Abstract
Bone tissue engineering through seeding of stem cells in three-dimensional scaffolds has greatly improved bone regeneration technology, which historically has been a constant challenge. In this study, we researched the use of adipose-derived stem cell (ADSC)-laden layer-by-layer paper-stacking polycaprolactone/gelatin electrospinning nanofibrous membranes for bone regeneration. Using this novel paper-stacking method makes oxygen distribution, nutrition, and waste transportation work more efficiently. ADSCs can also secrete multiple growth factors required for osteogenesis. After the characterization of ADSC surface markers CD29, CD90, and CD49d using flow cytometry, we seeded ADSCs on the membranes and found cells differentiated, with significant expression of the osteogenic-related proteins osteopontin, osteocalcin, and osteoprotegerin. During 4 weeks in vitro, the ADSCs cultured on the paper-stacking membranes in the osteogenic medium exhibited the highest osteogenic-related gene expressions. In vivo, the paper-stacking scaffolds were implanted into the rat calvarial defects (5 mm diameter, one defect per parietal bone) for 12 weeks. Investigating with microcomputer tomography, the ADSC-laden paper-stacking membranes showed the most significant bone reconstruction, and from a morphological perspective, this group occupied 90% of the surface area of the defect, produced the highest bone regeneration volume, and showed the highest bone mineral density of 823.06 mg/cm3. From hematoxylin and eosin and Masson staining, the new bone tissue was most evident in the ADSC-laden scaffold group. Using quantitative polymerase chain reaction analysis from collected tissues, we found that the ADSC-laden paper-stacking membrane group presented the highest osteogenic-related gene expressions of osteocalcin, osteopontin, osteoprotegerin, bone sialoprotein, runt-related transcription factor 2, and osterix (two to three times higher than the control group, and 1.5 times higher than the paper-stacking membrane group in all the genes). It is proposed that ADSC-laden layer-by-layer paper-stacking scaffolds could be used as a way of promoting bone defect treatment.
Introduction
Bone defects, especially large ones, still present a significant challenge in orthopedic tissue engineering, due to the limited self-repair capacity of bone tissues.Citation1,Citation2 Three-dimensional (3D) scaffolds play a vital role in bone regeneration and offer a promising approach to bone repair.Citation3,Citation4 Although perfectly imitative of bone structure, and providing necessary support for cells and new tissue in-growth, these scaffolds still have some limitations: how to imitate oxygen distribution and nutrient and waste transportation and how to signal transduction and spatiotemporal chemical gradients, which are important for cell growth in the scaffold and internal environment homeostasis in the living tissues.Citation5,Citation6 With these problems in view, Whitesides et alCitation7 proposed a paper-supported scaffold and demonstrated that oxygen and nutrients can distribute to the center layers of the scaffold to form spatiotemporal chemical gradients for cell growth if thin (30–1,500 μm) and permeable papers are used. As a well-designed chemical gradient, this paper-stacking scaffold can be used to either promote the living tissue growth or inhibit cancer and infection.Citation8–Citation10 In addition, this paper-stacking strategy provides a versatile approach to fabricate 3D scaffolds, which, in turn, provide chemical gradients with different polymers. Moreover, by using different polymers with a variety of layers and thicknesses, these scaffolds can supply a range of properties vital for tissue functions, especially for bone or cartilage.Citation11
Recently, therapeutics based on stem cells, such as bone marrow-derived mesenchymal stem cells, embryonic stem cells, and adipose-derived stem cells (ADSCs), have gained extensive attention in bone tissue engineering. By virtue of their abundance, easy isolation, and culture, which induces the release of multiple growth factors required for bone regeneration, ADSCs have been recommended for osteoblast differentiation and bone regeneration.Citation12–Citation15 Combined with biomaterials, many researchers have incorporated ADSCs into 3D scaffold for bone repair and achieved great success. For instance, ADSCs have been incorporated into coral scaffold-enhanced bone reconstruction.Citation12,Citation14
Inspired by these studies, we constructed a novel and versatile 3D paper-stacking scaffold composed of gelatin/polycaprolactone (PCL) electrospun nanofibrous membranes for bone regeneration. This scaffold has both excellent biocompatibility and biomechanical stability due to the superlative quality of gelatin for cell adhesion and the good mechanical character of PCL.Citation16,Citation17 The electrospun membranes are thin (approximately 50 μm) and permeable, which facilitate a good distribution of oxygen and nutrients into the center of the paper-stacking scaffold.Citation18 In addition, we laid ADSCs into the scaffold not only because they supply a large amount of stem cells that differentiate into osteogenic lineages but also because they secrete certain growth factors and extracellular matrix (ECM), which provide a biomimetic environment for tissue regeneration.Citation19 When the ADSCs are cultured in a layer-by-layer paper-stacking scaffold, they create a spatiotemporal chemical gradient of the secreted factors and improve the signaling transduction inside the scaffold.Citation20,Citation21 The ADSC-laden scaffold promotes the cell-to-cell and cell-to-tissue interactions that are crucial for cell amplification, differentiation, and tissue regeneration.Citation22–Citation24 Moreover, this ADSC-laden paper-stacking scaffold constructs a layer-by-layer structure that is unique to bone tissue or subcartilage bone.
In this study, we first used surface markers to characterize ADSCs. We then seeded ADSCs on the membrane in the osteogenic medium (OM) and detected the secreted osteogenic proteins. We also investigated cell viability and osteogenic relative gene expressions in vitro in paper-stacking scaffolds. Finally, ADSC-laden paper-stacking membranes were implanted into the rat parietal bone defect model. After 12 weeks, the regenerated bone tissue was detected using micro-computed tomography (micro-CT), histological staining, and a real-time quantitative polymerase chain reaction (RT-qPCR) test.
Materials and methods
Materials
PCL, gelatin, and trifluoroethanol were all purchased from Sigma-Aldrich (Saint Louis, MO, USA). An electrospun machine was made by Beijing Ucalery Technology Development Co., Ltd (Beijing, People’s Republic of China). Live/dead cell staining kits were purchased from Invitrogen (Carlsbad, CA, USA). A Cell Counting Kit-8 (CCK-8) was obtained from Biotium Inc (Hayward, CA, USA). Low glucose Dulbecco’s Modified Eagle’s Medium (DMEM), fetal bovine serum, penicillin (100 units/mL), and streptomycin (100 units/mL) were all ordered from Gibco (Grand Island, NY, USA). Dexamethasone (100 nM), ascorbate-2-phosphate (50 μM), and β-glycerophosphate (10 mM) were obtained from Sigma-Aldrich. Trizol reagent and a SuperScript First-Strand Synthesis System kit for RT-PCR were from Invitrogen. Rat glyceraldehyde-3-phosphate dehydrogenase (GAPDH), osteocalcin antibody (OCN), osteoproteoglycan antibody (OPG), and osteopontin antibody (OPN) were purchased from Abcom (Cambridge, MA, USA). Scanning electron microscopy (SEM) figures were obtained using a microscope (JSM-5510LV, JEOL, Tokyo, Japan). Flow cytometry tests were conducted with FAC-SCalibur cytometry (Becton Dickinson, Franklin Lakes, NJ, USA). Images were taken using a confocal laser scanning microscope (Olympus FV1000, Tokyo, Japan), and the micro-computed tomography (micro-CT) was Letheta 200 (Alcoa Company, Tokyo, Japan). The absorbance was measured with an enzyme-linked immunosorbent assay (ELISA) reader (Hiperion MPR4, Germany). Ribonucleic acid (RNA) concentration was detected with an RNA reading machine (Thermo Fisher Scientific, Waltham, MA, USA). A StepOnePlus RT-qPCR system machine for qPCR tests was purchased from Life Technologies.
