Abstract
Early cancer detection is a major factor in the reduction of mortality and cancer management cost. Here we developed a smart and targeted micelle-based contrast agent for magnetic resonance imaging (MRI), able to turn on its imaging capability in the presence of acidic cancer tissues. This smart contrast agent consists of pH-sensitive polymeric micelles formed by self-assembly of a diblock copolymer (poly(ethyleneglycol-b-trimethylsilyl methacrylate)), loaded with a gadolinium hydrophobic complex (tBuBipyGd) and exploits the acidic pH in cancer tissues. In vitro MRI experiments showed that tBuBipyGd-loaded micelles were pH-sensitive, as they turned on their imaging capability only in an acidic microenvironment. The micelle-targeting ability toward cancer cells was enhanced by conjugation with an antibody against the MUC1 protein. The ability of our antibody-decorated micelles to be switched on in acidic microenvironments and to target cancer cells expressing specific antigens, together with its high Gd(III) content and its small size (35–40 nm) reveals their potential use for early cancer detection by MRI.
Introduction
The recent developments in imaging techniques such as computed tomography, fluorescence imaging, positron emission spectroscopy, and magnetic resonance imaging (MRI) have led to major advances in the field of biomedical diagnosis and therapy. Currently, MRI is one of the most powerful diagnostic techniques for molecular imaging, mainly due to its noninvasive nature allied to other benefits, such as the ability to obtain direct multiplanar images, differentiate soft tissues, and obtain anatomical data while avoiding ionizing radiation. In addition, MRI allows sub-millimeter spatial resolution and has an excellent contrast resolution.Citation1 However, in terms of sensitivity, MRI still lags behind other imaging tools. For example, MRI has a sensitivity of approximately 10−3 mol L−1 to 10−5 mol L−1, which compared to other types of imaging, can be very limiting. Also, because healthy and pathological tissues as well as distinct diseases show similar magnetic moments, they produce a poor image contrast. Thus, in order to overcome these shortcomings and obtain more informative images, a substantial quantity of contrast agent (CA) needs to be injected.Citation2 CA molecules are paramagnetic chemical substances that act by shortening the longitudinal and transversal relaxation times of water protons, T1 and T2, respectively. The most frequently used in clinical MRI are small molecules of Gd(III) chelates, which are T1 agents that due to their low molecular weight can distribute uniformly to all perfused tissues throughout the vasculature and can diffuse across endothelial wall vessels into the extracellular spaces.Citation1,Citation2 Free Gd(III) is toxic because its diameter is close to that of calcium ions and, for this reason, it has to be chelated to a ligand. These ligands reduce Gd(III) toxicity significantly and influence the pharmacokinetics of the complex.Citation3 Then again, these low-molecular weight CAs suffer from some weaknesses such as lack of selectivity toward particular organs or pathological areas, absence of response toward cellular microenvironment, short half-life in blood (a large part of administered CA is eliminated before images are taken), and potential toxic side effects for some patients.Citation4
Recently, scientists have developed new classes of medical imaging probes with low molecular weight called MRI smart CAs to overcome the classic CAs limitations. Unlike standard CAs, which enhance MR image constantly, smart CAs change between two states, depending on the type of the stimulus.Citation5,Citation6 This change is detected as an alteration in signal, because one of the states has low or no enhancement effect (“off state”), while the other induces a high enhancement (“on state”). These stimulus-sensitive agents can be detected when switching from one state to another after exposure to a metabolic or physiological event in a specific molecular target. A variety of smart CAs have been developed that become activated in response to changes in pH,Citation7–Citation13 to the presence of metal ionsCitation14–Citation16 through enzymatic activation,Citation17 and to the presence of oxygenated hemoglobinCitation18 or radicalsCitation19 or upon exposure to ultrasoundsCitation20 or changes in temperature.Citation21,Citation22 These smart CAs have proven to be excellent MRI tools for very specific targets in vitro and in vivo.Citation7,Citation8 On the other hand, targeted nanoparticles such as liposomes,Citation23 dendrimers,Citation24 and micelles provide several unique features and capabilities such as to house a large number or different types of functional groups that can be used in multiple diagnostic (eg, gadolinium complex) and therapeutic (eg, anticancer) agents. Moreover, nanoparticles with the size ranging from 10 nm to 100 nm accumulate preferentially at tumor sites through an effect called enhanced permeability and retention.Citation25 Finally, the capability of these nanoparticles of encapsulating a large amount of molecules (eg, gadolinium complex) inside their core will allow the detection of cancer at early stages (0, I, and II) due to the signal amplification generated by the large number of CAs released from the nanoparticles at the cancer tissue sites.
