299156381564312434019
- SatoMNagayamaKKataokaNSasakiMHaneKLocal mechanical properties measured by atomic force microscopy for cultured bovine endothelial cells exposed to shear stressJ Biomech20003312713510609525
- DvorakJANagaoEKinetic analysis of the mitotic cycle of living vertebrate cells by atomic force microscopyExp Cell Res199824269749665803
- NagaoEDvorakJAPhase imaging by atomic force microscopy: analysis of living homoiothermic vertebrate cellsBiophys J1999763289329710354454
- MathurABTruskeyGAReichertWMAtomic force and total internal reflection fluorescence microscopy for the study of force transmission in endothelial cellsBiophys J2000781725173510733955
- YamaneYShigaHHagaHKawabataKAbeKItoEQuantitative analyses of topography and elasticity of living and fixed astrocytesJ Electron Microsc200049463471
- HendersonEHaydonPGSakaguchiDSActin filament dynamics in living glial cells imaged by atomic force microscopyScience1992257194419461411511
- RotschCBraetFWisseERadmacherMAFM imaging and elasticity measurements on living rat liver macrophagesCell Biol Int1997216856969817809
- WuHWKuhnTMoyVTMechanical properties of L929 cells measured by atomic force microscopy: effects of anticytoskeletal drugs and membrane crosslinkingScanning1998203893979737018
- ElsonELCellular mechanics as an indicator of cytoskeletal structure and functionAnnu Rev Biophys Biophys Chem1988173974303293593
- LangangerGMoeremansMDaneelsGSobieszekADe BrabanderMDe MeyJThe molecular organization of myosin in stress fibers of cultured cellsJ Cell Biol19861022002093510218
- SipkemaPvan der LindenPJWWesterhofNYinFCPEffect of cyclic axial stretch of rat arteries on endothelial cytoskeletal morphology and vascular reactivityJ Biomech20033665365912694995
- WhiteGEFujiwaraKExpression and intracellular distribution of stress fibers in aortic endotheliumJ Cell Biol198610363703722269
- PasternakCWongSElsonELMechanical function of dystrophin in muscle cellsJ Cell Biol19951283553617844149
- PorterJDMerriamAPKhannaSConstitutive properties, not molecular adaptations, mediate extraocular muscle sparing in dystrophic mdx miceFASEB J20031789389512670877
- SalterDMMillward-SadlerSJNukiGWrightMODifferential responses of chondrocytes from normal and osteoarthritic human articular cartilage to mechanical stimulationBiorheology20023994108
- WuZZZhangGLongMWangHBSongGBCaiSXComparison of the viscoelastic properties of normal hepatocytes and hepatocellular carcinoma cells undercytoskeletal perturbationBiorheology20003727929011145074
- NashGBJohnsonCSMeiselmanHJMechanical properties of oxygenated red blood cells in sickle cell (HbSS) diseaseBlood19846373826689955
- BallasSKDoverGJCharacheSEffect of hydroxyurea on the rheological properties of sickle erythrocytes in vivoAm J Hematol1989321041112757007
- SokolovIAtomic force microscopy in cancer cell researchNalwaHNWebsterTCancer NanotechnologyValencia, CAAmerican Scientific Publishers2007117
- PellingAESehatiSGrallaEBValentineJSGimzewskiJKLocal nanomechanical motion of the cell wall of Saccharomyces cerevisiaeScience20043051147115015326353
- CrossSEJinYSRaoJGimzewskiJKNanomechanical analysis of cells from cancer patientsNat Nanotech20072780783
- CrossSEJinYSTondreJWongRRaoJGimzewskiJKAFM-based analysis of human metastatic cancer cellsNanotechnology200819384003 doi: 10.1088/0957-4484/19/38/384003.
- WautierJLSchmid-SchonbeinGWNashGBMeasurement of leukocyte rheology in vascular disease: clinical rationale and methodology: International society of clinical hemorheologyClin Hemorheol Microcirc19992172410517484
- DobbeJGGHardemanMRStreekstraGJStrackeeJInceCGrimbergenCAAnalyzing red blood cell deformability distributionsBlood Cells Mol Dis20022837338412367581
- DufrêneYFTowards nanomicrobiology using atomic force microscopyNat Rev Microbiol2008667468018622407
- IngberDETensegrity: the architectural basis of cellular mechanotransductionAnnu Rev Physiol1997595755999074778
- OhashiTIshiiYIshikawaYMatsumotoTSatoMExperimental and numerical analyses of local mechanical properties measured by atomic force microscopy for shear endothelial cellsBiomed Mater Eng20021231932712446947
- WeisenhornALMaivaldPButtHJHansmaPKMeasuring adhesion, attraction, and repulsion between surfaces in liquids with an atomic force microscopePhys Rev B1992451122611232
- ButtHJSiedlePSeifertKScan speed limit in atomic force microscopyJ Microsc19931697584
- ClevelandJPManneSBocekDHansmaPKA nondestructive method for determining the spring constant of cantilevers for scanning force microscopyRev Sci Instrum199364403405
- RadmacherMMeasuring the elastic properties of living cells by the atomic force microscopeMethods Cell Biol200268679012053741