Rat ADSC isolation, culture, passage, and identification
ADSCs were isolated from a 1-month-old Sprague Dawley (SD) rat’s fat, according to the procedure outlined as follows. After perfect anesthesia with 10% chloral hydrate (0.2 mL/100 g), the SD rats were sterilized with the 75% ethyl alcohol three times, and the inguinal fat pad area was explored. The inguinal fat pad was harvested and minced in phosphate-buffered saline (PBS), adding two times the volume of 0.2% type I collagenase, and put in a 37°C water-bath shaker for 40 minutes, then centrifuged at 800 g for 5 minutes. The supernatant was discarded to obtain the ADSC pellets.Citation13,Citation25,Citation26 Then, the cell pellets were suspended and seeded in 75 cm2 cell cultured flasks in DMEM, supplemented with 10% fetal bovine serum and 1% antibiotics. Next, the cells were cultured at 37°C under 5% CO2. Unattached cells were removed after 24 hours. The medium was changed every 2 days until cells reached 80% confluence. Subculture and passage used a ratio of 1:3, and cells were used on passage 3 or 4.Citation15,Citation27
ADSCs were identified by flow cytometry. Passage 4 ADSCs were collected with Trypsin-EDTA, then were resuspended in PBS and counted. All the cells were divided into five aliquots of 106 cells in a 15 mL tube. The cells were centrifuged and the cell pellets were obtained. A volume of 100 μL PBS was added to each tube, and the cells were resuspended. The cells were stained with cell surface marker antibodies: antirat monoclonal CD90, CD49d-(fluorescein isothiocyanate), CD106, CD45-(phycoerythrin) and CD11b, and CD29-(allophycocyanin).Citation28 CD29 antibody was added in sample 1; CD90 in sample 2; CD90, CD11b, and CD49d in sample 3; CD106, CD29, and CD49d in sample 4; and no antibody in sample 5. All the antibodies were added at a concentration of 2 μg/mL at 4°C and incubated for 30 minutes. Then, we added 5% fetal bovine serum PBS 5 mL, and the cells were resuspended and centrifuged at 800 rpm for 5 minutes. Then, 300 μL 5% fetal bovine serum was added and washed with PBS. The unstained cells served as the control group. Then, the samples were examined using flow cytometry using CellQuest software.Citation29
Fabrication of electrospinning nanofibrous membranes and the gelatin crosslinking procedure in the membrane
The PCL and the gelatin were dissolved separately in the trifluoroethanol and stirred overnight to make solutions at a concentration of 10 wt%, respectively. Three milliliters of each solution was poured into 10 mL glass syringes. Then, the electrospinning was performed in the electrospun machine for about 2–3 hours in a gelatin:PCL ratio of 1:1 to manufacture the nanofibrous membranes to a thickness of approximately 70 μm. The electrospinning rate was set to 1.0 mL/hr. In the electrical field of 15 kV, the syringe tip injected the materials solution to the tuning axis, which was wound with a 6×8 cm piece of aluminum foil and tuned at a speed of 5 m/min.Citation30–Citation32 The distance between the syringe tip and the foil was 12 cm.
PCL/gelatin electrospun membranes were incubated in the glutaraldehyde vapor at room temperature for 2 hours for the purpose of crosslinking.Citation33 The amine groups of gelatin were crosslinked with the aldehyde groups of glutaraldehyde and became insoluble in water.Citation34
ADSCs were cultured on the membranes
The PCL/gelatin electrospun nanofibrous membranes were first fabricated using the method described in the section “Fabrication of electrospinning nanofibrous membranes and the gelatin crosslinking procedure in the membrane”, then the membrane was punched into 6 mm diameter circular pieces. The membranes were sterilized under ultraviolet light for 5 hours and turned over every 30 minutes. Then, the membranes were immersed in sterilized PBS for 24 hours. The PBS was changed every 4 hours. The cells’ culture dishes were first coated with 1% sterilized agarose. ADSCs at passage 4 were harvested, seeded on one side of the membranes (using sterilized filter paper to absorb surplus water), and cultured in an incubator heated to 37°C in 5% CO2 for 1 hour, allowing the cells to attach to the membrane. After 1 hour, the membranes were turned over and the cells were seeded on another side and cultured in the incubator for another hour, then fresh culture medium was added to them. They were then put back into the incubator. The medium was changed every 2 days.
SEM view of the membrane and ADSC-laden membrane
To analyze the structure and morphology of gelatin/PCL nanofiber electrospun membranes, we used the SEM at an acceleration voltage of 15 kV. Three days after the cells were cultured, all the samples (membrane and ADSC-laden membrane) were rinsed twice with PBS, fixed with 2% glutaraldehyde, and dehydrated in increased concentration gradient alcohol (50%, 70%, 80%, 90%, 95%, and 100% ethanol solution). Then, the samples were dried, coated with a layer of gold, and scanned with the SEM.
ADSC osteogenic differentiation on a single membrane
ADSCs were cultured on the membrane as shown in section “ADSCs were cultured on the membranes” in the osteogenic culture medium, which contained dexamethasone (10 nM), ascorbate-2-phosphate (50 μM), and β-glycerophosphate (10 mM). Then, the ADSC osteogenesis differentiation was detected with immunofluorescence technique and antibodies such as OPN, OCN, and OPG. ADSCs (2×103) were seeded on the membrane and cultured in the six-well cell culture plate, and the culture medium was changed every 2 days. After 2 weeks and 4 weeks, the samples were fixed with 4% formalin (pH 7.4) for 15 minutes, immersed in 2 mL 0.5% Triton X-100, and left at room temperature for 10 minutes. The samples were rinsed with PBS three times. One percent bovine serum albumin was added to each well for 30 minutes, then the antibodies-I (OCN, OPN, OPG) were added to the samples separately and incubated at 37°C for 2 hours. The antibodies-II were added and incubated for 1 hour. 4′,6-diamidino-2-phenylindole (DAPI) at 5 μg/mL was added into to the well for 5–10 minutes. The samples were put on the slides. Images were taken with a confocal laser scanning microscope.Citation35
Fabrication of the ADSC-laden layer-by-layer paper-stacking membranes (three layers)
The membrane pieces were sterilized under ultraviolet light for 5 hours and turned over every 30 minutes. The cells’ culture dishes were first coated with 1% sterilized agarose. ADSCs at passage 4 were cultured as shown in the section “ADSC osteogenic differentiation on a single membrane”.
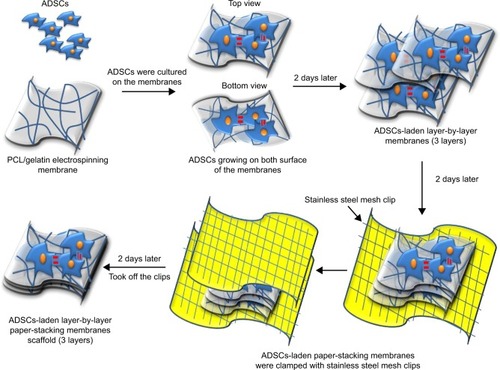
Two days later, the ADSCs were seeded on both sides of the membrane. After 2 more days, three pieces of ADSC-laden membrane were assembled in a layer-by-layer paper-stacking way to form the 3D scaffold, then the scaffold was clamped with two sterilized stainless steel mesh clips and cultured in 5% CO2 at 37°C for another 2 days.Citation36 The membranes interacted with each other and were developed into the 3D scaffold of ADSC-laden layer-by-layer paper-stacking membranes (three layers). The fabrication procedure is shown in .
Viability test of ADSCs in the layer-by-layer paper-stacking membrane scaffold
Three days after the cells were seeded, the ADSC-laden paper-stacking membrane scaffolds were harvested and immersed in 0.5% Triton X-100 solution at room temperature for 5 minutes, and then immersed in 1% bovine serum albumin PBS solution at room temperature for another 10 minutes. The cells in the scaffold were stained with the live/dead kit and DAPI staining kit, then embedded in the optimal cutting temperature compound. Then, the sections were cut into 5 μm thickness with a cryostat sectioning machine. Finally, images were taken using the fluorescence microscope.
