 ?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.
?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.Abstract
Transgene transfection techniques using cationic polymers such as polyethylenimines (PEIs) and PEI derivatives as gene vectors have shown efficacy, although they also have shortcomings. PEIs have decent DNA-binding capability and good cell internalization performance, but they cannot deliver gene payloads very efficiently to cell nuclei. In this study, three hyperbranched polyglycerol-polyethylenimine (PG6-PEI) polymers conjugated with myo-inositol (INO) molecules were developed. The three resulting PG6-PEI-INO polymers have an increased number of INO ligands per molecule. PG6-PEI-INO 1 had only 14 carboxymethyl INO (CMINO) units per molecule. PG6-PEI-INO 2 had approximately 130 CMINO units per molecule. PG6-PEI-INO 3 had as high as 415 CMINO units approximately. Mixing PG6-PEI-INO polymers with DNA produced compact nanocomposites. We then performed localization studies using fluorescent microscopy. As the number of conjugated inositol ligands increased in PG6-PEI-INO polymers, there was a corresponding increase in accumulation of the polymers within 293T cell nuclei. Transfection performed with spherical 293T cells yielded 82% of EGFP-positive cells when using PG6-PEI-INO 3 as the vehicle. Studies further revealed that extracellular adenosine triphosphate (eATP) can inhibit the transgene efficiency of PG6-PEI-INO polymers, as compared with PEI and PG6-PEI that were not conjugated with inositol. Our work unveiled the possibility of using inositol as an effective ligand for transgene expression.
Introduction
Gene transfection techniques have been widely used in functional genomics and gene therapy research.Citation1,Citation2 Virus-derived vectors are known for their high gene delivery efficiency; however, these vectors also pose potential risks.Citation3–Citation5 Owing to the convenience in preparation and tailoring design, nonviral vectors based on natural or synthetic biomaterials have become highlighted.Citation2,Citation6–Citation11
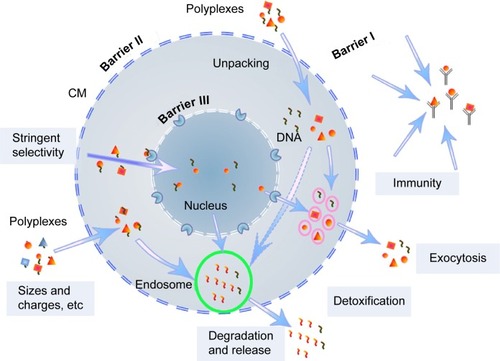
Various barriers, including the nuclear membrane as the main barrier, exist before efficient transgene expression is achieved (). Versatile gene carriers, including a class of intensively studied cationic polymers, such as branched polyethylenimine (PEI), poly-N-(2-hydroxypropyl) methacrylamide (poly-HPMA), poly(amido amine) dendrimers and chitosan, have been developed for transport of large nucleic acid payloads.Citation2–Citation8 These polymers can compact the nucleotides and help them escape from endosomal degradation through a “proton-sponge effect”.Citation6,Citation12 Gene vectors with reduced cytotoxicity and immune recognition can be achieved through various modifications, for example, with poly-HPMA, polycaprolactone (PCL), and polyethylene glycol (PEG).Citation13–Citation17 Target approaches can enhance cell internalization of the gene delivery systems,Citation18–Citation21 but the cell-membrane-targeted ligands are not ideal for improving transgene expression, which is strictly limited by the cell-nuclear membrane ().Citation22,Citation23 Just a few of the DNA isolates are able to enter the cell nucleus once the complexes are unpacked within the cell cytoplasm. A successfully studied target ligand was the chlorotoxin (CTX) peptide, which targets the nanovectors to cancer cells.Citation21 However, in the case of tumor therapy and analysis of gene functions, a superhigh ratio of transfection is required. In this regard, study and targeting of gene vectors to cell nuclei remains a big challenge.
Figure 1 Schematic illustration of self-protection by mammalian cells during cationic polymers-mediated transfection.
Abbreviations: CM, cell membrane; DNA, deoxyribonucleic acid.
Recently, cell-nuclear transfer technology has been receiving much attention as a promising strategy to meet the increased requirement for effective transgene expression therapies.Citation19 Frequently reported elements include proteins/peptides that are able to localize to mammalian cell nuclei, for example, nuclear localization signals (NLS).Citation3,Citation24–Citation27 Melittin protein derived from nonviral organisms was used to enhance nuclear access of nonviral gene delivery vectors.Citation28 Novel approaches were developed using polymeric gene carriers conjugated with all-trans-retinoic acid (ATRA).Citation29 Target ligands that are stable, small, and have cell-nuclear localization ability are of interest to researchers.
Myo-inositol (INO) is a biomolecule with crucial functions in eukaryotic cells.Citation30–Citation37 The H+-Myo-inositol receptors were reported in cancer stem cells (CSC) from various tumors.Citation38 The presence of free myo-inositol has been demonstrated in cell nuclei.Citation39 A series of inositol-related receptors/factors were reported to localize on the nuclear membrane, for example, the inositol 1,4,5-trisphosphate-sensitive Ca2+ pool, the InsP receptors, and the nucleocytoplasmic shuttling protein inositol 5-phosphatase SHIP1.Citation40,Citation41 Myo-inositol serves as the structural basis for various derivatives, including a number of essential secondary messengers. Myo-inositol and its derivatives, such as phosphatidylglycerol and phosphatidylinositol derivatives, inositol phosphate isoforms, and inositol polyphosphates had metabolism links and are involved in various metabolic pathways in mammalian cells.Citation42–Citation46 Among the inositol derivatives, inositol polyphosphates (IP2–IP6) play crucial roles in cellular functions including cell growth, endocytosis, migration, differentiation, and apoptosis.Citation30,Citation31 IP3, IP5, and IP6 are also involved in gene expression. Inositol(1,4,5)P3 receptors were revealed at the nuclear envelope of mammalian cells. The PI3K signaling pathway has been widely found in cancer cells, and nuclear transportation of phosphatidylinositol 3-kinase has been observed.Citation32,Citation34–Citation37 Importantly, inositol 1,4,5-trisphosphate receptors (IP3R) have been shown to mediate extracellular adenosine triphosphate (eATP) induced Ca2+ transients in cell nuclei rather than in the cytosol.Citation47
In our previous study, inositol conjugated PEI800-FA was detected on the cell-nuclear membrane or in cell nuclei with variant levels in the tested cell lines, and transfection efficiency of INO-PEI800 was as high as HMW PEI for some carcinoma cell lines.Citation22 To consolidate INO as a biocompatible ligand for efficient transgene expression, increased ratios of inositols were conjugated to PG6-PEI25k, which is derived from the hyperbranched polyglycerol core and branched HMW PEI25k grafts. As determined, PG6-PEI25k shows enhanced transgene activity and biocompatibility relative to PEI25k but does not enter cell nuclei. The resultant vectors, PG6-PEI-INOs with low cytotoxicity, achieved different degrees of enhanced transfection activity as compared with PG6-PEI25k. For PG6-PEI-INO 3 containing a much higher INO ratio at approximately 415 ligands per molecule, transgene expression resulted in >80% of EGFP-positive 293T cells, which demonstrated further enhancement relative to PG6-PEI-INO 1 and 2 that respectively contained 14 and 130 INO ligands per molecule only. Because some cell-nuclear activity, such as the nuclear Ca2+ transient, can be specifically induced by eATP,Citation47 we then investigated eATP-induced inhibition on EGFP expression, which shows evident differences when comparing the effects of PG6-PEI-INO and PG6-PEI polymers.
