Abstract
Radiotherapy is one of the main strategies for cancer treatment but has significant challenges, such as cancer cell resistance and radiation damage to normal tissue. Radiosensitizers that selectively increase the susceptibility of cancer cells to radiation can enhance the effectiveness of radiotherapy. We report here the development of a novel radiosensitizer consisting of monodispersed ceria nanoparticles (CNPs) covered with the anticancer drug neogambogic acid (NGA-CNPs). These were used in conjunction with radiation in MCF-7 breast cancer cells, and the efficacy and mechanisms of action of this combined treatment approach were evaluated. NGA-CNPs potentiated the toxic effects of radiation, leading to a higher rate of cell death than either treatment used alone and inducing the activation of autophagy and cell cycle arrest at the G2/M phase, while pretreatment with NGA or CNPs did not improve the rate of radiation-induced cancer cells death. However, NGA-CNPs decreased both endogenous and radiation-induced reactive oxygen species formation, unlike other nanomaterials. These results suggest that the adjunctive use of NGA-CNPs can increase the effectiveness of radiotherapy in breast cancer treatment by lowering the radiation doses required to kill cancer cells and thereby minimizing collateral damage to healthy adjacent tissue.
Introduction
Breast cancer mortality has been decreasing over the last decade owing to widespread adoption of surgery as the primary treatment option and more effective pre- and postoperative radiotherapy; nonetheless, the incidence of breast cancer is still on the rise.Citation1 Radiotherapy plays an important role in preventing metastasis and the regeneration of tumor tissue; however, normal tissue close to the tumor is inevitably exposed to radiation during administration, especially chest and abdominal tissue moving because of respiration.Citation2 Nanomaterials have been used in conjunction with radiotherapy in order to enhance the sensitivity of breast cancer cells to the effects of radiation and reduce damage to surrounding tissue;Citation3 some of these materials can be used for tumor-targeted drug delivery via enhancement of permeability and retention.Citation4 Cerium oxide has various applications, including as an oxygen sensor, as an automotive catalytic converter, and as solid oxide fuel cells.Citation5 Ceria nanoparticles (CNPs) have received considerable attention for their excellent catalytic capability derived from the rapid alteration of the oxidation state from Ce3+ to Ce4+.Citation6 CNPs have emerged as important and lucrative materials in biological fields such as neuroprotection, radiotherapy, ocular protection, bioanalysis, biomedicine, and antioxidant therapy;Citation7 in vitro and in vivo experiments suggest that CNPs have antioxidant and anti-inflammatory functionsCitation8–Citation10 but can also induce lipid peroxidation, lung damage, reactive oxygen species (ROS) production, and autophagy.Citation11 Other groups have previously investigated the potential of CNPs to protect normal cells against radiation-induced damage.Citation12
Apoptosis is the main mode of radiation-induced cell death, but another important mechanism is autophagy,Citation13 a conserved process of protein degradation in which double-membrane vesicles known as autophagosomes engulf intracellular contents such as endoplasmic reticulum, mitochondria, and ribosomes and fuse with lysosomes for cargo degradation.Citation14 Autophagy and consequent cell death can be induced by anticancer drugs in various types of tumor cells,Citation15 while radiotherapy stimulates both apoptosis and autophagy.Citation16 Previous studies have shown that gamboges have therapeutic effects on breast and skin cancer and pancreatic adenocarcinoma;Citation17 the chemotherapeutic agent neogambogic acid (NGA), an active component of gamboges, demonstrates antitumor potential in vitro and in vivo.Citation18–Citation20 NGA increased the death of S180-Lewis lung cancer, CNE-1 human nasopharyngeal carcinoma, and ascites carcinoma cells in a dose-dependent manner.Citation21 In the present study, we synthesized CNPs modified with NGA (NGA-CNPs) for increased toxicity and targeting, and NGA-CNPs were tested for their ability to enhance the effectiveness of radiation in killing cancer cells. MCF-7 breast cancer cells were treated with NGA-CNPs in conjunction with radiation; the combined treatment induced cell death to a greater extent than radiation alone, and also activated autophagy and led to cell cycle arrest at the G2/M phase. Pretreatment with NGA or CNPs did not potentiate radiation-induced cell death. These results suggest that adjunctive use of NGA-CNPs can increase the effectiveness of radiotherapy in breast cancer treatment.
Materials and methods
Cells and reagents
MCF-7 breast carcinoma cells were obtained from the School of Biological Science and Medical Engineering of Southeast University (Nanjing, People’s Republic of China). NGA was purchased from Shanghai Shifeng Biotechnology Co. (Shanghai, People’s Republic of China), and the Cyto-ID Autophagy Detection kit was purchased from Enzo Life Sciences (Plymouth Meeting, PA, USA). All other reagents were from Beyotime Institute of Biotechnology (Shanghai, People’s Republic of China) or Sigma-Aldrich (St Louis, MO, USA). No ethical approval was required for this study.