In order to discover the ADSCs’ proliferation in the paper-stacking membrane scaffold, we used a CCK-8 to investigate. ADSCs were seeded in the paper-stacking membrane scaffold (three layers of membrane) at a density of 104 cells/well in a 96-well culture plate system. The samples were divided into three groups as follows: an ADSCs group in which ADSCs were cultured in the plate wells, an ADSC-laden paper-stacking membranes group in which the ADSCs were cultured in the paper-stacking membrane scaffold, and paper-stacking membranes without ADSCs in the culture medium were used as a control group. At the designated time points of 6, 12, 24, and 72 hours, 10 μL of CCK-8 reagent was added to each well and incubated for 1 hour in 5% CO2 at 37°C. Then, all the liquid in the sample well was transferred to a new 96-well plate. The absorbance was measured at 570 nm using an ELISA reader (n=3).
Osteogenic gene expressions in vitro
We introduced the RT-qPCR technique to test the gene expression in the ADSC-laden paper-stack 3D scaffolds. Four groups were investigated as follows: in Group I, ADSCs were cultured in a regular medium (cells group); in Group II, ADSC-laden paper-stacking scaffolds were cultured in a regular medium (cells-paper-stacking membranes group); in Group III, ADSCs were cultured in OM (cells-OM group); and, in Group IV, ADSC-laden paper-stacking scaffolds were cultured in OM (cells-paper-stacking membranes-OM group, n=3). At the designated time points (1, 2, 3, and 4 weeks), three samples of each group were harvested. The total RNA was isolated following a standard RNA isolating protocol. RNA pellets were dissolved in 15 μL ribonuclease-or deoxyribonuclease-free water. The RNA concentration of each sample was detected with an RNA reading machine on A495, and then the same amount of RNA from the samples was reversed and transcribed into first-strand complementary deoxyribonucleic acid (cDNA). First-strand cDNA (800 ng) was amplified by RT-qPCR using the oligonucleotide primers fabricated by GeneCopoeia. Forty-two cycles were used to amplify the first-strand cDNA in all samples, and the DNA products were analyzed in the StepOnePlus RT-qPCR system. The housekeeping gene GAPDH was used as an internal control. The osteogenic genes were examined, including OCN, OPN, OPG, bone sialoprotein (BSP), runt-related transcription factor 2 (RUNX2), and osterix (OSX). Primer sequences (forward primer and reverse primer) for RT-qPCR analysis were as follows:
GAPDH: F-AGGTCGGTGTGAACGGATTTG
R-TGTAGACCATGTAGTTGAGGTCA
OCN: F – ATTGTGACGAGCTAGCGGAC
R-TCGAGTCCTGGAGAGTAGCC
OPN: F – CTACGACCATGAGATTGGCAG
R-CATGTGGCTATAGGATCTGGG
OPG: F – CAGTGTGCAACGGCATATCG
R-CCAGGCAAGCTCTCCATCAA
BSP: F – AGCTGACCAGTTATGGCACC
R-TTCCCCATACTCAACCGTGC
RUNX2: F-CAACCGAGTCAGTGAGTGCT
R-AAGAGGCTGTTTGACGCCAT
OSX: F – CTTCGTGCCAGACCTCTTGA
R-GGACTGGAGCCATAGTGAGC
The ADSC-laden paper-stacking scaffolds for bone regeneration in vivo
The ADSC-laden layer-by-layer paper-stacking 3D scaffolds were investigated in a rat calvarial defect model.Citation37,Citation38 Twenty-seven male SD rats (weight range 250–300 g, 2 months old) were fed with a standard diet and housed in a standard laboratory for 1 week before the experiments. All animals were purchased from the Southern Medical University Experiment Animal Center (Guangzhou, People’s Republic of China) and were approved by the Southern Medical University Animal Ethics Committee, in accordance with the Regulations for the Administration of Affairs Concerning Experimental Animals.
Implantation procedure of the paper-stacking scaffolds
Two 5 mm diameter circular acute calvarial bone defects were created in each calvarial bone, each in the middle of the parietal bones, and three groups were designed as follows: in Group I, two bone defects were created without any treatment (control group, n=9); in Group II, the paper-stacking membrane scaffolds (three layers of membranes for each scaffold and one scaffold per bone defect) were implanted into the bone defects (paper-stacking membranes group, n=9); and, in Group III, ADSC-laden paper-stacking membrane scaffolds were implanted in the parietal bone defects (ADSC-laden paper-stacking membranes group, n=9).
All the rats were anesthetized with chloral hydrate (5%, 0.2 mL/100 g) using an intraperitoneal injection. Once anesthetized, a medial front–back incision was made on the rat’s head, penetrating the scalp, subcutaneous tissue, and periosteum and exposing the middle line of the cranium. Then, the scalp and subcutaneous tissue was carefully pulled to the side.Citation38 With a sharp trephine, a 5 mm diameter cylindrical defect totally penetrating the calvarial bone was made without any damage to the dura mater.Citation14,Citation39–Citation41 The PBS was injected into the defect to wet the operation site. The PCL/gelatin membranes were punched into 6 mm diameter circular pieces and sterilized for usage. ADSC-laden layer-by-layer paper-stacking membrane scaffolds were fabricated following the method described in the section “Fabrication of the ADSC-laden layer-by-layer paper-stacking membranes (three layers)”. The ADSCs (105, passage 4) were suspended in a 10 μL medium, seeded on each surface of the 6 mm diameter circular membranes, and cultured in the incubator. After 2 days, three layers of membrane were overlaid, clamped with two sterilized clips, and cultured in the incubator in 5% CO2 at 37°C. Two days later, ADSC-laden layer-by-layer paper-stacking membrane scaffolds were implanted into the defect (ADSC-laden paper-stacking membranes group).Citation36 Finally, the incision was sutured in separate layers and the wound was cleaned. The layer-by-layer paper-stacking membrane scaffolds without cells (three layers of membranes) were fabricated and planted into each defect (paper-stacking membranes group), and the incision closed in the same way. For the control group, the defect was made without any treatment. The regeneration tissue samples were harvested at 12 weeks postoperation, and the micro-CT test, histological analysis, and qPCR detection were performed.
Micro-CT scanning
For the bone regeneration investigation, we introduced the micro-CT to scan and show a morphological view of the defect site. During the scanning procedure, the parameters used for scanning were as follows: voltage source 85 kV, current source 120 mA, and a tomographic rotation of 180°. The results were reconstructed with Mimics (10.0) software, using a global threshold of 250–3,071 (low–high Hounsfield units).
Histological analysis
All the rats were sacrificed. The defect regenerated tissue samples were washed with PBS and then were fixed in 4% phosphate-buffered formalin (pH 7.4) for 48 hours and decalcified in 20% EDTA for 4 weeks, with the EDTA solution changed every 3 days. The tissue samples were dehydrated through a graded series of ethanol treatments and embedded in paraffin. Finally, sections of 5 μm thickness were made. To analyze bone regeneration, hematoxylin and eosin (HE) and Masson staining were used.
Osteogenic gene expressions analysis in vivo
The collected bone regenerated samples for RT-qPCR analysis were frozen in liquid nitrogen and then milled to powder. Total RNA isolation, cDNA synthesis, and RT-qPCR were conducted with the same standard method described in the section “Osteogenic gene expressions in vitro”.
Statistical analysis
The mean and standard deviations of the data were presented and were analyzed statistically using one-way analysis of variance with 95% confidence interval. A value of P<0.05 was considered to be statistically significant.