In this work, we aimed to develop a new type of smart CAs based on polymeric nanoparticles capable of amplifying the MRI signal chemically and at the same time have the ability to remain silent during circulation and active at the target site. These newly developed nanoparticle-based CAs consist of pH-sensitive polymeric stealth PEGylated micelles formed by self-assembly of a diblock copolymer, poly(ethyleneglycol-b-trimethylsilyl methacrylate) (PEG-b-PTMSMA), loaded with the hydrophobic complex tetraaquodichloro(4,4′-di-t-butyl-2,2′-bipyridine) gadolinium(III) chloride (tBuBipyGd). To enhance the targeting ability of these polymeric nanoparticles toward cancer cells, we decorated micelles with monoclonal antibody (mAb) against the human MUC1 protein, which is aberrantly expressed in many epithelial cancers, including breast cancer.Citation26,Citation27
Materials and methods
Reagents and equipment
Ethanol (96%) was from AGA, tetrahydrofuran (THF), HPCL grade, from Ridel-de-Haën, hydrochloric acid (37%) from Merck, N,N-dimethylformamide (DMF) and isopropanol from BDH Prolabo, methoxypolyethylene glycol N-hydroxysuccinimide ester and paraformaldehyde from Sigma-Aldrich, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT) from Applichem, and Trizol reagent from Invitrogen. For cell culture, complete Dulbecco’s Modified Eagle’s Medium or RPMI1640 culture media (DMEM or RPMI1640 basic medium [Lonza] supplemented with 10% fetal bovine serum [PAA], 2 mM glutamine, 100 units mL−1 penicillin, and 100 μg mL−1 streptomycin) were prepared. We purchased mAbs against the human MUC1 protein, C595 (Santa Cruz Biotechnology) and SM3 (eBioscience), and phycoerythrin-conjugated goat anti-mouse secondary antibody (Biolegend). Oligonucleotide primers were synthesized by Sigma-Aldrich. Milli-Q water (Millipore) was used in all procedures. For dialysis, cassettes were purchased from Pierce Biotech, 10-kDa MWCO. For 1H nuclear magnetic resonance (NMR) experiments, a Bruker ARX 400 (400 MHz) spectrometer was used, and MR images were obtained in a Signa 1.5T SYS#GEMSOW, GE Healthcare, clinical MRI. Fourier transform infrared spectroscopy spectrum was recorded in a KBr pellet within the range of 4,000–600 cm−1 in a Bruker, Tensor 27 spectrophotometer. Elemental analysis was performed on a Truspec 630-200-200 apparatus. Steady-state fluorescence spectra were recorded on a SPEX Fluoromax-3 spectrofluorometer (Jobin Yvon–Horiba). All emission spectra were carried out in 1-cm quartz cuvettes (Hellma Quartz-Suprasil), using λexc =334 nm, collecting the emission from 360 nm to 510 nm (increment of 1 nm), using 1-nm slit widths in excitation and emission (wavelength resolution of 1 nm), and corrected for nonlinear instrument response. For MTT assays, a Tecan Infinite M200 monochromator-based multifunction microplate reader was used. Nucleic acid concentration was determined on a NanoDrop 2000c Spectrophotometer (Thermo Scientific). Polymerase chain reaction (PCR) amplification was performed on a CFX96 Real-Time PCR Detection System (Bio-Rad). Flow cytometry was performed on a FACS Calibur analyzer, and data were processed with Cell Quest software (BD Biosciences). White light and fluorescence observation and image acquisition of living cells was performed with an inverted fluorescence microscope (Leica DM IL) coupled to a digital CCD camera (Evolution MP-5.1, Media Cybernetics). Observation and image acquisition of fixed cells was performed on an Axio Observer Z2 Fluorescence microscope (Zeiss), coupled to a digital AxioCam camera (Zeiss). Images were processed using AxioVision software (Zeiss). All light-scattering experiments were performed using Brookhaven Instruments BI-200SM Multiangle DLS & SLS Light Scattering System.
Diblock co-polymer synthesis
The macro-CTA weighing 0.8 g was placed in a Schlenk tube, previously charged with nitrogen and dissolved in 10 mL of THF. A volume of 3.5 mL (45 eq) of trimethylsilyl methacrylate and 0.030 g (1 mol% of the monomer) of AIBN were added. After three freeze–pump–thaw cycles, the mixture was incubated in an oil bath at 60°C, under stirring, for 24 hours. The flask was then cooled to room temperature, 10 mL of THF added to dilute the mixture, which had become very viscous, and its content transferred to a round-bottomed flask. After evaporation of the solvent, the white residue was dissolved in methanol and the polymer precipitated by the addition of water. The polymer was separated from the supernatant by centrifugation at 15,344× g during 30 minutes, followed by decantation (). This procedure was repeated one more time, affording a white solid that was vacuum dried, at 60°C, overnight, yielding 1.47 g (40%) 1H NMR (DMSO) δ 0.056 (s, (CH3)3Si), 1.086 (s, CH3C–C=O), 1.177 (s, CH3C–C=O), 1.878 (s, CH2C–C=O), 1.992 (s, CH2C–C=O), 3.356 (s, CH3O–(CH2CH2O)n), 3.639 (s, (CH2CH2O)n), and 4.660 (q, CH3CH2O–C=S); DPn (NMR) =45. Gel permeation chromatography (GPC) analysis (PSSNa standards) reveals a monomodal molecular weight distribution, from which a value of Mn =9 kDa was predicted.Citation28
Figure 1 (A) Synthesis strategy of the co-polymer poly(ethyleneglycol-b-trimethylsilyl methacrylate); (B) synthesis strategy of tBuBipyGd complex; and (C) encapsulation of contrast agent during the self-assembly of polymeric nanoparticles (micelles).
Abbreviations: THF, tetrahydrofuran; h, hour; AIBN, azobisisobutyronitrile; DMSO, dimethyl sulfoxyde.
Preparation of tBuBipyGd complex
The preparation of this complex () was based on a well-described method.Citation29,Citation30 A mixture of 0.500 g (0.830 mmol) of GdCl3·4H2O and 0.223 g (0.830 mmol) of tBuBipy in 25 mL of ethanol was stirred and refluxed overnight. The resulting solution was concentrated, and the final product was obtained by the addition of diethyl ether. The white solid was washed with diethyl ether to remove unreacted tBuBipy and dried under reduced pressure to afford 0.450 g (90%) of tBuBipyGd complex. Elemental analysis theoretical values are as follows: 35.77% C; 5.34% H; 4.64% N; and the obtained values were as follows: 35.99% C; 5.59% H; 4.69% N. Fourier transform infrared spectroscopy (KBr) values were as follows: 2,963 cm−1 (s), 2,866 cm−1 (s), 1,594 cm−1 (vs), 1,545 cm−1 (vs), 1,374 cm−1 (s), 837 cm−1 (s), 605 cm−1 (s). To obtain a 50 mM stock solution, 5.1 mg of the complex was dissolved in 169 μL of ethanol. The solution was kept at −20°C.
Preparation of fluorophore-loaded and tBuBipyGd complex-loaded polymeric micelles
Polymeric micelles loaded with tBuBipyGd complex were obtained as follows: 30 mg of PEG-b-PTMSMA and 0.5 mg of either 1-methylpyrene or tBuBipyGd complex was dissolved in 0.3 mL of DMF. The mixture was stirred for 3 hours at room temperature. Subsequently, 0.7–1.0 mL of pure Milli-Q water was added at a rate of one drop every 10 seconds to induce micellization. The resulting micelle solution was then dialyzed against 4 L of Milli-Q water for 3 days at room temperature, using a 10-kDa cut-off dialysis cassette to exclude unencapsulated compound or nonself-assembled polymer molecules. The Milli-Q water was replaced twice a day.