Synthesis and characterization of CNPs
CNPs 3–5 nm in size were synthesized by a microemulsion method as previously described.Citation22–Citation24 Briefly, surfactant sodium bis(2-ethylhexyl) sulfosuccinate was dissolved in 100 mL toluene, followed by the addition of 5 mL of 0.1 M aqueous cerium nitrate solution. The reaction mixture was stirred for 45 minutes before 10 mL of 1.5 M ammonium hydroxide aqueous solution was added dropwise. The reaction proceeded for 1 hour, and the mixture was allowed to separate into two layers, with the upper layer consisting of toluene containing non-agglomerated CNPs. CNPs were washed six times with acetone and distilled water to remove the surfactant and other impurities, and size distribution and morphology were examined using a JEM-2100 high-resolution transmission electron (TE) microscope (JEOL Ltd., Tokyo, Japan) equipped with an energy-dispersive analyzer by depositing drops of suspended particle solution onto a carbon-coated copper grid.Citation25 Broad peaks in the X-ray diffraction spectrum (Ultima-3; Rigaku, Tokyo, Japan) confirmed the crystallinity of the CNPs.Citation26
Amine functionalization of CNPs
CNPs were resuspended in 0.1 M sodium hydroxide solution and stirred for 5 minutes before adding 2.5 mL distilled epichlorohydrin followed by 0.25 mL of 2 M sodium hydroxide solution. The mixture was stirred at room temperature for 6–8 hours. The CNPs were then recovered by centrifugation and washed with distilled water several times until the pH value of the discarded water was approximately 7.0. The nanoparticles (NPs) were resuspended in distilled water, and 12.5 mL of 30% ammonium hydroxide solution was added followed by stirring for 14 hours. After centrifugation, the CNPs were washed with water several times and dried under vacuum.Citation27,Citation28 Fourier transform infrared spectroscopy (FTIR) with a Bruker Vector-22 instrument (Bruker Daltonics, Billerica, MA, USA) was used to confirm amine functionalization.
Preparation of NGA-CNPs
NGA was dissolved in 5 mL dimethyl formamide (DMF) and 1.5 mL dichloromethane; 120 μL of N-methyl morpholine (NMM) was then added, followed by 147.5 mg benzotriazol-1-yloxytris-(dimethyl amino) phosphonium hexafluorophosphate (BOP) reagent. The mixture was stirred for 10 minutes at room temperature before adding the amine-modified CNPs, followed by stirring for approximately 20 hours. The reaction was terminated by adding 1 mL distilled water.Citation29 The particles were recovered by centrifugation and washed sequentially with DMF, acetone, and water several times to remove unconjugated NGA and other impurities. The surface functionalization of CNPs was verified by X-ray photo-electron spectroscopy (XPS) using a PHI5000 VersaProbe instrument (Thermo Fisher Scientific, Waltham, MA, USA). The base pressure during XPS analysis was approximately 10−10 Torr with Mg Kα X-radiation (1,253.6 eV) delivered at a power of 200 W.
Cell culture and in vitro radiation
Cells were cultured in Roswell Park Memorial Institute (RPMI) 1640 medium containing 10% fetal bovine serum (FBS), 100 U/mL penicillin-G, and 100 U/mL streptomycin. Cells were maintained at 37°C in a humidified incubator with 5% CO2/95% air. NGA, CNPs, or NGA-CNPs were dissolved in dimethyl sulfoxide (DMSO) and diluted to desired concentrations with complete culture medium.Citation30 Control cells were treated with medium containing <0.05% DMSO (v/v). Radiotherapy was administered in vitro using a Clinac IX 6 MeV beam linear accelerator (Varian Medical Systems, Palo Alto, CA, USA).
Clonogenic survival assay
Radiosensitivity was evaluated with the clonogenic survival assay.Citation2 Cells (3–5×103) were seeded in six-well plates. After allowing 1 day for attachment, cells were pretreated with vehicle, NGA, CNPs, or NGA-CNPs for 24 hours before exposure to radiation at doses of 0 Gy, 2 Gy, 4 Gy, 6 Gy, or 8 Gy. After 7–10 days of culture, colonies were washed with phosphate-buffered saline (PBS) and stained with Giemsa dye, and the surviving fraction was determined by counting colonies consisting of >50 cells.Citation31
Determination of apoptosis and survival rates
To assess the role of apoptosis in the cell death induced by combined treatment with NGA, CNPs, or NGA-CNPs and radiation, cells were subjected to annexin V/propidium iodide (PI) staining and analyzed by flow cytometry.Citation32 Briefly, cells were trypsinized, resuspended in 10% FBS-containing medium, centrifuged and washed twice in cold PBS, counted, and resuspended in annexin-binding buffer; 5 μL annexin V–fluorescein isothiocyanate (FITC) and 10 μL PI were added to a 100 μL cell suspension, and the mixture was stored in the dark for 15 minutes at room temperature. A 400 μL volume of annexin-binding buffer was added to the cells, followed within 1 hour by flow cytometry analysis.