Results and discussion
In this study, we constructed a 3D paper-stacking gelatin/PCL nanofiber scaffold. The electrospun nanofibrous membranes made of gelatin and PCL were stacked using a layer-by-layer method ().
ADSC identification with flow cytometry technique
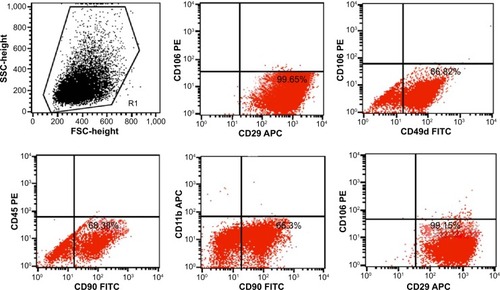
Rat ADSCs were first identified by flow cytometry. CD29, CD90, and CD49d are characteristic ADSC surface markers and are commonly used for ADSC identification.Citation42,Citation43 As shown in , the ADSC-positive antibodies, such as CD29 (99.65%), CD90 (68.38%), and CD49d (86.82%), are at high levels, while the negative antibodies, such as CD45 (0.06%), CD11b (0.24%), and CD106 (0.84%), are low. The results showed that the cells that we harvested from the rat’s fat were mainly ADSCs.
Figure 2 Flow cytometry identification of adipose-derived stem cell (ADSCs) using the surface antibody markers CD29 (99.65%), CD90 (68.38%) and CD49d (86.82%), CD45 (0.06%), CD11b (0.24%), and CD106 (0.84%).
Abbreviations: APC, allophycocyanin; FITC, fluorescein isothiocyanate; FSC, forward scatter; PE, phycoerythrin; SSC, side scatter.
Fabrication of electrospun nanofiber membranes
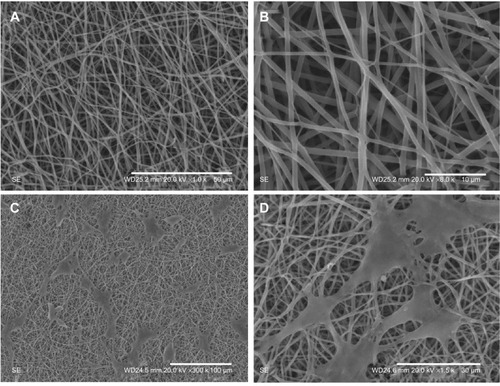
Gelatin and PCL are both biodegradable and biocompatible polymers that endow the membranes with good mechanical properties and cell adhesion.Citation44–Citation46 The nanofiber membranes were formed by electrospinning. The nanofibrous structure has a large surface area for cells to grow and adhere to the membrane and is highly porous, facilitating the transportation of the cells’ nutrients and waste.Citation47,Citation48 As shown in , a uniform membrane composed of random nanofibers was fabricated via electrospinning. The membrane contains interconnected pores, which are favorable for cell growth, proliferation, and migration. demonstrate that cells grew and adhered to the membrane to form a lamellar shape and stretched further into the pores. The results suggest that the gelatin/PCL membranes provide space for cells to stretch inside.
Figure 3 The scanning electron microscopy (SEM) images of the polycaprolactone (PCL)/gelatin electrospun membranes at low magnification (A) and at high magnification (B), followed by the SEM images of adipose-derived stem cells (ADSCs) cultured on the electrospun membranes under low magnification (C) and at high magnification (D).
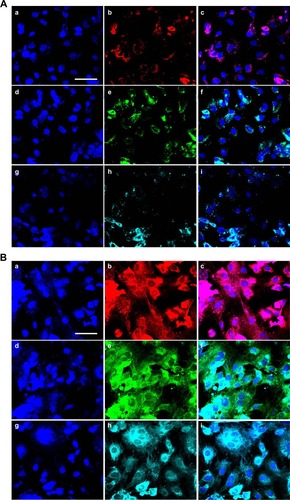
Osteogenic differentiation of ADSCs seeded on the single membrane
The ADSCs were seeded on the membranes in the OM. After 2 weeks and 4 weeks, the osteogenic protein secretion was examined with immunofluorescence technique. As shown in , the OCN in (a, b, and c) and (a, b, and c), OPG in (d, e, and f) and (d, e, and f), and OPN in (g, h, and i) and (g, h, and i) can be found around the cells as the ECM. The osteogenic proteins secreted after 4 weeks were found to be at a much higher level than they were after 2 weeks.
Figure 4 Adipose-derived stem cell osteogenic differentiation on the membranes in vitro.
Notes: (A) Adipose-derived stem cell (ADSC) osteogenic differentiation on the membranes after 2 weeks: a, b, and c show osteocalcin (OCN) protein staining; d, e, and f show osteoproteoglycan (OPG) protein staining; g, h, and i show osteopontin (OPN) protein staining; a, d, and g show 4′,6-diamidino-2-phenylindole (DAPI) staining; b, e, and h show antibody staining; and c, f, and i are the merged images. Scale bar, 50 μm. (B) ADSC osteogenic differentiation on the membranes after 4 weeks: a, b, and c show OCN protein staining; d, e, and f show OPG protein staining; g, h, and i show OPN protein staining; a, d, and g show DAPI staining; b, e, and h show antibody staining; and c, f, and i are the merged images. Scale bar, 50 μm.
The results show that the ADSCs grew well on the membranes, differentiated into osteoblasts, and secreted osteogenic proteins for bone regeneration. Used as an ECM, the membranes can provide support for cell adhesion and proliferation, as well as providing a porous structure for the transportation of nutrients and wastes. The ADSCs in paper-stacking membrane scaffolds secrete proteins and other factors that produce a biomimetic environment for cell growth and osteogenic differentiation.
Live/dead and DAPI staining of ADSC-laden paper-stacking membrane scaffold
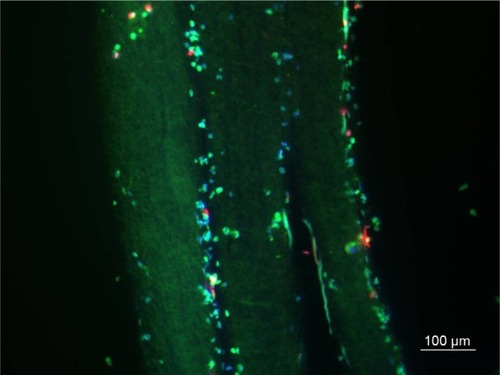
ADSC-laden layer-by-layer paper-stacking gelatin/PCL electrospun membranes were harvested, stained with live/dead assay, and cut into sections of 5 μm thickness. shows that the cells can be seen on and in between the membrane layers. All the nuclei were stained in blue (DAPI staining). The cells are mainly green stained, showing that most of the cells are live, and some red-stained cells were also observed, indicating that the cells in the paper-stacking membranes were viable.
Viability of ADSCs cultured on the paper-stacking scaffold
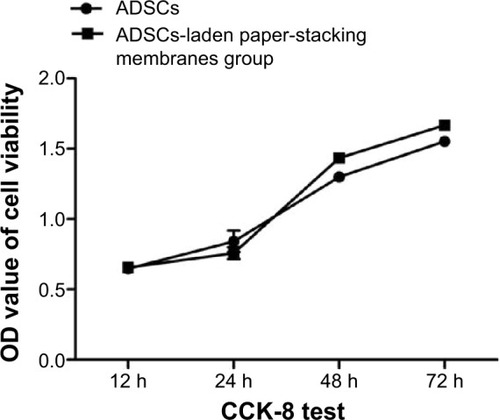
Proliferation and viability of the ADSCs were estimated using the CCK-8 kits. At specified time points, the CCK-8 reagent was added and samples were tested under an ELISA reader. As shown in , in the first 24 hours, the cells proliferated slightly slower in the scaffold group than in the cells group, possibly due to the time required for the cells to adhere onto the membranes. But after 24 hours, the cells in the scaffold group proliferated faster than those in the cells group. The results suggest that the paper-stacking membrane scaffold may provide more space for the cells to grow, thereby promoting cell proliferation and migration.