Measurement of Gd(III) complex and fluorophore concentrations loaded in micelles
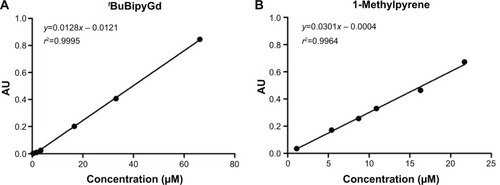
A 6.62×10−5 M stock solution of Gd(III) complex in ethanol was diluted to obtain standards with the following concentrations: 3.31×10−5 M, 1.66×10−5 M, 3.31×10−6 M, 1.66×10−6 M, and 3.31×10−7 M, and then obtain a Gd(III) complex calibration curve (). The corresponding absorbance values were recorded at 282 nm, and the molar absorption coefficient was 1.28×104 dm3 mol−1 cm−1. From a 2.17×10−5 M stock solution of 1-methylpyrene in THF, five standards of concentrations 1.63×10−5 M, 1.09×10−5 M, 8.68×10−6 M, 5.43×10−6, and 1.09×10−6 M were prepared and a 1-methylpyrene calibration curve was obtained (). The absorbance values were measured at 344 nm and a value of 3.01×104 dm3 mol−1 cm−1 for the molar absorption coefficient was obtained. For the determination of Gd(III) complex or fluorophore concentrations in loaded micelles, each solution was diluted (30 μL of solution to a total volume of 3 mL) in a spectrophotometer cuvette containing the appropriate solvent (ethanol for the former and THF for the latter) and the absorbance was read at 282 nm or 344 nm, respectively. The average concentration of tBuBipyGd complex in the micelles was 0.065 mM, which represents 70% yield of contrast encapsulation. The average concentration of 1-methylpyrene in micelle solution was found to be 0.96 mM.
Bioconjugation of polyethylene glycol with MUC1 antibodies
Bioconjugation of PEG with C595 or SM3 mAb was performed as follows: 3 mg of methoxypolyethylene glycol N-hydroxysuccinimide ester was added to a solution containing 3 μg of mAb in 0.1 M bicarbonate buffer (500 μL, pH 8.3). The mixture was gently stirred for 5 days in slow tilt rotation at 4°C.
Preparation of fluorophore-loaded bioconjugated micelles
Fluorophore-loaded bioconjugated micelles were prepared by mixing 30 mg of PEG-b-PTMSMA with 0.5 mg of 1-methylpyrene and dissolved in 0.4 mL of DMF. The mixture was stirred for 3 hours at room temperature. Subsequently, 0.5 mL PEG-Anti-MUC1 aqueous solution was added at a rate of one drop every 10 seconds and, immediately after, 0.5 mL of cold Milli-Q water was added at the same rate to induce micellization. The resulting micelle solution was then placed in a dialysis cassette with a 10-kDa cut-off, to exclude unencapsulated compounds (either 1-methylpyrene or Gd complex) or possibly nonself-assembled polymer molecules and dialyzed against cold Milli-Q water for 3 days at 4°C. The recovered solution was then centrifuged to remove the free PEG-Anti-MUC1 and then the pellet was diluted once again with Milli-Q water.
Assessment of micelle stability in water and cell culture medium
1-Methylpyrene–loaded micelle solution (200 μL) was added to a cuvette containing 800 μL of either water or complete DMEM culture medium, and the fluorescence spectrum was acquired. The spectrum of 1-methylpyrene–loaded micelles bioconjugated with antibody in culture medium was also obtained.
Determination of the influence of tBuBipyGd complex in the T1 value of water
tBuBipyGd complex–loaded micelle aqueous solution (10 μL) was placed in an NMR tube containing deuterated water, and T1 of water protons was measured using the inversion recovery method. The sequence was [180°x′ (16 μs) − τ (0.05, 0.25, 0.5, 1, 1.5, 2, 3, 5, and 10 seconds) − 90°x′ (8 μs) − Ta (equal to τ) − D (12 seconds)]8. Subsequently, approximately 10 μL of acidic solution (pH 5.5) was added to the tube to induce micelle bursting, and thus tBuBipyGd complex release. The above procedure was repeated. Data were fit to an exponential, from which T1 values were derived, using Bruker Topspin 2.0 software.
Determination of the tBuBipyGd complex imaging ability in vitro
To image tBuBipyGd complex released from micelles, the same volume of tBuBipyGd complex–loaded micelles in serum was dispensed in two microfuge tubes. Next, hydrochloric acid was added to only one of them to obtain approximately pH 5.5. Both tubes were positioned on top of an MRI phantom for calibration. MR images were subsequently acquired with a Spin Echo sequence (TR/TE=300/12 ms; slice thickness, 3 mm; field of view, 18×18 cm; matrix, 256×256).
Cell lines
The MCF-7 and MDA-MB-468 (human breast adenocarcinoma), HEK293T (Human Embryonic Kidney), and S17 (mouse bone marrow stroma) cell lines were maintained in complete DMEM culture medium. The Jurkat (human T-cell leukemia) cell line was maintained in complete RPMI1640 culture medium. Cells were cultured at 37°C under humidified atmosphere of 5% CO2, unless stated otherwise.
In vitro cytotoxicity tests
Jurkat or MCF-7 cells were seeded on 96-well culture plates. Different concentrations (5 μM, 10 μM, 15 μM, and 20 μM) of tBuBipyGd complex buffer solution was added to the culture medium and incubated for 3 days. In addition, different concentrations of free and encapsulated tBuBipyGd complex (at concentrations of 10 μM or 50 μM) were added to the cells and incubated for 3 days. These assays included tBuBipyGd-loaded micelles dialyzed for 3 days and also nonencapsulated micelles. After incubation, toxicity was evaluated by the MTT assay. Briefly, 10 μL of 5 mg mL−1 MTT solution was added to each well followed by further incubation for 4 hours. Then, 100 μL of 0.04 N HCl in isopropanol was added to the wells and formazan crystals were allowed to solubilize for 1 hour at room temperature. The formazan concentration was quantified by measuring the absorbance at 570 nm and 630 nm. The final optical density was calculated following the formula: optical density = L570 nm–L630 nm, where L was the obtained absorbance value for each wavelength. The toxicity was assessed by calculating the percentage of cell survival in relation to the control untreated group. The results were statistically compared by Student’s t-test using GraphPad Prism software.