Material and methods
Materials
Polyglycerol PG6 (Mn =6,170 g/mol) was obtained from Hyperpolymers GmbH (Freiburg, Germany). Branched PEI25k (Mw =25,100 g/mol), N-hydroxysuccinimide (NHS), N,N′-dicyclohexylcarbodiimide (DCC), and 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide hydrochloride (EDC) were from Sigma-Aldrich (St Louis, MO, USA). N-dimethyl-formamide (DMF) was dried over CaH2 and distilled under reduced pressure prior to use. 4′,6-diamidino-2-phenylindole (DAPI) was purchased from Roche (Branford, CT, USA). 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT), trypsin, and penicillin/streptomycin were purchased from Invitrogen (Carlsbad, CA, USA). All other reagents were purchased from Sinopharm Chemical Reagent Co., Ltd (Shanghai, People’s Republic of China). Human embryonic kidney cell line 293T was purchased from the China Center for Typical Culture Collection (Wuhan, People’s Republic of China). The cells were cultivated in Dulbecco’s Modified Eagle’s Medium (DMEM) supplemented with 10% fetal bovine serum, 2 mg/mL NaHCO3, and 100 units/mL penicillin/streptomycin. Cells were incubated at 37°C in humidified air with 5% CO2. The reporter plasmid pEGFP-C1 was obtained from Invitrogen.
Preparing plasmid
Plasmid DNA pEGFP-C1 was amplified in Escherichia coli, extracted with E.Z.N.A. FastFilter Endo-Free Plasmid Maxi Kit (Omega, Norcross, GA, USA). The purified plasmid DNA was stored in TE buffer (supplied with the kit) or deionized (DI) water at a final concentration of 1 mg/mL.
Polymer conjugation
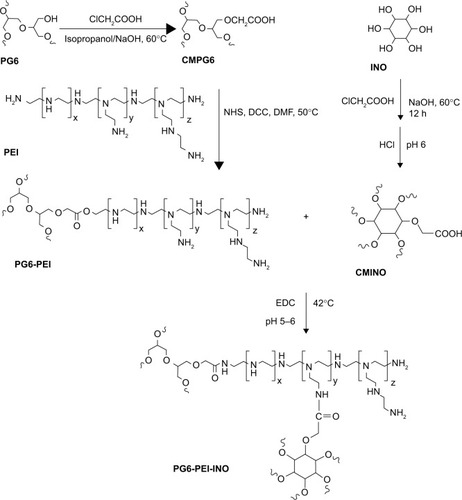
Hyperbranched polymer PG6-PEI25k, which was derived from hyperbranched polyglycerol core (Mw =6,170 g·mol−1, had an average of 90 hydroxyl end-groups per molecule) and branched PEI25k grafts, was synthesized and characterized according to our previous method.Citation13 INO (22.5 mg) was added to a 14.3 M NaOH solution (100–200 μL), and the mixture was stirred at room temperature (22°C) for 1 hour. The chloroacetic acid (CAA) (0.3 mg/mL) was added dropwise to the mixture, and the suspension was stirred at 60°C for 12 hours to carboxylate hydroxyls of INO. The solution was then adjusted to acidic (pH 6) with HCl (1 M) to convert–COONa to –COOH. The product was cooled and filtrated to obtain INO-COOH (carboxymethyl INO [CMINO]). The CMINOs were washed with ethanol and distilled under reduced pressure. CMINO was conjugated to PG6-PEI with EDC as the coupling reagent (). The molar ratios of introduced CMINO to PG6-PEI25k units were approximately 1, 10, and 100 for the three reaction systems. The mixtures were dissolved in DI water, and pH was adjusted to 5–6. The reactions were performed at 42°C for 18 hours, and the reaction mixtures were dialyzed with a dialysis membrane (MWCO: 8,000–12,000 g/mol) against DI water, respectively, and freeze dried under vacuum to obtain PG6-PEI-INO polymers.
Table 1 Weight ratio of the reactants and molecular weights of PG6-PEI-INO polymers
FT-IR spectroscopy
Infrared analysis (KBr) of the samples was studied on FT-IR (Fourier transform infrared) spectrometer (Spectrum One; Perkin Elmer, Waltham, MA, USA). Spectra were run in the 4,000–400 cm−1 region.
1H NMR spectroscopy
1H nuclear magnetic resonance (NMR) spectroscopy was performed on a Varian Inova 600 MHz spectrometer; the solvent was deuterium oxide.
Gel permeation chromatographic ([GPC]/SEC) analysis
The molecular weights of the polymers were determined by combined size-exclusion chromatography and multiangle laser light scattering (SEC–MALLS) analysis. A dual detector system, consisting of a MALLS device (DAWN EOS; Wyatt Technology, Santa Barbara, CA, USA) and an interferometric refractometer (Optilab DSP, Wyatt Technology), was used. PG6-PEI-INO polymers were dissolved in 600 mM of ammonium acetate with a final polymer concentration of 1 mg/mL, and analyzed in ammonium acetate (600 mM) mobile phase. The MALLS detector was operated at a laser wavelength of 690.0 nm. The samples were then determined at a flow rate of 0.3 mL/min.
Agarose gel electrophoresis retardation assays
PG6-PEI-INO polymers were dissolved in 150 mM (or 0.9%) NaCl. PG6-PEI-INO/pDNA (pEGFP-C1) complexes were prepared by adding 10 μL of PG6-PEI-INO solutions at serial concentrations to 20 ng of pDNA (20 ng/μL in 150 mM NaCl). The complexes were incubated at 37°C for 30 minutes, and sampled for electrophoresis on the 0.7% (w/v) agarose gel containing GelRed™ with Tris-acetate-EDTA (TAE) electrophoresis buffer at 80 V. Plasmid DNA bands were visualized on a V-transilluminator with a Vilber Lourmat imaging system (Marne La Valée, France).
TEM analysis of PG6-PEI-INO/pDNA polyplexes
PG6-PEI-INO/pDNA (pEGFP-C1) particles were composited in DI water, and the morphology was determined by transmission electronic microscopy (TEM) on a JEM-100CXII transmission electronic microscope (JEOL, Tokyo, Japan). Aqueous suspensions (3 μL) were placed on copper grids with Formvar film. The samples were stained by phosphotungstic acid, vacuum-dried and visualized.