Expression of osteogenic genes in vitro
Bone tissues can generally be divided into two major parts: the mineral part is mostly composed of hydroxyapatite, while the organic part, which accounts for 20% of the dry weight, includes OPN, OCN, OPG, BSP, type I collagen, and alkaline phosphatase. Type I collagen accounts for the major portion (nearly 90%) of the proteins in bone tissues, but it can also be secreted in other tissues. OCN, OPN, OPG, and BSP are characteristic genes for bone regeneration.Citation49,Citation50
OPN is a glycoprotein secreted by osteoblasts for bone synthesis.Citation51,Citation52 It is a prominent component of bone tissues and plays a vital part in bone formation, biomineralization, remodeling, and maintenance. OCN is a hormone secreted by osteoblasts and has a special role in the body’s metabolic regulation and pro-osteoblastic modification.Citation53–Citation55 OPG can protect the bone by reducing the osteoclasts and inhibiting precursor cells from differentiating into osteoclasts.Citation56 BSP can promote vascular formation.Citation57 RUNX2 and OSX are two important factors in the upward pathway and regulate the differentiation of ADSCs into osteoblasts.Citation58–Citation60 RUNX2 is an indispensable transcription factor for stem cell differentiation and the osteoblast maturation process during both endochondral and intramembranous ossification. It is important to maintain the progenitors of the stem cells and make the cells differentiate into osteoblasts. OSX is in the downstream of RUNX2 and is important in the process to regulate the stem cell differentiations.
In order to investigate the impact of paper-stacking membranes’ osteogenic differentiation in vitro, RT-qPCR experiments were introduced to investigate OCN, OPN, OPG, BSP, RUNX2, and OSX gene expressions in four groups. As shown in , after 1 week, OCN, OPN, OPG, and BSP gene expressions were low, while RUNX2 and OSX expressions were higher in the cell-laden paper-stacking membranes group, the cells-OM group, and the cells-paper-stacking membranes-OM group. After 2, 3, and 4 weeks, OCN, OPN, OPG, BSP, RUNX2, and OSX gene expressions in the cell-laden paper-stacking membranes-OM group were significantly higher than in other groups. In the cells-paper-stacking membranes group, the relative gene expressions (OCN, OPN, OPG, BSP, RUNX2, and OSX) were significantly higher when compared with the cells group and cells-OM group.
Figure 7 The osteogenic related gene expressions (osteocalcin [OCN], osteopontin [OPN], osteoproteoglycan [OPG], bone sialoprotein [BSP], runt-related transcription factor 2 [RUNX2], and osterix [OSX]) in different groups after 1, 2, 3, and 4 weeks.
Notes: The cells-paper-stacking membranes-osteogenic medium (OM) group has the highest gene expressions on all the genes and at the 4-week point. ***P<0.05, and it shows the different between each groups in the same time point. #P<0.05, and it shows the different between time points.
![Figure 7 The osteogenic related gene expressions (osteocalcin [OCN], osteopontin [OPN], osteoproteoglycan [OPG], bone sialoprotein [BSP], runt-related transcription factor 2 [RUNX2], and osterix [OSX]) in different groups after 1, 2, 3, and 4 weeks.Notes: The cells-paper-stacking membranes-osteogenic medium (OM) group has the highest gene expressions on all the genes and at the 4-week point. ***P<0.05, and it shows the different between each groups in the same time point. #P<0.05, and it shows the different between time points.](/cms/asset/02934b20-f157-497a-9327-07ca95b08643/dijn_a_77118_f0007_b.jpg)
The results showed that the scaffold group (cells-paper-stacking membranes group, cells-paper-stacking membranes-OM group) had higher osteogenic gene expressions. The layer-by-layer paper-stacking 3D scaffold has a highly porous structure and good biocompatibility for ADSCs to grow inside. The 3D scaffold produces a biomimetic environment for cell growth and promotes cell amplification, differentiation, and osteogenic gene expressions for bone regeneration.Citation61–Citation66 When the ADSCs grow in the layer-by-layer paper-stacking 3D scaffolds, they have more osteogenic gene expressions.
Micro-CT scanning of regenerated bone
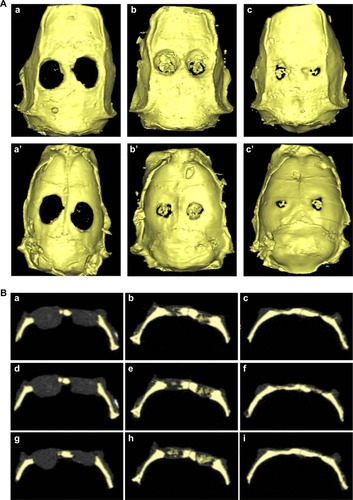
Twelve weeks postoperation, all the animals were sacrificed for micro-CT scanning. As shown in , the large cavity defect in the control group still remains, indicating that there had been little bone regeneration. In , a, b, and c are the outside–inside view, a’, b’, and c’ are the inside–outside view, a and a’ are the control group, b and b’ are the paper-stacking membranes group, and c and c’ are the ADSC-laden paper-stacking membranes group. In the paper-stacking membranes group, more bone tissue with higher density had been detected in the defect site than in the control group, and the regenerated bone tissues covered most of the defect. However, there was a big gap in the defect site, and the edge of the gap was not smooth. The largest amount of regenerated bone tissues and the highest bone density were found in the ADSC-laden paper-stacking membranes group, with the regenerated bone tissues filling almost 90% of the defect and producing a smooth edge at the defect site. The result showed that the ADSC-laden paper-stacking membranes group obtained the best results in bone regenerations. shows the coronal view of the samples of the three groups in three different section planes (along the sagittal diameter of the defect, sections on 25%, 50%, and 75% of the defect diameter, one sample from each group). In , a, d, and g are different section planes of the control group; b, e, and h are different section planes of the paper-stacking membranes group; and c, f, and i are different section planes of the ADSC-laden paper-stacking membranes group. In the ADSC-laden paper-stacking membranes group, the defect site was filled with new bone tissue to almost 90% of the defect, and the density of the new bone is similar to the host bone. In the paper-stacking membranes group, there is a larger gap in the defect site, and the density of the new bone is lower than in the former group. There is little bone growth in the defect site in the control group.
Figure 8 Micro-computed tomography scanning detection.
Notes: (A) Micro-computed tomography (micro-CT) scanning shows the morphology of the regenerated bones: the control group (a and a’), the paper-stacking membranes group (b and b’), the adipose-derived stem cell (ADSC)-laden paper-stacking membranes group (c and c’); a, b, and c are the outside–inside view; and a’, b’, and c’ are the inside–outside view. (B) Micro-CT scanning shows the coronal view of the samples in the three groups in three different section planes: a, d, and g are different section planes of the control group; b, e, and h are different section planes of the paper-stacking membranes group; and c, f, and i are different section planes of the ADSC-laden paper-stacking membranes group. (C) Bone mineral density (BMD) of regenerated bone in the defect site in the control group, paper-stacking membranes group, and ADSC-laden paper-stacking membranes group. **P<0.05, ***P<0.01.
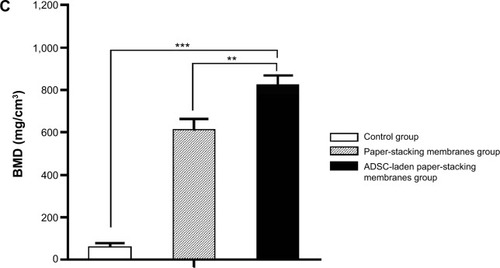
The quantitative results of the regenerated bone was examined by micro-CT, and the bone mineral density (BMD) was investigated. As shown in , the results showed a significantly enhanced BMD in the ADSC-laden paper-stacking membranes group. The BMD value is 823.06 mg/cm3. In the paper-stacking membranes group, the BMD is 610.91 mg/cm3, and in the control group (defect group), the BMD value is around 58.54 mg/cm3. The results showed that the ADSC-laden paper-stacking membranes group generated the bone in a highest BMD, which is close to the BMD value of host bone.