Detection of MUC1 expression by reverse-transcriptase PCR and immunofluorescent flow cytometry
Total RNA was extracted from cultured cell lines using Trizol following the manufacturer’s instructions. The concentration and purity of isolated RNA was measured by spectrophotometry, and its quality assessed by agarose gel migration. RNA treated with RNase-free DNAse I (Fermentas) was reversed transcribed using the RevertAid First Strand cDNA Synthesis Kit (Fermentas), and quantitative PCR was performed using the iQ SYBR-Green Supermix kit (Bio-Rad). Primers to detect expression of MUC1 and the ACTB reference gene were used at a 200 mM concentration: MUC1-Forward, 5′-GTG CCC CCT AGC AGT ACC G-3′; MUC1-Reverse, 5′-GAC GTG CCC CTA CAA GTT GG-3′; ACTB-Forward, 5′-AGG CCA ACC GCG AGA AGA TGA C-3′; and ACTB-Reverse, 5′-AGG TCC AGA CGC AGG ATG GCA T-3′. Relative mRNA expression levels were calculated through the 2−ΔΔCt method. For MUC1 protein detection by flow cytometry, HEK293T, MCF-7, and MDA-MB-468 cells were dissociated by 0.02% EDTA treatment, washed, and labeled with SM3 mAb (1/500 dilution), C595 mAb (1/100 dilution), or an isotypic nonspecific antibody and phycoerythrin-conjugated goat anti-mouse secondary antibody. Each antibody incubation was performed for 1 hour on ice and was followed by PBS/3% fetal bovine serum/5 mM NaN3 washes, before flow cytometry analysis.
Assessment of polymeric micelle cellular uptake by microscopy
MCF-7 or MDA-MB-468 cells were seeded in culture plates and allowed to attach for 24 hours at 37°C. Micelles bioconjugated with C595 or SM3 antibody and loaded with 1-methylpyrene were added to the cells and incubated for 1 hour at 37°C. As a control, we used non-conjugated 1-methylpyrene–loaded polymeric micelles. Excess micelles were removed by washing three times with PBS. For mixed cell cultures, MDA-MB-468 and S17 cells were either grown together (proportion of 1–5, respectively) or alone on glass cover slips. The cells were allowed to adhere for 24 hours at 37°C, and then, anti-MUC1–conjugated 1-methylpyrene–loaded micelles were added to the cells and incubated for 1 hour at 37°C. Cells were then washed three times with PBS and fixed with 4% paraformaldehyde for 30 minutes. After washing, the cover slips were mounted on microscope slides and observed on a fluorescence microscope.
Results
Polymeric micelle preparation
Poly(ethylene glycol-b-trimethylsilyl methacrylate), which contains silicon moieties that can be cleaved under mildly acidic conditions (pH between 5 and 6), was synthesized by Reversible Addition-Fragmentation chain Transfer method (). The amphiphilic diblock co-polymer (PEG-b-PTMSMA) micelles were designed to accommodate the hydrophobic CA (tBuBipyGd) within its core and to release its load at the site of interest, via the hydrolysis of its silyl ester groups located in the hydrophobic moiety of the copolymer in a mildly acidic medium, such as cancer tissue.
The tBuBipyGd complex was obtained from the reflux of GdCl3·4H2O and tBuBipy in ethanol. The structure [GdtBuBipy(H2O)4(Cl)2]Cl was assessed based on the literature data for the complex [GdBipy(H2O)4(Cl)2]Cl (). Elemental analysis and Fourier transform infrared spectroscopy spectrum were in accordance with the proposed structure. We chose this complex as a model compound because of its hydrophobicity, conferred by the tert-butyl group in the bipyridine core, as well as the expected strong MR signal due to the high number of coordinated water molecules.
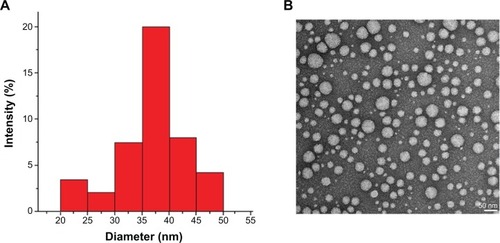
In the encapsulation process, tBuBipyGd complex and PEG-b-PTMSMA were co-dissolved in DMF and self-assembly was induced by slow addition of water. Due to its hydrophobic nature, the complex tends to lay within the micelle core (). Organic solvent was then removed by dialysis to yield micelles with the size ranging from 35 nm to 40 nm (). The tBuBipyGd complex concentration in micelles determined by spectrophotometry was on average of 0.065 mM.
Figure 3 (A) Dynamic light-scattering results for loaded polymeric micelles after the encapsulation of the contrast agent. (B) TEM of contrast-loaded polymeric micelles after the self-assembly of poly(ethylene glycol-b-trimethylsilyl methacrylate).
Note: Magnification: ×200,000. Bar scale: 50 nm.
Abbreviation: TEM, transmission electron microscopy.
The approximate micelle aggregation number (Nagg) can be calculated from the ratio of the average molecular weight of the micelle to the molecular weight of the block copolymer. Light-scattering experiments (SLS and DLS) indicated that the average molecular weight of the micelle is 8.1×104 Da (individual co-polymer weight of around 9 kDa). Thus, we estimated an aggregation number of 9. Accordingly, the maximum number of micelles formed in a solution of 1 mL was found to be approximately 4.2×1010. By correlating the dye concentration with its absorbance at 344 nm, we quantified the total number of dye molecules as 6.3×1015 after they were released into the solution. Because we washed off any unencapsulated dye in the solution before the destruction of micelles, we can estimate that one micelle with an average diameter (Rh=30 nm, Nagg~9) can load at least 150,000 dye molecules and around 75,000 Gd(III) complex if we assume that the volume of Gd(III) doubles the volume of 1-methylpyrene. Therefore, the release of these dye molecules into solution should provide a significant signal amplification effect for better resolution and sensitivity.