Cell viability measurement
Cells were seeded in a 96-well plate at a density of 3×103 cells/well in 100 μL of DMEM culture media (10% serum) containing polymers or polymer/pEGFP-C1 complexes (1.3 μg of pEGFP-C1 per mL medium) at varied feed ratios. After 52 hours of cultivation, the culture media were replaced with fresh DMEM medium (100 μL) plus 20 μL of MTT (5 mg/mL), and the plate was incubated in the incubator at 37°C for 4 hours. Then the supernatants were replaced with 150 μL of DMSO. After incubation for 15 minutes at 37°C, the absorbance of 50 μL of sample solution was measured in a microplate reader (Bio-Rad 550; Bio-Rad Laboratories Inc., Hercules, CA, USA) at 570 nm. The cell viability was calculated as follows:
In vitro transfection
293T cells were seeded in a 24-well plate at a density of 3×104 cells/well in 1 mL of DMEM culture media (10% serum), followed by cultivation for 4 hours. Prior to transfection, 1 μL of pEGFP-C1 solution (1.3 μg/μL in DI water) was mixed with 1 μL of varied concentrations of PG6-PEI-INO aqueous solutions and diluted with 20 μL of filtrated NaCl (150 mM) solution, followed by vortex and incubation at 37°C for 30 minutes. The complexes were then supplemented to the cell suspension, and coincubated with the cells for 52 hours. The EGFP-positive cell ratio was calculated on a counting chamber with fluorescent phase-contrast microscopy (Olympus IX 70; Olympus Corporation, Tokyo, Japan; at 400×), after the cell suspensions were prepared with tryptic digestion to prevent miscounting of the undispersed cells.
Influence of eATP on cell viability and transgene expression
Optimized ratios of PEI25k/pEGFP-C1 (w/w =1.3), PG6-PEI25k/pEGFP-C1 (w/w =7), and PG6-PEI-INO 3/pEGFP-C1 (w/w =7) with fixed dosage of pEGFP-C1 (1.3 μg per mL medium) were supplemented with serial concentrations of ATP, respectively, to compare the response of transgene activity of the materials to ATP supplements. The mixtures were incubated at 37°C for 30 minutes before transgene experiments. Detailed MTT assay and transfection procedure were performed in 24-well plates according to the descriptions above. The relative level of transgene expression was calculated as follows:
Fluorescence tracking
The PG6-PEI-INO polymers were labeled with Rhodamine B, which is usually used to stain cytosol instead of cell nuclei. For the three PG6-PEI-INO systems with different INO ratios, identical numbers of amino groups per microgram of polymers were labeled. In brief, PG6-PEI-INO polymers (0.36 mg) and Rhodamine B (2 μg) were dissolved in 5 mL DMF, using NHS and DCC as carboxyl activator and coupling reagent, respectively. The reactions were carried out at 45°C. The mixtures were dialyzed against deionized water at room temperature, and dried in a freeze drier (VirTis, Warminster, PA, USA) to purify PG6-PEI-INO-Rhodamine (Rh)s.
293T cells were seeded in a 6-well plate at a density of 8×104 cells/mL media and cultivated for 12 hours. PG6-PEI-INO-Rh (5 μg/mL) was supplemented to the cell cultures and incubated with the cells for 48 hours. The cells were washed with PBS (pH 7.4), stained with DAPI, and washed with PBS. Confocal laser scanning microscopy (CLSM, Leica TCS SP2AOBS; Leica Microsystems, Wetzlar, Germany) was used to record fluorescence within the cells.
Results and discussion
Synthesis and characterization of PG6-PEI-INO polymers
GPC (SEC) analysis showed that PG6-PEI-INO 1, 2, and 3 () had weight average molecular weights (Mw) of 455, 465, and 542 kDa, respectively ().
Figure 2 Synthesis route of PG6-PEI-INO polymers.
Abbreviations: CMPG6, carboxymethyl polyglycerol; CMINO, carboxymethyl inositol; DCC, N,N’-dicyclohexylcarbodiimide; DMF, N-dimethyl-formamide; EDC, 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide hydro chloride; INO, myo-inositol; NHS, N-hydroxysuccinimide; PEI, polyethylenimine; PG6, polyglycerol.
FT-IR spectrum showed the peak at 1,724.82 cm−1, depicting–COOH of CMINO (). The intensity of this peak declined for PG6-PEI-INOs because–COOH of CMINO reacted with –NH2 of PEI25k. With the increase in the grafted INO ratio, an increase in the O-H deformation vibration (1,261.96 cm−1) and the fingerprint region of CMINO (799.95 cm−1) was detected in the order of PG6-PEI-INO 1, 2, and 3. The peak (1,462.87 cm−1) indicating the in-plane bending vibration of –CH2 groups from PG6-PEI and CMINO increased in PG6-PEI-INO 1, 2, and 3. The results indicated successful conjugation of INO to PG6-PEI.
Figure 3 Characterization of the PG6-PEI-INO polymers.
Notes: (A) FT-IR spectra of PG6-PEI-INO 1, PG6-PEI-INO 2, and PG6-PEI-INO 3. (B) 1H NMR spectra of the polymers. Deuterium oxide was used as the solvent.
Abbreviations: CMINO, carboxymethyl inositol; CMPG6, carboxymethyl polyglycerol; FT-IR, Fourier transform infrared spectroscopy; INO, myo-inositol; NMR, nuclear magnetic resonance; PEI, polyethylenimine; PG6, polyglycerol.
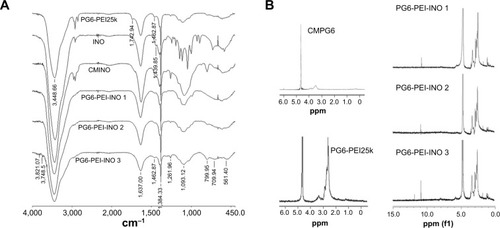
1H NMR results showed the unreacted –COOH of CMINO units (10.8 ppm), characteristic PEI proton deviation peaks (2.4–3.0 ppm), and characteristic proton deviation peaks of PG6 and INO (3.0–4.0 ppm) (). With CMINO grafts increased, the ratio of the integral of the 3.0–4.0 ppm peak to that of the 2.0–3.0 ppm peak increased, indicating that an increased number of CMINO molecules were conjugated to PG6-PEI. The molar ratio of PG6 to PEI25k is 1:1, as previously characterized. The ratio of CMINO to PG6-PEI25k units was approximately 1:1, 10:1, and 35:1 in PG6-PEI-INO 1, 2, and 3, respectively. According to the weight average molecular weight (Mw) of PG6-PEI-INOs (), a PG6-PEI-INO 1 molecule had approximately 14 PEI25k and 14 CMINO units; PG6-PEI-INO 2 had approximately 13 PEI25k and 130 CMINO units; PG6-PEI-INO 3 had approximately 12 PEI25k and 415 CMINO units.