Histological analysis
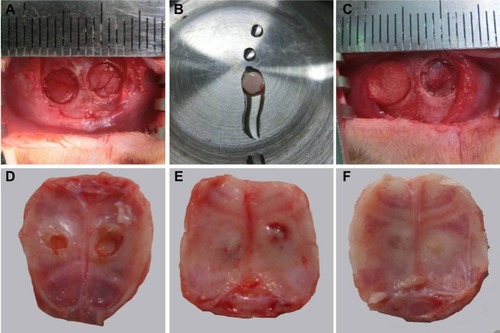
The layer-by-layer paper-stacking electrospun 3D scaffolds were planted into the parietal bone defects of the rats. Two circular defects 5 cm in diameter were created on the calvarial bone of each rat, and the paper-stacking scaffold was implanted into the defect site, which is shown in (). During the procedure, all rats survived without dura mater damage. The scaffolds were implanted without any infection. After 12 weeks postoperation, all the rats remained alive and did not show obvious immunoreaction or infectious complications. The animals were sacrificed and the morphology of the regenerated tissues in the parietal defect area was examined. In the control group, two holes of 2–3 mm in diameter were still visible in the parietal defect sites and the holes were covered with soft tissue (). In the paper-stacking membranes group, two small holes remained and the color of the regenerated tissues was different from that of the host bone. There was a gap between the new and host bones, and the edge of the gap was not smooth (). However, in the ADSC-laden paper-stacking membranes group, the regenerated tissues substantially filled the parietal defect and integrated smoothly with the host bone. The color of the regenerated tissues was only slightly whiter than that of the host bone (), showing that the ADSC-laden paper-stacking membranes group produced more regenerated bone tissue and repaired the parietal defect more effectively.
Figure 9 The rat calvarial bone defect model and the morphology of the calvarial bone defect 12 weeks postoperation.
Notes: (A, B, C) The rat calvarial bone defect model: (A) the rat calvarial bone defect model, (B) the layer-by-layer paper-stacking scaffold, and (C) the scaffold was implanted into the defect. (D, E, F) The morphology of the regenerated tissues in the parietal defect area 12 weeks postoperation: (D) the control group, (E) the paper-stacking membranes group, and (F) the adipose-derived stem cell (ADSC)-laden paper-stacking membranes group.
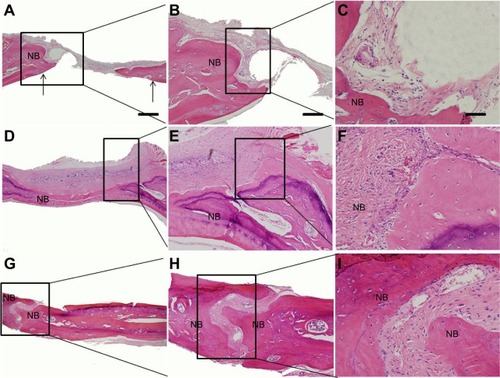
The regenerated tissues in the parietal defect area were harvested, decalcified, embedded, cut into sections, and stained. In HE staining, collagen showed red. As indicated in , the defect area of the control group shows largely white, indicating that there is little new tissue in the defect area (). In the paper-stacking membranes group, the staining is uniformly red in the bottom part of the defect area, indicating a significant amount of regenerated tissue. The regenerated tissue has the same color and density as the host bone, but it is more disordered when compared with the host bone. Also, no layered structure was identified in the regenerated tissue. The top part of the regenerated tissue is loose and lightly stained, and there are many cells growing within it (). In the ADSC-laden paper-stacking membranes group, the regenerated tissue was much more extensive than in the control group or the paper-stacking membranes group. The uniformly red-stained and fibrous-layered structure tissue in the defect site suggests that most of the defect area was covered by dense regenerated bone tissue. The regenerated bone has similar color and density to the host bone. The new bone in this group is very similar to the host bone but is thicker ().
Figure 10 Hematoxylin and eosin staining.
Notes: (A, B, C) control group; the arrows in (A) show the defect edge; (D, E, F) paper-stacking membranes group; (G, H, I) adipose-derived stem cell (ADSC)-laden paper-stacking membranes group; and NB is new bone. The scale bar of (A), (D), and (G) is 500 μm. The scale bar of (B), (E), and (H) is 200 μm. The scale bar of (C), (F), and (I) is 50 μm.
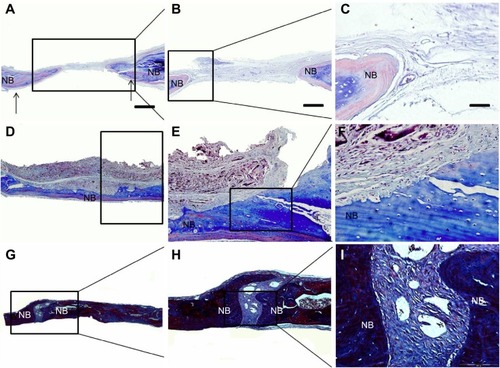
In Masson trichrome staining, collagen was stained blue, nuclei were stained black, and the muscle, cytoplasm, and keratin were stained red. Among them, collagen accounts for 90% of bone proteins. Therefore, it is a good indicator of regenerated bone. As shown in , in the control group, there was very limited collagen and regenerated tissues formed in the defect site, as indicated by the thin layer of blue stain around the edge of the defect (). In the paper-stacking membranes group, much more tissue was regenerated than in the control group, as indicated by the blue stain at the bottom part of the defect (). Some blue stain was also identified at the top part of the defect, indicating that it was undergoing the process of bone regeneration. In the ADSC-laden paper-stacking membranes group, the largest amount of tissue was regenerated using a layered structure, as indicated by the blue stain evenly distributed over the entire defect area (). This shows that the ADSC-laden paper-stacking membrane scaffolds can support more bone tissue regeneration and can repair the bone defect better. The 3D scaffold may activate the growth factors for bone regeneration and promote the differentiation of ADSCs to form the bone tissues. These results agree well with the RT-qPCR results in the in vitro experiments.
Figure 11 Masson staining.
Notes: (A, B, C) control group; the arrows in (A) show the defect edge; (D, E, F) paper-stacking membranes group; (G, H, I) the adipose-derived stem cell (ADSC)-laden paper-stacking membranes group; and NB is new bone. The scale bar of (A), (D), and (G) is 500 μm. The scale bar of (B), (E), and (H) is 200 μm, and the scale bar of (C), (F), and (I) is 50 μm.
Expressions of osteogenic genes in vivo
For further quantitative measurement of osteogenesis, we introduced an RT-qPCR technique to analyze the osteogenic gene expressions in the rat parietal bone defect model. After 12 weeks postoperation, regenerated tissues in the parietal bone defect were harvested (n=4). As shown in , gene expressions of OCN, OPN, OPG, BSP, RUNX2, and OSX were significantly higher in the paper-stacking membranes group and the ADSC-laden paper-stacking membranes group when compared with the control group.
Figure 12 The real-time quantitative polymerase chain reaction results of osteogenic gene expressions in different groups.
Notes: The adipose-derived stem cell (ADSC)-laden paper-stacking membranes group had the highest osteogenic gene expressions (osteocalcin [OCN], osteopontin [OPN], osteoproteoglycan [OPG], bone sialoprotein [BSP], runt-related transcription factor 2 [RUNX2], and osterix [OSX]). **P<0.05, ***P<0.01.