Polymeric micelle stability and selectivity study
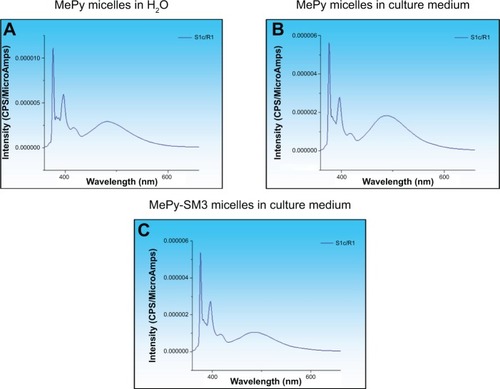
To test the in vitro stability and uptake of micelles by cells, 1-methylpyrene fluorophore was loaded into micelles instead of tBuBipyGd complex, because of the feasibility of fluorescence detection and quantification in cells. The concentration of 1-methylpyrene in these micelles, as determined by spectrophotometry, was on average of 0.96 mM. In the fluorescence spectrum of 1-methylpyrene–loaded micelles (), the observation of both monomer (375 nm) and excimer (480 nm) emission bands clearly confirms the encapsulation of the fluorophore.Citation28 When nonconjugated and bioconjugated micelles were diluted in cell culture medium, there were no significant changes in the fluorophore fluorescence spectrum (), which is indicative of maintenance of micelle integrity. However, and in spite of being stable in aqueous solutions for weeks, when the solution pH was lowered to values between 5 and 6, a progressive release of the fluorophore was observed.Citation28
Figure 4 1-Methylpyrene (MePy) fluorophore-loaded polymeric micelle stability in water and cell culture medium.
Notes: Fluorescence intensity spectra of 1-methylpyrene monomer (375 nm) and excimer (480 nm) was measured to detect disintegrated and intact micelles, respectively, and these were found to be stable in (A) water and (B) culture medium. (C) Micelle bioconjugation with the SM3 MUC1 monoclonal antibody did not alter stability in culture medium.
Determination of PEG-b-PTMSMA micelle-based CA imaging ability
The paramagnetic Gd(III) ions in complexes used as CA in MRI shorten the relaxation time T1 of the water protons in their proximity, leading to an increase of the image contrast. Therefore, restricting water access to those chelates should prevent them from decreasing the T1 of the surrounding water molecules. As a result, the CA remains switched-off in neutral pH but switched-on at acidic pH, which is a typical characteristic of malignant tumors. This feature was first demonstrated in vitro using 1H NMR, where the T1 of water protons was initially measured in a sample containing intact tBuBipyGd-loaded micelles and it was found to be 3.4 seconds, but upon micelle disruption, by the addition of an acid solution (pH 5.5), the T1 value dropped to 1.2 seconds ().
Figure 5 (A) Scheme of 1H NMR relaxometric experiment to measure T1 of water before and after the release of the contrast agent from the polymeric micelles. (B) Magnetic resonance image of microfuge tubes containing tBuBipyGd complex–loaded micelles, at pH 7 (square a) and at pH 5.5 (square b).
Abbreviation: NMR, nuclear magnetic resonance.
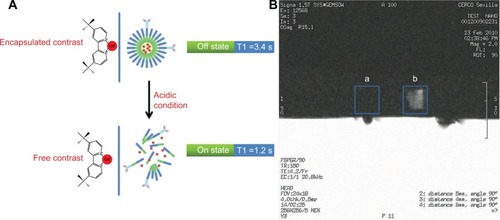
Further testing using a clinical MRI was performed. This experiment consisted of imaging two microfuge tubes containing tBuBipyGd complex–loaded micelle in water, one at neutral pH and the second with pH 5.5. The first sample, with neutral pH, did not change the background image signal and no contrast was observed (, square a), while the second tube presented a significant signal enhancement visible as a bright area (, square b). These results indicate that the micelles disassembled at low pH and released the tBuBipyGd complex. Consequently, the complex interacted with the surrounding water molecules and decreased the T1 relaxation time, which leads to an increase in the image signal intensity from that specific area. Together, these experiments show the ability of these tBuBipyGd-loaded micelles to respond to an acidic condition by changing their states (on and off), as indicated by the alteration in T1 time and image signal intensity.
In vitro cytotoxicity tests
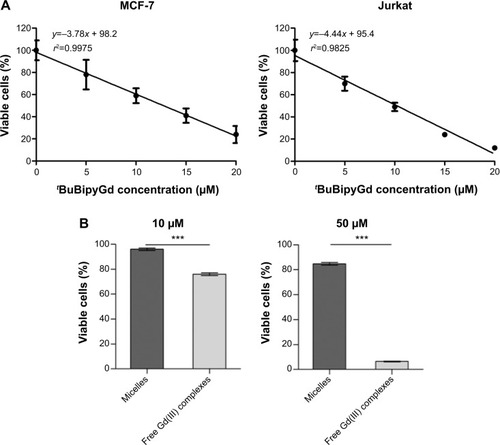
To assess the cytotoxicity of free tBuBipyGd complex, we tested its effect on two different human cell lines, the Jurkat leukemia cell line and the MCF-7 breast cancer cell line. By performing MTT assays, we observed that the median lethal dose (LD50) of tBuBipyGd complex for the MCF-7 and Jurkat cell lines was, respectively, 12.8 μM and 10.2 μM (), indicating that the tBuBipyGd complex is toxic at the micromolar range. Further cytotoxicity studies demonstrated that encapsulation of tBuBipyGd complex inside polymeric PEG-b-PTMSMA micelles greatly decreased its toxicity (). The difference in cytotoxicity was most evident at 50 μM of tBuBipyGd, the highest concentration tested, where most cells treated with encapsulated complex for 3 days were still viable, while most cells treated with free tBuBipyGd complex were dead (). The observation that Gd(III) complex–loaded micelles were not cytotoxic corroborated their observed stability in vitro (), because neither during dialysis nor during contact with cell culture medium there appeared to occur significant release of the complex and consequent increased cell death. Cytotoxicity tests also showed that up to 1 mg mL−1 of PEG-b-PTMSMA micelles alone were not toxic to Jurkat cells (data not shown), further supporting the notion that encapsulation of tBuBipyGd complex within PEG-b-PTMSMA micelles prevents cytotoxicity. The observed protection from content toxicity provided by our polymeric micelles, an important property for their in vivo administration as nanocarriers, should lead to reduced or no toxicity to healthy tissues, at least until the micelles reach the appropriate acidic microenvironment and disassemble. This important feature will open the door to new types of dual-function CAs that detect and kill at the same time the cancer cells in early stages, these dual-function contrasts are currently under investigation in our group.