PG6-PEI-INOs composite DNA efficiently
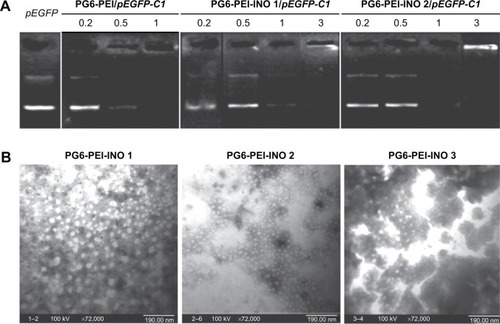
Gel electrophoresis retardation of pEGFP-C1 demonstrated the DNA-binding activity of PG6-PEI-INOs (). TEM analysis showed that all PG6-PEI-INO polymers could compact plasmid DNA to polyplexes with a diameter of less than 30 nm (). This compacted nanostructure could protect DNA against enzyme degradation and meanwhile benefit cell internalization. With respect to the small particle sizes, it has been reported that the diameter of the nuclear pore complex (NPC) was up to 120 nm and permitted molecules or complexes with diameters of 39 nm to pass through.Citation34,Citation48 Therefore, we subsequently determined the transgene expression mediated by PG6-PEI-INO polymers and the cell-nuclear localization of the PG6-PEI-INOs.
Figure 4 DNA-binding ability of PG6-PEI-INO polymers.
Notes: (A) Agarose gel electrophoresis of PG6-PEI-INOs/pEGFP-C1 complexes at varied weight ratios. (B) Morphologic study of PG6-PEI-INO/pEGFP-C1 (w/w =5) complexes using transmission electron microscopy.
Abbreviations: INO, myo-inositol; PEI, polyethylenimine; PG6, polyglycerol.
Inositol improves biocompatibility of HMW PEI-based vectors
Viability assays showed that both PG6-PEI-INOs/pDNA (w/w =5–9) () and an identical weight of PG6-PEI-INOs () achieved decent biocompatibility (70%–80%) when they were used at high dosage. A previous study indicated that the 293T cell viability was much lower when using an identical dosage of PG6-PEI/pDNA or PG6-PEI.Citation15 Instead, cells treated with an identical dosage of PEI25k/pEGFP-C1 or unmodified PEI25k had a viability of less than 10% or 8%, respectively. These results demonstrated that INO could further enhance biocompatibility of PG6-PEI25k.
Figure 5 Biocompatibility of the PG6-PEI-INO/plasmid or PG6-PEI-INO polymers.
Notes: (A) MTT analysis of cell viability was performed after 293T cells were cocultivated for 52 hours with PG6-PEI-INOs/plasmid (pEGFP-C1), which had weight ratios of 5 (a), 7 (b), and 9 (c), respectively. (B) MTT analysis of cell viability was performed after 293T cells were cocultivated with identical dosages of PG6-PEI-INOs for 52 hours. Cells were treated with PEI25k/pEGFP-C1 at a weight ratio of 6, or with an identical dosage of PEI25k as the control. DNA was used at 1.3 μg of pEGFP-C1 per mL medium.
Abbreviations: INO, myo-inositol; PEI, polyethylenimine; PG6, polyglycerol; p, plasmid (pEGFP-C1).
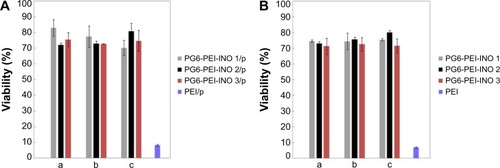
When the weight ratio of PG6-PEI-INO/DNA is 9, the corresponding N/P ratio for PG6-PEI-INO 1, 2, and 3 to DNA has reached 53, 44, and 35, respectively, which is far higher than the permitted dose range of HMW PEI25k/DNA. These results offer an explanation for why PG6-PEI-INO has good biocompatibility. To the question of potential toxicity at the higher doses, for example, whether they produce toxicity after partial degradation, and how organisms respond, will be addressed by in vivo experiments in future.
Using inositol as efficient ligands for transgene expression
The ability of PG6-PEI-INOs to mediate transgene expression was evaluated in 3D 293T cells because they are sensitive indicators. Results showed that the PG6-PEI-INO polymers-mediated EGFP expression effectively without causing cell injury (). The expression level was higher with the increase of PG6-PEI-INOs. Generally, PG6-PEI-INO 3 containing the highest INO component achieved the highest transgene expression level, as compared with PG6-PEI and PG6-PEI-INO 1 and 2. PG6-PEI-INO 3/pEGFP-C1 at w/w of 5 and 7 achieved 75% and 82% of positive cells, respectively, which was approximately 19% and 23% higher than that achieved with PG6-PEI/pEGFP-C1 at identical weight ratios, and was 27% and 34% higher than that achieved by PEI25k/pEGFP-C1 at the optimal ratio (w/w =1.3 or N/P =10).Citation15,Citation49 For PEI25k/pEGFP-C1 at weight ratios exceeding 6, cells were severely distorted and/or detached. PG6-PEI-INO 1 with only one INO moiety per molecule showed similar activity as compared with PG6-PEI in general. Compared with PG6-PEI-INO 1/pEGFP-C1, PG6-PEI-INO 2/pEGFP-C1 with more INO grafts achieved more EGFP-positive cells.
Figure 6 Transfection activities of PG6-PEI-INO polymers.
Notes: (A) Transgene expression reported by protein EGFP in 293T cell line treated with (a) PG6-PEI-INO 1/pEGFP-C1, (b) PG6-PEI-INO 2/pEGFP-C1, and (c) PG6-PEI-INO 3/pEGFP-C1 complexes at weight ratio of 7, respectively. Plasmid pEGFP-C1 was used at 1.3 μg per mL culture medium. The cells were cultivated for 52 hours. The adherent cells and the cell suspensions achieved with trypsin treatment were analyzed by phase-contrast fluorescent microscopy. Scale bar: 15 μm. (B) EGFP-positive cell ratios achieved by transgene expression mediated by PG6-PEI-INO polymers. The 293T cells were transfected using the following agents: (a) PG6-PEI-INO 1, (b) PG6-PEI-INO 2, (c) PG6-PEI-INO 3, (d) PG6-PEI at varied weight ratios to pEGFP-C1, and (e) the PEI25k control at its optimal weight ratio (1.3) to pEGFP-C1 (1.3 μg of pEGFP-C1 per mL cell culture).
Abbreviations: INO, myo-inositol; PEI, polyethylenimine; PG6, polyglycerol; EGFP, enhanced green fluorescent protein.
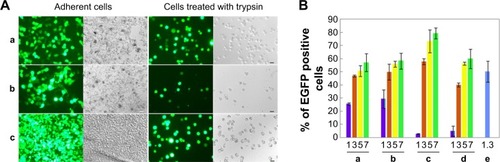
When the weight ratios to DNA were 1 and 3, the transfection activity increased in the order of PG6-PEI < PG6-PEI-INO 1 < PG6-PEI-INO 2. When the weight ratios were 5 and 7, no apparent increase was observed among the three systems. Compared with PG6-PEI-INO 3, which had approximately 12 PEI25k and 415 CMINO units and showed the highest transfection activity, a PG6-PEI-INO 1 molecule had approximately 14 PG6, 14 PEI25k, and only 14 CMINO units, and PG6-PEI-INO 2 had approximately 13 PG6, 13 PEI25k, and 130 CMINO units. Conjugation with INO will reduce –NH2 termini of PEI25k and therefore lower the DNA-binding ability of PEI25k. On the other hand, conjugation with INO increases biocompatibility of PEI25k and accumulation of PG6-PEI-INOs in cell nuclei, as will be illustrated in the subsequent experiment. Therefore, we deem that a balance exists between the two aspects. When the number of INO is high enough to achieve a high accumulation of gene vectors in cell nuclei and meet the requirement for DNA binding/protection simultaneously, then transgene expression levels tended to increase as a result of transcription from DNA within cell nuclei and the subsequent translation from mRNA in cytoplasm. From these results, we observed that as the proportion of the INO increased, the efficiency of identical dosages of PG6-PEI-INOs to composite DNA was similar (), indicating that DNA-binding ability might be primarily related to polymer structure. PG6-PEI-INO 3 at identical dosages with the lowest N/P ratios showed the highest transgene expression efficiency at most tested weight ratios to DNA, without obvious impairment in cell viability, demonstrating the potentiality of using inositol as an efficient ligand for transgene expression.