![Figure 12 The real-time quantitative polymerase chain reaction results of osteogenic gene expressions in different groups.Notes: The adipose-derived stem cell (ADSC)-laden paper-stacking membranes group had the highest osteogenic gene expressions (osteocalcin [OCN], osteopontin [OPN], osteoproteoglycan [OPG], bone sialoprotein [BSP], runt-related transcription factor 2 [RUNX2], and osterix [OSX]). **P<0.05, ***P<0.01.](/cms/asset/0bb5b939-11a9-46dd-9581-591c256bdcc8/dijn_a_77118_f0012_b.jpg)
OSX and RUNX2 both are key steps for osteogenic differentiation and bone formation in the bone formation signal pathways, such as Hedgehog signaling, Notch signaling, WNT signaling, BMP signaling, and FGF signaling. As shown in , the high gene expressions of RUNX2 and OSX suggest the active differentiation of ADSCs to osteoblasts. Meanwhile, as shown in vitro, the osteogenic gene expressions investigation () and the increase of RUNX2 and OSX gene expressions were found to occur during the whole process of bone regeneration.
OCN, OPN, OPG, and BSP are important, special proteins secreted by osteoblasts. They form the ECM in the bone and play vital roles in all stages of bone formation, including osteogenic differentiation, biomineralization remodeling, and maintenance. As shown in , all gene expression levels were significantly higher in the ADSC-laden paper-stacking membranes group than those in the control group, with OCN two times higher, OPN 12 times higher, OPG eight times higher, BSP eight times higher, RUNX2 nine times higher, and OSX 20 times higher, respectively (P<0.05 for all gene expressions). Compared with the paper-stacking membranes group, the ADSC-laden paper-stacking membranes group showed significantly higher gene expressions, with OCN one time higher, OPN one time higher, OPG almost one time higher, BSP six times higher, RUNX2 five times higher, and OSX four times higher, respectively (P<0.05 for all genes). The results suggest that the increase of RUNX2 and OSX gene expressions promotes the differentiation of ADSCs into osteoblasts, thus encouraging the formation of more bone.
Conclusion
In this study, we fabricated a novel layer-by-layer paper-stacking nanofibrous scaffold composed of the biocompatible polymers, gelatin, and PCL. Osteogenic differentiation capability and ADSC viability on the layer-by-layer paper-stacking membranes were evaluated. The results showed that the paper-stacking membrane scaffold has a good biocompatibility and outstanding osteogenic differentiation capability. The osteogenic protein staining showed that ADSCs have a good osteogenic differential capability in an osteogenic differential environment (eg, in an OM). The ADSC-laden paper-stacking membrane scaffold cultured in the OM showed a significant osteogenic phenotype, with the highest osteogenic gene expressions and a rich osteogenic proteins secretion.
In vivo, the paper-stacking membrane scaffold group and the ADSC-laden paper-stacking membranes group healed the calvarial bone defect better than the control group, and the ADSC-laden paper-stacking membranes group healed the defect best. Overall, the results of the experiments suggest that the ADSC-laden paper-stacking membrane scaffold played a pivotal role in osteogenic differentiation and bone regeneration. Also, they provided a 3D biomimetic microenvironment for ADSC growth, forming spatiotemporal chemical gradients (which are crucial for cell amplification, differentiation, and tissue regeneration), assisting signal transduction, and improving the cell-to-cell, cell-to-scaffold, and cell-to-matrix interactions, thus demonstrating osteoconduction and osteoinduction capacities for bone regeneration.
The limitation of this study is that we did not have a longer time point (6 months or 1 year) to examine the long-term result of bone regeneration in the in vivo part.
Acknowledgments
This work was supported by the National Basic Research Program of China (Grant No 2012CB619100), the NSERC Discovery Grant and NSERC RTI Grant, Manitoba Health Research Council Establishment Grant, Dr Moore House Fellowship, Manitoba Diabetes Foundation, Manitoba Institute of Child Health, China 863 Project (Grant No 2012AA020504), and National Natural Science Foundation of China (No 21304098).
Disclosure
The authors report no conflicts of interest in this work.
References
- PetiteHViateauVBensaidWTissue-engineered bone regenerationNat Biotech200018959963
- WangHLiYZuoYLiJMaSChengLBiocompatibility and osteogenesis of biomimetic nano-hydroxyapatite/polyamide composite scaffolds for bone tissue engineeringBiomaterials2007283338234817481726
- LutolfMPHubbellJASynthetic biomaterials as instructive extracellular microenvironments for morphogenesis in tissue engineeringNat Biotech2005234755
- KimSSSun ParkMJeonOYong ChoiCKimBSPoly(lactide-co-glycolide)/hydroxyapatite composite scaffolds for bone tissue engineeringBiomaterials2006271399140916169074
- EngGLeeBWParsaHAssembly of complex cell microenvironments using geometrically docked hydrogel shapesProc Natl Acad Sci U S A20131104551455623487790
- KhetanSBurdickJAPatterning network structure to spatially control cellular remodeling and stem cell fate within 3-dimensional hydrogelsBiomaterials2010318228883420674004
- DerdaRLaromaineAMammotoAPaper-supported 3D cell culture for tissue-based bioassaysProc Natl Acad Sci U S A2009106184571846219846768
- PampaloniFReynaudEGStelzerEHKThe third dimension bridges the gap between cell culture and live tissueNat Rev Mol Cell Biol2007883984517684528
- GriffithLGSwartzMACapturing complex 3D tissue physiology in vitroNat Rev Mol Cell Biol2006721122416496023
- YamadaKMCukiermanEModeling tissue morphogenesis and cancer in 3DCell200713060161017719539
- CostaRRCustodioCAAriasFJRodriguez-CabelloJCManoJFLayer-by-layer assembly of chitosan and recombinant biopolymers into biomimetic coatings with multiple stimuli-responsive propertiesSmall201172640264921809443
- CuiLLiuBLiuGRepair of cranial bone defects with adipose derived stem cells and coral scaffold in a canine modelBiomaterials2007285477548617888508
- GimbleJMKatzAJBunnellBAAdipose-derived stem cells for regenerative medicineCirc Res20071001249126017495232
- LiuGZhangYLiuBSunJLiWCuiLBone regeneration in a canine cranial model using allogeneic adipose derived stem cells and coral scaffoldBiomaterials2013342655266423343633
- LinCYLinKJKaoCYThe role of adipose-derived stem cells engineered with the persistently expressing hybrid baculovirus in the healing of massive bone defectsBiomaterials2011326505661421669458
- FrohberghMEKatsmanABottaGPElectrospun hydroxyapatite-containing chitosan nanofibers crosslinked with genipin for bone tissue engineeringBiomaterials2012339167917823022346
- LiuWThomopoulosSXiaYElectrospun nanofibers for regenerative medicineAdv Healthc Mater20121102523184683
- AlameinMALiuQStephensSNanospiderwebs: artificial 3D extracellular matrix from nanofibers by novel clinical grade electrospinning for stem cell deliveryAdv Healthc Mater2013270271723184860
- OriveGSantosEPedrazJLHernandezRMApplication of cell encapsulation for controlled delivery of biological therapeuticsAdv Drug Deliv Rev201467–8314
- FischbachCChenRMatsumotoTEngineering tumors with 3D scaffoldsNat Meth20074855860