Figure 6 Encapsulation of Gd(III) complex in polymeric micelles prevents cytotoxicity.
Notes: (A) A linear regression line was calculated from data obtained by MTT assay, to obtain the LD50 of tBuBipyGd complex for MCF-7 and Jurkat cell lines. (B) Comparison of cytotoxicity of low (10 μM) and high (50 μM) concentrations of free and micelle-encapsulated tBuBipyGd complex using MTT assay on the Jurkat cell line, after 3 days of incubation. Data represent the means of quadruplicate + standard error of the mean (***P≤0.0001, unpaired t-test).
Abbreviation: MTT, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide; LD50, median lethal dose.
Targeting of MUC1-expressing breast cancer cells by bioconjugated micelles
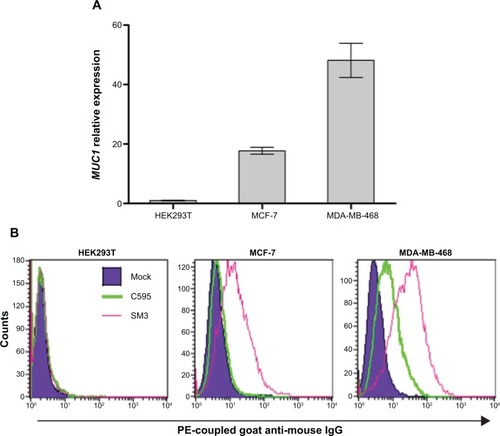
An important desired feature of the designed micelles, besides the pH-sensitive release mechanism, is the ability to specifically target cancer cells. To obtain this capability we bioconjugated micelles with an antibody against a cell surface tumor antigen. For this work, the MUC1 glycoprotein was chosen as a target for our new MRI CA because it is often overexpressed in human cancers, notably in breast cancer.Citation27 Furthermore, MUC1 has shown a great potential for early detection and staging of tumors, as well as for the assessment of tumor responses to therapy and detection of disease recurrence.Citation27 In addition MUC1 has been tested as target for quantum dot and gold nanoparticle–mediated cancer diagnosis or therapy.Citation31–Citation33 Confirming previously published data,Citation34 we verified that the MDA-MB-468 and MCF-7 human breast adenocarcinoma cell lines, but not the unrelated HEK293T cell line, expressed high levels of MUC1 mRNA () and MUC1 protein at the cell surface (), indicating their suitability as cancer models to test our bioconjugated micelles. Of two different mAbs tested by flow cytometry, SM3 showed higher affinity for surface MUC1 protein in both cell lines, indicating its potential to be more suitable for breast cancer cell targeting ().
Figure 7 (A) Detection of MUC1 mRNA expression in different cell lines by quantitative RT-PCR analysis. (B) Detection of MUC1 cell surface expression on MCF-7 and MDA-MB-468 cell lines by fluorescent flow cytometry using the C595 (green line) and SM3 (pink line) mAbs.
Note: The dark blue shaded region corresponds to the signal obtained with a control nonspecific isotypic antibody.
Abbreviations: mRNA, messenger RNA; RT-PCR, real-time polymerase chain reaction.
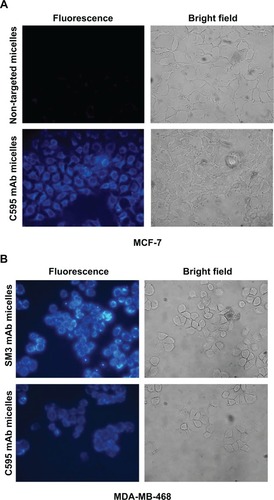
To test the ability of bioconjugated micelles to target breast cancer cells, we loaded micelles with the 1-methylpyrene fluorophore instead of tBuBipyGd complex, because of the fast and ready detection by microscopy of internalized fluorescence in targeted cells. After 1 hour incubation with micelles loaded with this fluorophore, followed by unbound micelle removal, cells were observed on a fluorescence microscope. Thus, MCF-7 cells incubated with C595 anti-MUC1–conjugated micelles presented more fluorescent staining than cells incubated with nontargeted micelles, in which no specific fluorescence was observed (). C595-conjugated micelles targeted both MCF-7 and MDA-MB-468 breast cancer cells, but micelles conjugated with the higher affinity SM3 mAb conferred increased fluorescence to target cells (). Together, these results demonstrated the specificity of the targeted micelles to MUC1-expressing cells and demonstrated that micelles released their content within cells.
Figure 8 Targeting of anti-MUC1 bioconjugated micelles to breast cancer cell lines.
Notes: (A) Specific binding of C595 anti-MUC1 1-methylpyrene–loaded micelles to MCF-7 breast cancer cells. After washing three times, high 1-methylpyrene fluorescence signal was found in cells incubated with polymeric micelles conjugated with C595 monoclonal antibody, while a low level of background fluorescence was visible in MCF-7 cells incubated with nontargeting micelles. (B) Targeting of C595 and SM3 anti-MUC1 1-methylpyrene–loaded micelles to MDA-MB-468 breast cancer cells.