In our previous studies, INO-PEI800-FA was constructed and demonstrated efficient transgene activity. However, on the basis of low molecular weight PEI without any transgene activity, the transfection should still be limited. Partially attributed to the greatly enhanced biocompatibility compared with PEI25k, PG6-PEI-INO developed in this study can be used at high dosages without apparent injury to 293T cells, and this may allow the consequent generation of much higher transgene expression levels in the 293T cell line. It has been revealed that PEI800-INO performs better in carcinoma cells than in 293T. The performance of PG6-PEI-INO in carcinoma cells is the goal of our next study.
It is noteworthy that, along with the increase in myo-inositol from 1 to 35 per PEI25k unit within polymeric molecule, PG6-PEI-INO polymers showed a corresponding increase in the transgenic efficiency. As compared with the current commercial reagent Lipofectamine (20%–60% transfection rate depending on cell types),Citation50,Citation51 PG6-PEI-INO polymers based on HMW PEI can obtain a higher transgene expression rate in the presence of serum and antibiotics. For 293T cell lines, this high transfection rate (>80%) achieved with PG6-PEI-INO 3 is higher than commercial Lipofectamine, and the efficiency can be realized in a considerably higher and wider range of dosages with reasonable viability. Lipofectamine can achieve higher efficiency when used for transient RNA delivery. This is because the RNA does not need to pass through the nuclear membrane. Whether the myo-inositol group can improve the transfection activity of vectors like Lipofectamine 3000, Fugene 6, and chitosan-based vectors is a topic worthy of study, and this work is currently ongoing in our laboratory.
Extracellular ATP increases viability of cells treated with PG6-PEI-INOs/pDNA
Since eATP can influence cell-nuclear activity and transport (including transmembrane or transnuclear transport) and because the metabolism of inositol and its derivatives are related to ATP in some essential aspects,Citation31,Citation47,Citation52–Citation54 we investigated the influence of eATP on cell viability and transgene expression with PG6-PEI-INO 3 containing the highest contents of INO as a representative medium for transfection. PEI25k and PG6-PEI without nuclear entrance ability were set as negative controls.
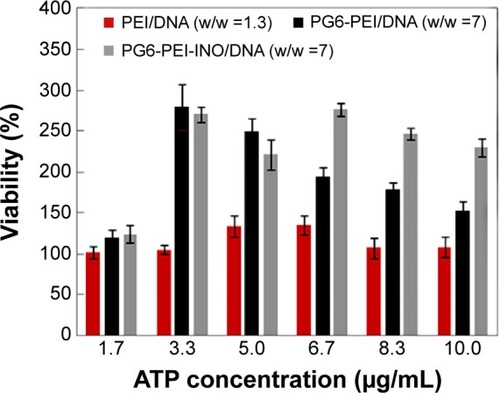
Results indicated that eATP clearly stimulated propagation of 293T cells transfected with PG6-PEI-INO 3/pEGFP-C1, compared with those transfected with PEI25k/pEGFP-C1 or PG6-PEI/pEGFP-C1 (). PG6-PEI/pEG-FP-C1 transfected cells doubled in growth at 3.3 μg/mL of eATP, and then decreased with further increases in eATP. The viability of PEI25k/pEGFP-C1 treated cells was not significantly affected by eATP. Therefore, with respect to cell viability, response levels to eATP differed because of the biocompatible PG6 and INO moieties. Since polyglycerol and inositol can be metabolized more efficiently when ATP is increased, this may increase the cell viability. In addition, derivatives of the INO ligands may also participate in PI3K signaling pathways that are required by ATP-stimulated cell proliferation.Citation55
Figure 7 Effect of extracellular ATP on the growth rate of 293T cells transfected with various polymer/complexes (at optimal weight ratio for transfection).
Notes: MTT analyses of cell viability were performed after the 293T cells were cocultivated with the materials for 44 hours. About 1.3 μg of plasmid pEGFP-C1 was used per microliter of cell culture.
Abbreviations: ATP, adenosine triphosphate; INO, myo-inositol; PEI, polyethylenimine; PG6, polyglycerol.
Extracellular ATP inhibited transgene activity of PG6-PEI-INO
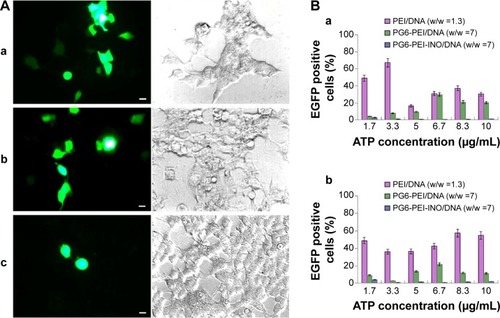
The influence of eATP on transfection with PG6-PEI-INO 3/pEGFP-C1, PG6-PEI/pEGFP-C1, and PEI25k/pEGFP-C1 at optimized weight ratios was analyzed. With the supplementation of eATP, cells transfected with the PG6-PEI-INO 3/pEGFP-C1 reagent declined to <1%. Transfection with PEI25k/pEGFP-C1 was not apparently influenced by eATP (). Compared with PG6-PEI-INO, the influence of eATP on PG6-PEI was much smaller. Since eATP did not reduce the transfection activity of PEI25k or PG6-PEI apparently, we deduced that the transgene activity of PG6-PEI-INO 3 should be related to certain intracellular pathways, rather than weakening of DNA-binding activity.
Figure 8 Inhibition of extracellular ATP on transgene activity of PG6-PEI-INO.
Notes: (A) The 293T cells were treated with (a) PEI25k/pEGFP-C1 (w/w =1.3), (b) PG6-PEI/pEGFP-C1 (w/w =7), and (c) PG6-PEI-INO 3/pEGFP-C1 (w/w =7) for 96 hours, respectively. Extracellular ATP was used at 1.7 μg per mL culture medium. Scale bar: 10 μm. (B) EGFP-positive 293T cell ratios mediated by polymers/DNA/ATP. Plasmid DNA (pEGFP-C1) was used at 1.3 μg per mL culture medium. The cells were treated with the polymers/DNA/ATP for 44 hours (a) and 96 hours (b), respectively.