- ZhangHLinCYHollisterSJThe interaction between bone marrow stromal cells and RGD-modified three-dimensional porous polycaprolactone scaffoldsBiomaterials2009304063406919487019
- Di MaggioNPiccininiEJaworskiMTrumppAWendtDJMartinIToward modeling the bone marrow niche using scaffold-based 3D culture systemsBiomaterials20113232132920952054
- KumarGTisonCKChatterjeeKThe determination of stem cell fate by 3D scaffold structures through the control of cell shapeBiomaterials2011329188919621890197
- LevenbergSHuangNFLavikERogersABItskovitz-EldorJLangerRDifferentiation of human embryonic stem cells on three-dimensional polymer scaffoldsProc Natl Acad Sci U S A2003100127411274614561891
- CowanCMShiYYAalamiOOAdipose-derived adult stromal cells heal critical-size mouse calvarial defectsNat Biotechnol200422560715077117
- FrohlichMGimbleJMGraysonWLKregar-VelikonjaNMaroltDVunjak-NovakovicGBone grafts engineered from human adipose-derived stem cells in perfusion bioreactor cultureTissue Eng Part A20101617919678762
- YoungDAChoiYSEnglerAJChristmanKLStimulation of adipogenesis of adult adipose-derived stem cells using substrates that mimic the stiffness of adipose tissueBiomaterials2013348581858823953825
- KatsudaTTsuchiyaRKosakaNHuman adipose tissue-derived mesenchymal stem cells secrete functional neprilysin-bound exosomesSci Rep20133119723378928
- EstesBTDiekmanBOGimbleJMGuilakFIsolation of adipose-derived stem cells and their induction to a chondrogenic phenotypeNat Protoc201051294131120595958
- JiWYangFMaJIncorporation of stromal cell-derived factor-1alpha in PCL/gelatin electrospun membranes for guided bone regenerationBiomaterials20133473574523117215
- LeeJYooJJAtalaALeeSJThe effect of controlled release of PDGF-BB from heparin-conjugated electrospun PCL/gelatin scaffolds on cellular bioactivity and infiltrationBiomaterials2012336709672022770570
- XueJHeMLiuHDrug loaded homogeneous electrospun PCL/gelatin hybrid nanofiber structures for anti-infective tissue regeneration membranesBiomaterials2014359395940525134855
- BigiAPanzavoltaSRubiniKRelationship between triple-helix content and mechanical properties of gelatin filmsBiomaterials2004255675568015159084
- XueJFengBZhengREngineering ear-shaped cartilage using electrospun fibrous membranes of gelatin/polycaprolactoneBiomaterials2013342624263123352044
- ZhaoYLinHZhangJCrosslinked three-dimensional demineralized bone matrix for the adipose-derived stromal cell proliferation and differentiationTissue Eng Part A200915132118652539
- HaraguchiYShimizuTSasagawaTFabrication of functional three-dimensional tissues by stacking cell sheets in vitroNat Protoc2012785085822481530
- BeneschJManoJFReisRLProteins and their peptide motifs in acellular apatite mineralization of scaffolds for tissue engineeringTissue Eng Part B Rev20081443344518826338
- SpicerPPKretlowJDYoungSJansenJAKasperFKMikosAGEvaluation of bone regeneration using the rat critical size calvarial defectNat Protoc201271918192923018195
- DeganoIRVilaltaMBagoJRBioluminescence imaging of calvarial bone repair using bone marrow and adipose tissue-derived mesenchymal stem cellsBiomaterials20082942743717950456
- PieriFLucarelliECorinaldesiGDose-dependent effect of adipose-derived adult stem cells on vertical bone regeneration in rabbit calvariumBiomaterials2010313527353520170950
- DeclercqHADe CaluweTKryskoOBachertCCornelissenMJBone grafts engineered from human adipose-derived stem cells in dynamic 3D-environmentsBiomaterials2013341004101723146435
- LiW-JLaurencinCTCatersonEJTuanRSKoFKElectrospun nanofibrous structure: a novel scaffold for tissue engineeringJ Biomed Mater Res20026061362111948520
- LangerRVacantiJPTissue engineeringScience19932609209268493529
- AwadHAWickhamMQLeddyHAGimbleJMGuilakFChondrogenic differentiation of adipose-derived adult stem cells in agarose, alginate, and gelatin scaffoldsBiomaterials2004253211322214980416
- WangHHansenMBLowikDWOppositely charged gelatin nanospheres as building blocks for injectable and biodegradable gelsAdv Mater201123H119H12421394793
- LoverdeSMKleinMLDischerDENanoparticle shape improves delivery: rational coarse grain molecular dynamics (rCG-MD) of taxol in worm-like PEG-PCL micellesAdv Mater2012243823383022105885
- BadamiASKrekeMRThompsonMSRiffleJSGoldsteinASEffect of fiber diameter on spreading, proliferation, and differentiation of osteoblastic cells on electrospun poly(lactic acid) substratesBiomaterials20062759660616023716
- LiangDHsiaoBSChuBFunctional electrospun nanofibrous scaffolds for biomedical applicationsAdv Drug Deliv Rev2007591392141217884240
- MurakamiTSaitoAHinoS-iSignalling mediated by the endoplasmic reticulum stress transducer OASIS is involved in bone formationNat Cell Biol2009111205121119767743
- MendoncaGMendoncaDBSimoesLGThe effects of implant surface nanoscale features on osteoblast-specific gene expressionBiomaterials2009304053406219464052
- WeilbaecherKNGuiseTAMcCauleyLKCancer to bone: a fatal attractionNat Rev Cancer20111141142521593787
- Opsahl VitalSGaucherCBardetCTooth dentin defects reflect genetic disorders affecting bone mineralizationBone20125098999722296718
- SmithLBSaundersPTThe skeleton: the new controller of male fertility?Cell201114464264321376228
- FulzeleKRiddleRCDiGirolamoDJInsulin receptor signaling in osteoblasts regulates postnatal bone acquisition and body compositionCell201014230931920655471
- FerronMWeiJYoshizawaTInsulin signaling in osteoblasts integrates bone remodeling and energy metabolismCell201014229630820655470
- SimonetWSLaceyDLDunstanCROsteoprotegerin: a novel secreted protein involved in the regulation of bone densityCell1997893093199108485
- MatsubaraHHoganDEMorganEFMortlockDPEinhornTAGerstenfeldLCVascular tissues are a primary source of BMP2 expression during bone formation induced by distraction osteogenesisBone20125116818022391215
- YeJHXuYJGaoJCritical-size calvarial bone defects healing in a mouse model with silk scaffolds and SATB2-modified iPSCsBiomaterials2011325065507621492931
- HamannCKirschnerSGuntherK-PHofbauerLCBone, sweet bone – osteoporotic fractures in diabetes mellitusNat Rev Endocrinol2012829730522249517
- GiulianiNCollaSMorandiFMyeloma cells block RUNX2/CBFA1 activity in human bone marrow osteoblast progenitors and inhibit osteoblast formation and differentiationBlood20051062472248315933061
- ShiJOuyangJLiQWangLWuJZhongWXingMMCell-compatible hydrogels based on a multifunctional crosslinker with tunable stiffness for tissue engineeringJ of Mater Chem2012222395223962
- ChenJQiuXWangLZhongWKongJXingMMFree-Standing Cell Sheet Assembled with Ultrathin Extracellular Matrix as an Innovative Approach for Biomimetic TissuesAdv Funct Mater2014241522162223
- ChenJZhouBLiQOuyangJKongJZhongWXingMMPLLA-PEG-TCH-labeled bioactive molecule nanofibers for tissue engineeringInt J Nanomedicine201162533254222072887
- WangZXingMOjoOMussel-inspired ultrathin film on oxidized Ti–6Al–4V surface for enhanced BMSC activities and antibacterial capabilityRSC Adv201445579055799
- LiXZhouJLiuZChenJLüSSunHA PNIPAAm-based thermosensitive hydrogel containing SWCNTs for stem cell transplantation in myocardial repairBiomaterials201435225679568824746964
- GeLLiQHuangYYangSPolydopamine-coated paper-stack nanofibrous membranes enhancing adipose stem cells’ adhesion and osteogenic differentiationJ Mater Chem B2014269176923