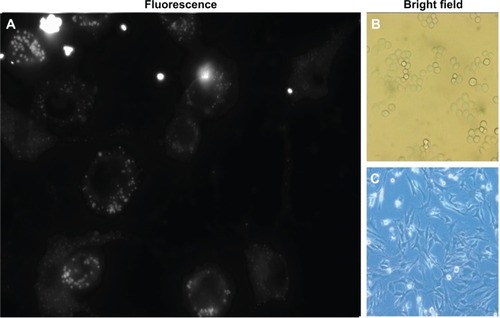
Next we used another approach to test the targeting specificity of anti-MUC1 micelles. The goal was to observe whether anti-human MUC1 micelles were able to specifically target MDA-MB-468 cells, even when these cells were mixed in a lower proportion (1:5) with unrelated cells from another species, the mouse S17 bone marrow stromal cell line. By incubating this mixed culture with anti-MUC1 polymeric micelles, we observed that MDA-MB-468 cells, which display spherical morphology, incorporated far more 1-methylpyrene fluorescence than S17 cells (). In this experiment little fluorescence of the elongated morphology cells (S17) was observed, in contrast to the larger and brighter spots in round-shaped MDA-MB-468 cells ().
Figure 9 Increased targeting of anti-MUC1 1-methylpyrene–loaded micelles to MDA-MB-468 breast cancer cells.
Notes: (A) MDA-MB-468 and S17 cells cultured together in a 1:5 proportion were incubated for 1 hour with C595 mAb–bioconjugated micelles. (B) Bright field microscopy of MDA-MB-468 cells. (C) Brigth field microscopy of S17 cells.
Discussion
The present study reports the development of a smart CA composed of pH-sensitive micelles containing a hydrophobic Gd(III) complex with the aim of specifically detecting cancer by MRI. Two main features confer high specificity for these CAs to detect cancer tissues: (i) pH sensitivity, which leads to a contrast signal only in acidic microenvironments, and (ii) antibody conjugation, which leads to preferential targeting to tumor tissues expressing the respective antigen.
Our experiments demonstrated that when encapsulated in these micelles, the CA remained in the off state, being activated only upon micelle disruption in an acidic medium. Several researchers have reported pH-sensitive particles such as liposomes with the aim of detecting cancer tissues or other acidic pathologic areas by MRI.Citation7–Citation13 However, pH sensitivity was the only feature in these tools for cancer targeting, besides the enhanced permeability and retention effect.Citation25 Other researchers have developed nanoparticles conjugated to biomolecules (eg, antibody or peptides) to target and image cancer.Citation35,Citation36 Our results with MUC1-expressing cell lines indicate that the conjugation of self-assembled polymeric micelles with a tumor-specific mAb increases significantly their affinity toward cancer cells. Therefore, our bioconjugated pH-sensitive micelles present two features enabling targeting and imaging of cancer. This characteristic is instrumental for the application of these smart micelles as carriers to deliver CAs to MUC1-expressing cancer tissues. In this work we have tested two mAbs targeting the MUC1 surface protein, often overexpressed in mammary and other adenocarcinomas, such as gastric, colorectal, lung, prostate, ovarian, pancreatic, and bladder cancers.Citation26,Citation27 The C595 mAb, which reacts with the tetrameric motif RPAP present on the tandem repeats in the extracellular region of the MUC1 protein, has already been proven to be an efficient and specific targeting vector in preclinical and clinical trials of radioimmunoscintigraphy and radioimmunotherapy.Citation37,Citation38 The SM3 mAb reacts against the APDTRP sequence, which is exposed only in underglycosylated MUC1 protein.Citation26 Because underglycosylated MUC1 is very frequent and specific for breast and other epithelial cancers,Citation26 the SM3 mAb should be highly suitable to conjugate the micelles to target cancer tissues. The possibility of conjugating our polymeric micelles with other antibodies specific for different tumor antigens should enable their application to a wide range of human cancers. The identification of cancer biomarkers is an area of intense research, so novel cell surface biomarkers may become potential targets for our nanoparticle-based smart CAs.
Conclusion
The presented work demonstrated that the developed polymeric micelle-based CA is able to turn on its imaging ability upon a pH decrease. The encapsulation of the active complex within micelles decreased its toxicity and allows the use of small amounts of CA (1.08 μmol kg−1) in the clinic, as compared to current protocols, to avoid potential side effects. These features allied with the finding that micelle conjugation with an antibody specific for a tumor antigen increased their uptake by cancer cells and with their small size, turns this polymeric nanoparticles into a promising new tool for cancer early detection by MRI.
Acknowledgments
We thank João Barata, Leonor Parreira (both from IMM, Lisbon), Raquel Seruca, Joana Paredes (both from IPA-TIMUP, Porto), and Guilherme Ferreira (IBB/CBME, Faro) for providing cell lines. We also thank Rosário Caras-Altas (REQUIMTE/FCT-UNL, Caparica) for performing the relaxation time measurements and José Mariano (Physics Department, University of Algarve) for helpful discussions concerning this experiment. We thank André Mozes, Maurícia Vinhas, and Cláudia Florindo (Biomedical Sciences and Medicine Department, University of Algarve) for invaluable technical assistance. We acknowledge the financial support was mostly from King Abdulaziz City for Science and Technology (KACST) Saudi Arabia (grant number NPST 11-BIO1578-02) and the Fundação para a Ciência e a Tecnologia, Portugal (projects PEst-OE/EQB/LA0023/2011 and PEst-OE/QUI/UI4023/2011).
Disclosure
The authors report no conflicts of interest in this work.