Abbreviations: ATP, adenosine triphosphate; INO, myo-inositol; PEI, polyethylenimine; PG6, polyglycerol.
Distribution of PG6-PEI-INO in cell nuclei
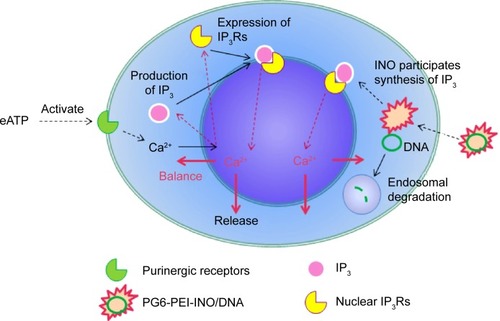
By 48 hours of incubation, all the INO conjugated vectors were internalized by 293T cells at high frequencies. Results of the MTT analysis indicated high viability of the cells. Compared with PG6-PEI-INO-Rh 1 with approximately one INO moiety grafted per PEI unit, PG6-PEI-INO-Rh 2 and 3 with increased number of INO grafts had increased accumulation within the cell nuclei (). More than 80% of the 293T cells showed distinct nuclear access of the PG6-PEI-INO-Rh 3 polymers. Of note, PG6-PEI-INO-Rh 3 polymers were concentrated to cell nuclei from the cytoplasm. We believe that this difference between PG6-PEI-INO-Rh 1, 2, and 3 should be related to the mechanisms relevant to the transport of nanoparticles mediated by the NPC. As reported, this NPC-mediated transport of various large molecules (such as DNAs, RNAs) is related to the Ca2+ pump.Citation56 In some mammalian cell nuclei, extracellular ATP activates purinergic receptors such as P2Y2 and P2X4, which induces an increase in the free Ca2+ concentration in the cell nuclei. To balance the Ca2+ transient, cADP-ribose and the intracellular messenger, inositol 1,4,5-triphosphate [Ins(1,4,5)P3, IP3], can trigger the release of partial nuclear Ca2+ through the actions of inositol 1,4,5-trisphosphate receptors (IP3R), indicating a role for extracellular ATP in regulating nuclear function, by increasing nuclear Ca2+ concentrations.Citation47,Citation56–Citation60 Since inositol is metabolically linked to IP3 within mammalian cells,Citation42–Citation46 this course may consume INO ligands in the PG6-PEI-INO vectors and result in unpacking of vector/pDNA complexes, and further inhibit transgene expression. Relevant mechanisms were predicted and shown in . This was further demonstrated by the less negative influence of eATP on PEI25k or PG6-PEI-mediated EGFP expression (). Although cells treated with PG6-PEI/pEGFP-C1 were inhibited by eATP more apparently than PEI25k/pEGFP-C1-treated cells, their variance is considerably smaller compared with cells treated with PG6-PEI-INO 3/pEGFP-C1, which approaches complete inhibition.
Figure 9 Fluorescence tracking of PG6-PEI-INO polymers in 293T cell nuclei.
Notes: Fluorescence was analyzed after the 293T cells were cocultivated for 48 hours with (A) PG6-PEI-INO-Rh 1, (B) PG6-PEI-INO-Rh 2, and (C) PG6-PEI-INO-Rh 3, which had increased inositol ligands per molecule (1:1, 1:10, and 1:35, respectively). Accumulation of PG6-PEI-INO-Rhs in cell nuclei was indicated with arrows. Scale bar in magnified images: 15 μm. Red fluorescence shows Rhodamine B (Emission: ~572 nm) labeled polymers. Blue fluorescence shows the cell nuclei stained with DAPI (Emission: ~461 nm when bound with DNA). Merged images in purple color show the PG6-PEI-INO-Rh polymers in cell nuclei.
Abbreviations: DAPI, (4′,6-diamidino-2-phenylindole); INO, myo-inositol; PEI, polyethylenimine; PG6, polyglycerol; Rh, Rhodamine.
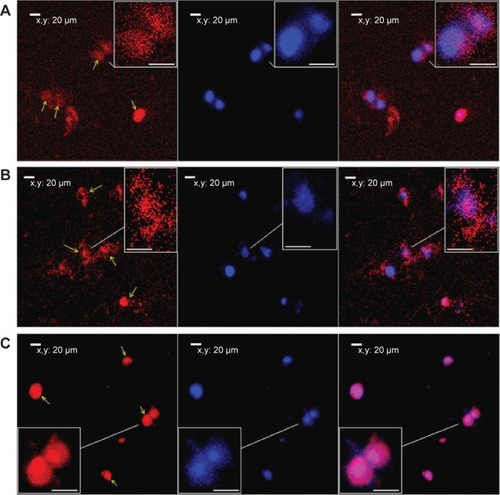
Figure 10 Prediction of eATP function during the delivery of plasmid DNA with PG6-PEI-INO as mediator.
Abbreviations: eATP, extracellular adenosine triphosphate; INO, myo-inositol; IP, inositol polyphosphate; IP3R, inositol 1,4,5-trisphosphate receptor; PEI, polyethylenimine; PG6, polyglycerol.
It is known that PEIs can help nucleotides escape from endosomal degradation through the “proton-sponge effect”.Citation6,Citation12 However, the ability of PEIs to mediate transgene expression was limited. LMW PEIs lack transfection activity, whereas mid-to-high concentration of HMW PEIs can damage cell membranes.Citation61 In general, current gene vectors lack the ability to locate in cell nuclei, and it was deemed that nuclear import of therapeutic DNA or RNA occurred after they were released from the PEI transgene systems.Citation62–Citation64 As commonly accepted, the barrier of nuclear membrane can largely limit transgene expression. Therefore, researchers are currently interested in investigating cationic gene vectors that can locate in cell nuclei.Citation29,Citation63,Citation64 Using PG6-PEI25k as the platform, the ability of the PG6-PEI-INO polymers to enter cell nuclei and to mediate efficient transgene expression was found to increase with their INO moiety numbers, as reported by fluorescent tracking and EGFP expression. Therefore, the INO might be a potential ligand for efficient transgene expression.
As compared with PEI800-INO-FA, the higher frequency exhibited by PG6-PEI25k-INO of entering cell nuclei of 293T should be related to the enhanced water solubility and improvement in cell internalization.Citation22
Conclusion
HMW PEI has high transfection activity, but its cytotoxicity and inability to pass through the nuclear barrier are deemed as shortcomings. In our previous studies based on LMW PEI without transfection activity but possessing good cell compatibility, myo-inositol was introduced. The resultant polymers (PEI-INOs) have obvious transfection activity, and in some experimental cell lines, they exhibit transfection activity near HMW PEI. In order to explore myo-inositol as a potential ligand for enhancing transfection efficiency and improving cell compatibility, and to further expand the range of applications for HMW PEI, we studied PG6-PEI. PG6-PEI was synthesized by grafting HMW PEI to the functional hydroxyl termini of the branched biocompatible polyglycerol, as a platform in this report. Previous research showed that LMW PEI conjugated with inositol can partially enter cell nuclei, and, therefore, the properties of myo-inositol with nuclear localization capability are a major topic of attention for our group.