References
- GoreJCManningHCQuarlesCCWaddellKWYankeelovTEMagnetic resonance in the era of molecular imaging of cancerMagn Reson Imaging20112958760021524870
- LiuYZhangNGadolinium loaded nanoparticles in theranostic magnetic resonance imagingBiomaterials2012335363537522521487
- ErsoyHRybickiFJBiochemical safety profiles of gadolinium-based extracellular contrast agents and nephrogenic systemic fibrosisJ Magn Reson Imaging2007261190119717969161
- StrattaPCanaveseCAimeSGadolinium-enhanced magnetic resonance imaging, renal failure and nephrogenic systemic fibrosis/nephrogenic fibrosing dermopathyCurr Med Chem2008151229123518473815
- ChengZAl ZakiAHuiJZMuzykantovVRTsourkasAMultifunctional nanoparticles: cost versus benefit of adding targeting and imaging capabilitiesScience201233890391023161990
- NieSXingYKimGJSimonsJWNanotechnology applications in cancerAnnu Rev Biomed Eng2007925728817439359
- CaoCYShenYYWangJDLiLLiangGLControlled intracellular self-assembly of gadolinium nanoparticles as smart molecular MR contrast agentsSci Rep201331024103023289066
- LiangGRonaldJChenYDPanditPMaMLControlled self-assembling of gadolinium nanoparticles as smart molecular magnetic resonance imaging contrast agentsAngew Chem Int Ed Engl2011506283628621618367
- HartmanKBLausSBolskarRDMuthupillaiRHelmLTothEGadonanotubes as ultrasensitive pH-smart probes for magnetic resonance imagingNano Lett2008841541918215084
- TorresEMaininiFNapolitanoRFedeliFCavalliRAimeAImproved paramagnetic liposomes for MRI visualization of pH triggered releaseJ Control Release201115419620221621569
- GaoGHImGHKimMSLeeJWYangJJeonHMagnetite-nanoparticle-encapsulated pH-responsive polymeric micelle as an MRI probe for detecting acidic pathologic areasSmall201061201120420449849
- JingLLiangXLiXYangYDaiZCovalent attachment of Mn-porphyrin onto doxorubicin-loaded poly(lactic acid) nanoparticles for potential magnetic resonance imaging and pH-sensitive drug deliveryActa Biomater201399434944123962645
- Van ButseleKMorilleMPassiraniCLegrasPBenoitJPVarshneySKStealth properties of poly(ethylene oxide)-based triblock copolymer micelles: a prerequisite for a pH-triggered targeting systemActa Biomater201173700370721704739
- HanaokaKKikuchiKUranoYNarazakiMYokawaTSakamotoSDesign and synthesis of a novel magnetic resonance imaging contrast agent for selective sensing of zinc ionChem Biol200991027103212323377
- LiWFraserSEMeadeTJA calcium-sensitive magnetic resonance imaging contrast agentJ Am Chem Soc199912114131414
- QueELChangCJA smart magnetic resonance contrast agent for selective copper sensingJ Am Chem Soc2006128159421594317165700
- BonnetCSTóthESmart MR imaging agents relevant to potential neurologic applicationsAm J Neuroradiol20103140140919833798
- AimeSDigilioGFasanoMPaolettiSArnelliAAscenziPMetal complexes as allosteric effectors of human hemoglobin: an NMR study of the interaction of the gadolinium(III) bis(m-boroxyphenylamide) diethylenetriaminepentaacetic acid complex with human oxygenated and deoxygenated hemoglobinBiophys J1999762735274310233088
- GløgårdCStensrudGAimeSNovel radical-responsive MRI contrast agent based on paramagnetic liposomesMagnet Resonann Chem200341585588
- LiuTYHuangTCA novel drug vehicle capable of ultrasound-triggered release with MRI functionsActa Biomater201173927393421745611
- YuanQVenkatasubramanianRHeinSMisraRDKA stimulus-responsive magnetic nanoparticle drug carrier: magnetite encapsulated by chitosan-grafted-copolymerActa Biomater200841024103718329348
- ZhangJMisraRDKMagnetic drug-targeting carrier encapsulated with thermosensitive smart polymer: core-shell nanoparticle carrier and drug release responseActa Biomater2007383885017638599
- KozlowskaDForanPMacMahonPShellyMJEustaceSO’KennedyRMolecular and magnetic resonance imaging: the value of immunoliposomesAdv Drug Deliv Rev2009611402141119796661
- BumbABrechbielMWChoykePMacromolecular and dendrimer-based magnetic resonance contrast agentsActa Radiol20105175176720590365
- MaedaHWuJSawaTMatsumuraYHoriKTumor vascular permeability and the EPR effect in macromolecular therapeutics: a reviewJ Control Release20006527128410699287
- SinghRBandyopadhyayDMUC1: a target molecule for cancer therapyCancer Biol Ther2007648148618027437
- KufeDWMucins in cancer: function, prognosis and therapyNat Rev Cancer2009987488519935676
- MouffoukFda CostaAMRMartinsJZourobMAbu-SalahKHAlrokayanSADevelopment of a highly sensitive bacteria detection assay using fluorescent pH-responsive polymeric micellesBiosens Bioelectron2011263517352321382708
- SemenovaLISkeltonBWhiteAHStructural systematics of rare earth complexes. XVIII (hydrated) mononuclear 1: 1 adducts of lanthanoid(III) chlorides with 2,2′-bipyridineAust J Chem199952551570
- GagoSPillingerMSá FerreiraRACarlosLDSantosTMGonçalvesISImmobilization of lanthanide ions in a pillared layered double hydroxideChem Mater2005758035809
- ChengAKSuHWangYAYuHZAptamer-based detection of epithelial tumor marker mucin 1 with quantum dot-based fluorescence readoutAnal Chem2009816130613919572710
- DanyshBPConstantinouPELukianova-HlebEYLapotkoDOCarsonDDThe MUC1 ectodomain: a novel and efficient target for gold nanoparticle clustering and vapor nanobubble generationTheranostics2012277778722916077
- YuCHuYDuanJNovel aptamer-nanoparticle bioconjugates enhances delivery of anticancer drug to MUC1-positive cancer cells in vitroPLoS One20116e2407721912664
- WalshMDLuckieSMCummingsMCAntalisTMMcGuckinMAHeterogeneity of MUC1 expression by human breast carcinoma cell lines in vivo and in vitroBreast Cancer Res Treat19995825526610718487
- KellyKABardeesyNAnbazhaganRGurumurthySBergerJAlencarHTargeted nanoparticles for imaging incipient pancreatic ductal adenocarcinomaPLoS Med20085e8518416599
- YangLMaoHWangYASingle chain epidermal growth factor receptor antibody conjugated nanoparticles for in vivo tumor targeting and imagingSmall2009523524319089838
- SongEYQuCFRizviSMARajaCBeretovJMorgensternABismuth-213 radioimmunotherapy with C595 anti-MUC1 monoclonal antibody in an ovarian cancer ascites modelCancer Biol Ther20087768018347423
- SimmsMSPerkinsACPriceMRScholfieldDPBishopMC99mTechnetium-C595 radioimmunoscintigraphy: a potential staging tool for bladder cancerBJU Int20018868669111890238