Since INO is a natural biomolecule that impacts many important physiological activities, it was introduced as a potential biological moiety to PG6-PEI in this study through covalent reaction. The resultant polymers, PG6-PEI-INO macromolecules, were characterized by FT-IR spectrometry, 1H NMR spectroscopy, and combined SEC-MALLS analysis. Gel electrophoresis retardation and TEM analysis demonstrated the DNA-binding activity of PG6-PEI-INOs, and all the polymers could compact plasmid DNA and generate nanoparticles with diameters of less than 30 nm. Enhancement of effective transgene reported by EGFP expression was achieved in an actively propagating 3D 293T cell line using PG6-PEI-INO systems as mediators. Meanwhile, PG6-PEI-INO polymers presented reduced cytotoxicity when compared with PG6-PEI and PEI25k polymers. In the presence of eATP, the transgene expression level was inhibited, while cell growth was stimulated. CLSM analysis demonstrated the ability of PG6-PEI-INO polymers to enter the cell nuclei of 3D 293T cells. With an increase in conjugated INO, both the level of transgene activity and the accumulation of the PG6-PEI-INO polymers in the cell nuclei were elevated.
In conclusion, this work showed the potentiality of INO to be used as a ligand for efficient transgene expression applied in a wide area including transgene therapy and phenotypic analysis of a cell population.
Acknowledgments
We thank the National Nature Science Foundation of China (31300650 and 31270790), the Key Project of Fujian Province (2013N0039, 2012Y0070), the Nature Science Foundation of Fujian Province (2013J01151), the Science Foundation for Post Doctorate Research of Wuhan University of China (203-180462), the National Thousand Talents Program of China, and the Scientific Research Starting Foundation for Returned Overseas Chinese Scholars of State Human Resource Ministry of China for support on this work.
Disclosure
The authors report no conflicts of interest in this work.
References
- VermaIMSomiaNGene therapy – promises, problems and prospectsNature199738966482392429305836
- PackDWHoffmanASPunSStaytonPSDesign and development of polymers for gene deliveryNat Rev Drug Discov20054758159316052241
- CarlisleRCBettingerTOgrisMHaleSMautnerVSeymourLWAdenovirus hexon protein enhances nuclear delivery and increases transgene expression of polyethylenimine/plasmid DNA vectorsMol Ther20014547348311708884
- KayMAAAV vectors and tumorigenicityNat Biotechnol200725101111111317921994
- FerberDGene therapy: safer and virus-free?Science200129455471638164211721029
- BoussifOLezoualchFZantaMAA versatile vector for gene and oligonucleotide transfer into cells in culture and in-vivo – polyethylenimineProc Natl Acad Sci U S A19959216729773017638184
- PutnamDPolymers for gene delivery across length scalesNat Mater20065643945116738681
- OhlfestJRFreeseABLargaespadaDANonviral vectors for cancer gene therapy: prospects for integrating vectors and combination therapiesCurr Gene Ther20055662964116457652
- GloverDJLippsHJJansDATowards safe, non-viral therapeutic gene expression in humansNat Rev Genet20056429931015761468
- OgrisMWagnerETargeting tumors with non-viral gene delivery systemsDrug Discov Today20027847948511965397
- WagnerETargeting tumors with non-viral gene delivery systemsJ Gene Med200353S7S8
- GodbeyWTWuKKMikosAGPoly(ethylenimine) and its role in gene deliveryJ Control Release1999602–314916010425321
- HuCHZhangLWuDQChengSXZhangXZZhuoRXHeparin-modified PEI encapsulated in thermosensitive hydrogels for efficient gene delivery and expressionJ Mater Chem2009192031893197
- KreppelFKochanekSModification of adenovirus gene transfer vectors with synthetic polymers: a scientific review and technical guideMol Ther2008161162917912234
- ZhangLHuCHChengSXZhuoRXPEI grafted hyperbranched polymers with polyglycerol as a core for gene deliveryColloids Surf B Biointerfaces201076242743320042319
- NeuMGermershausOBeheMKisselTBioreversibly cross-linked polyplexes of PEI and high molecular weight PEG show extended circulation times in vivoJ Control Release20071241–2698017897749
- Bivas-BenitaMRomeijnSJungingerHEBorchardGPLGA-PEI nanoparticles for gene delivery to pulmonary epitheliumEur J Pharm Biopharm20045811615207531
- ChiuSHUenoNTLeeRJTumor-targeted gene delivery via anti-HER2 antibody (trastuzumab, Herceptin®) conjugated polyethylemmineJ Control Release200497235736915196762
- KunathKMerdanTHegenerOHaberleinHKisselTIntegrin targeting using RGD-PEI conjugates for in vitro gene transferJ Gene Med20035758859912825198
- ZantaMABoussifOAdibABehrJPIn vitro gene delivery to hepatocytes with galactosylated polyethylenimineBioconjugate Chem199786839844
- VeisehOKievitFMGunnJWRatnerBDZhangMQA ligand-mediated nanovector for targeted gene delivery and transfection in cancer cellsBiomaterials200930464965718990439
- ZhangLFanYWuYInositol based non-viral vectors for transgene expression in human cervical carcinoma and hepatoma cell linesBiomaterials20143562039205024314555
- ZhangCGaoSJJiangWTargeted minicircle DNA delivery using folate-poly(ethylene glycol)-polyethylenimine as non-viral carrierBiomaterials201031236075608620488533
- ZantaMABelguise-ValladierPBehrJPGene delivery: a single nuclear localization signal peptide is sufficient to carry DNA to the cell nucleusProc Natl Acad Sci U S A199996191969874777
- ManickamDSOupickyDMultiblock reducible copolypeptides containing histidine-rich and nuclear localization sequences for gene deliveryBioconjugate Chem200617613951403
- JeonOLimHWLeeMSongSJKimBSPoly(L-lactide-co-glycolide) nanospheres conjugated with a nuclear localization signal for delivery of plasmid DNAJ Drug Target200715319019817454356
- YooHSJeongSYNuclear targeting of non-viral gene carriers using psoralen-nuclear localization signal (NLS) conjugatesEur J Pharm Biopharm2007661283317123797
- OgrisMCarlisleRCBettingerTSeymourLWMelittin enables efficient vesicular escape and enhanced nuclear access of nonviral gene delivery vectorsJ Biol Chem200127650475504755511600500
- ParkKMKangHCChoJKAll-trans-retinoic acid (ATRA)-grafted polymeric gene carriers for nuclear translocation and cell growth controlBiomaterials200930132642265219181377
- HuangHMToral-BarzaLThalerHTofel-GrehlBGibsonGEInositol phosphates and intracellular calcium after bradykinin stimulation in fibroblasts from young, normal aged and Alzheimer donorsNeurobiol Aging19911254694731770982
- ShenXTXiaoHRanalloRWuWHWuCModulation of ATP- dependent chromatin-remodeling complexes by inositol polyphosphatesScience2003299560311211412434013
- StegerDJHaswellESMillerALWenteSRO’SheaEKRegulation of chromatin remodeling by inositol polyphosphatesScience2003299560311411612434012
- JiaSDRobertsTMZhaoJJShould individual PI3 kinase isoforms be targeted in cancer?Curr Opin Cell Biol200921219920819200708
- BatrakouDGKerrARWSchirmerECComparative proteomic analyses of the nuclear envelope and pore complex suggests a wide range of heretofore unexpected functionsJ Proteomics2009721567018852071
- MissiroliSEtroDBuontempoFYeKQCapitaniSNeriLMNuclear translocation of active AKT is required for erythroid differentiation in erythropoietin treated K562 erythroleukemia cellsInt J Biochem Cell Biol200941357057718694847
- HumbertJPMatterNArtaultJCKopplerPMalviyaANInositol 1,4,5-trisphosphate receptor is located to the inner nuclear membrane vindicating regulation of nuclear calcium signaling by inositol 1,4,5-trisphosphate – discrete distribution of inositol phosphate receptors to inner and outer nuclear membranesJ Biol Chem199627114784858550605
- LaniniLBachsOCarafoliEThe calcium-pump of the liver nuclear-membrane is identical to that of endoplasmic-reticulumJ Biol Chem19922671611548115521317870
- LeeDGLeeJHChoiBKH+-myo-inositol transporter SLC2A13 as a potential marker for cancer stem cells in an oral squamous cell carcinomaCurr Cancer Drug Targets201111896697521861841
- NarumiKTsumitaTIsolation and identification of free myo-inositol and scyllo-inositol in cell nucleiJpn J Exp Med19693944094155308952
- MishraOPQayyumIDelivoria-PapadopoulosMHypoxia-induced modification of the inositol triphosphate receptor in neuronal nuclei of newborn piglets: role of nitric oxideJ Neurosci Res200374233333814515363
- NalaskowskiMMMetznerABrehmMAThe inositol 5-phosphatase SHIP1 is a nucleo-cytoplasmic shuttling protein and enzymatically active in cell nucleiCell Signal201224362162821864674
- CarrascoDAllendeCCAllendeJEThe incorporation of myo-inositol into phosphatidylinositol derivatives is stimulated during hormone-induced meiotic maturation of amphibian oocytesExp Cell Res199019123133182257883
- VoglmayrJKAlpha-chlorohydrin-induced changes in the distribution of free myo-inositol and prostaglandin F2alpha, and synthesis of phosphatidylinositol in the rat epididymisBiol Reprod19741155936004376969
- ChuSWGeyerRPmyo-Inositol action on gerbil intestine. Association of phosphatidylinositol metabolism with lipid clearanceBiochim Biophys Acta1982710163706173076
- AbelKAndersonRAShearsSBPhosphatidylinositol and inositol phosphate metabolismJ Cell Sci2001114Pt 122207220811493657
- MattinglyRRStephensLRIrvineRFGarrisonJCEffects of transformation with the v-src oncogene on inositol phosphate metabolism in rat-1 fibroblasts. D-myo-inositol 1,4,5,6-tetrakisphosphate is increased in v-src-transformed rat-1 fibroblasts and can be synthesized from D-myo-inositol 1,3,4-trisphosphate in cytosolic extractsJ Biol Chem19912662315144151531869546
- ChenZLiZZPengGExtracellular ATP-induced nuclear Ca2+ transient is mediated by inositol 1,4,5-trisphosphate receptors in mouse pancreatic beta-cellsBiochem Biophys Res Commun2009382238138419285037
- PanteNKannMNuclear pore complex is able to transport macromolecules with diameters of about 39 nmMol Biol Cell200213242543411854401
- HonoreIGrosseSFrisonNFavatierFMonsignyMFajacITranscription of plasmid DNA: influence of plasmid DNA/polyethylenimine complex formationJ Control Release2005107353754616087268
- LiuCXChenZJYuWYZhangNNovel cationic 6-lauroxyhexyl lysinate modified poly(lactic acid)-poly(ethylene glycol) nanoparticles enhance gene transfectionJ Colloid Interf Sci20113542528535
- TripathiSKGoyalRKumarPGuptaKCLinear polyethylenimine-graft-chitosan copolymers as efficient DNA/siRNA delivery vectors in vitro and in vivoNanomedicine20128333734521756861
- ArkhammarPHallbergAKindmarkHNilssonTRorsmanPBerggrenPOExtracellular ATP increases cytoplasmic free Ca-2+ concentration in clonal insulin-producing Rinm5f cells – a mechanism involving direct interaction with both release and refilling of the inositol 1,4,5-trisphosphate-sensitive Ca-2+ poolBiochem J199026512032112405836
- NagyRGrobHWederBThe Arabidopsis ATP-binding cassette protein AtMRP5/AtABCC5 is a high affinity inositol hexakisphosphate transporter involved in guard cell signaling and phytate storageJ Biol Chem200928448336143362219797057
- GerasimenkoOVGerasimenkoJVTepikinAVPetersenOHATP-dependent accumulation and inositol trisphosphate-mediated or cyclic ADP-ribose-mediated release of Ca2+ from the nuclear-envelopeCell19958034394447859285
- WildenPAAgazieYMKaufmanRHalendaSPATP-stimulated smooth muscle cell proliferation requires independent ERK and PI3K signaling pathwaysAm J Physiol19982754 Pt 2H1209H12159746468
- PaulilloSMPowersMAUllmanKSFahrenkrogBChanges in nucleoporin domain topology in response to chemical effectorsJ Mol Biol20063631395016962132
- BustamanteJOMicheletteERGeibelJPDeanDAHanoverJAMcDonnellTJCalcium, ATP and nuclear pore channel gatingPflugers Arch2000439443344410678739
- NicoteraPOrreniusSNilssonTBerggrenPOAn inositol 1,4,5-trisphosphate-sensitive Ca2+ pool in liver nucleiProc Natl Acad Sci U S A19908717685868622204067
- ElsingCGeorgievTHubnerCABogerRStremmelWSchlenkerTExtracellular ATP induces cytoplasmic and nuclear Ca2+ transients via P2Y2 receptor in human biliary epithelial cancer cells (Mz-Cha-1)Anticancer Res20123293759376722993317
- GuihardGProteauSRousseauEDoes the nuclear envelope contain two types of ligand-gated Ca2+ release channels?FEBS Lett1997414189949305738
- KunathKvon HarpeAFischerDLow-molecular-weight polyethylenimine as a non-viral vector for DNA delivery: comparison of physicochemical properties, transfection efficiency and in vivo distribution with high-molecular-weight polyethylenimineJ Control Release200389111312512695067
- KanazawaTTakashimaYMurakoshiMNakaiYOkadaHEnhancement of gene transfection into human dendritic cells using cationic PLGA nanospheres with a synthesized nuclear localization signalInt J Pharm2009379118719519555751
- HebertEImprovement of exogenous DNA nuclear importation by nuclear localization signal-bearing vectors: a promising way for non-viral gene therapy?Biol Cell2003952596812799061
- LedleyFDPharmaceutical approach to somatic gene therapyPharm Res19961311159516148956323