Cell cycle analysis
Cell cycle analysis was carried out by PI/RNase buffer staining as previously described.Citation33 Briefly, treated cells were collected, fixed in 70% ethanol at 4°C overnight, washed with cold PBS, stained with PI/RNase staining buffer for 15 minutes in the dark, and then sorted by flow cytometry using a FACSCalibur instrument (BD Biosciences, San Jose, CA, USA).
Autophagy analysis
Cells (5×105/well) were seeded on coverslips in six-well plates. After a 24-hour incubation, cells were treated with vehicle or NGA-CNPs for 24 hours at 37°C followed by exposure to different doses of radiation. The following day, cells were carefully washed with buffer, and dual detection reagent (prepared by diluting Hoechst 33342 nuclear stain and stock Cyto-ID green autophagy detection reagent) was applied for 20 minutes in the dark. Cells were then fixed and washed several times,Citation34,Citation35 and the coverslips were mounted and visualized using an Olympus FV1000 laser scanning confocal microscope (Olympus, Tokyo, Japan).
Samples were also analyzed by flow cytometry in the green (FL1) channel, which enables the visualization of the (fluorescent) autophagic fraction.
Detection of ROS
Intracellular ROS levels were determined by flow cytometry using the membrane-permeable fluorescent probe 2′,7′-dichlorofluorescin diacetate (DCFH-DA).Citation36 Cells were seeded in six-well plates; after attachment, the cells were treated with vehicle or NGA-CNPs for 24 hours and then exposed to different doses of radiation. The cells were collected 24 hours later, washed, incubated with DCFH-DA for 30 minutes, washed with serum-free RPMI 1640, and then sorted by flow cytometry. The intracellular ROS level is represented as mean fluorescence intensity.
Statistical analysis
Data are presented as mean ± standard deviation of at least three independent experiments and were processed with OriginPro 8.0 (Origin Lab, Northampton, MA, USA). One-way analysis of variance and a Bonferroni test were used to analyze data. A P-value <0.05 was considered statistically significant.
Results and discussion
Functionalization and characterization of NGA-CNPs
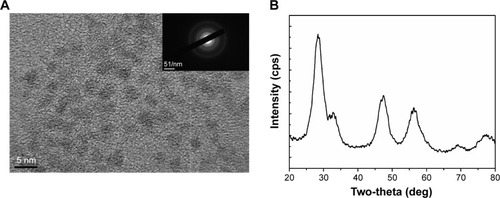
The size of CNPs synthesized by the microemulsion method ranged between 3 nm and 5 nm (). The selected-area electron diffraction pattern, which indicates the crystallinity and fluorite structure of the NPs, showed lattice planes at 111, 200, 220, and 311. The TE microscopy results were supported by the X-ray diffraction pattern ().
Figure 1 Structural analysis of CNPs.
Notes: (A) Particle size distribution (3–5 nm) of CNPs as determined by TE microscopy. The selected-area electron diffraction pattern (inset) revealed the crystallinity and fluorite structure of CNPs. (B) X-ray diffraction pattern of CNPs; 111, 200, 220, and 311 correspond to the different lattice planes of the CNP crystal structure.
Abbreviations: CNP, ceria nanoparticle; TE, transmission electron.
The structure of NGA and steps in the synthesis of the CNPs are shown in . NGA was conjugated by first attaching epichlorohydrin to the NP surface via a standard SN2 reaction, in which the oxygen atom of the NP replaced the chlorine atom of epichlorohydrin, creating an oxygen bond between the NP and the carbon of epichlorohydrin. Next, ammonia was used to open up the epoxide ring of epichlorohydrin to yield hydroxyl (−OH) and amine (−NH2) groups available for additional reactions. The FTIR spectrum confirmed amine functionalization ().
Figure 2 NGA-CNP synthesis.
Notes: (A) Structure of NGA. (B) Steps in the synthesis of NGA-CNPs.
Abbreviations: NGA-CNP, ceria nanoparticle modified with neogambogic acid; NGA, neogambogic acid; CNP, ceria nanoparticle; BOP, benzotriazol-1-yloxytris-(dimethyl amino) phosphonium hexafluorophosphate; NMM, N-methyl morpholine; DMF, dimethyl formamide.
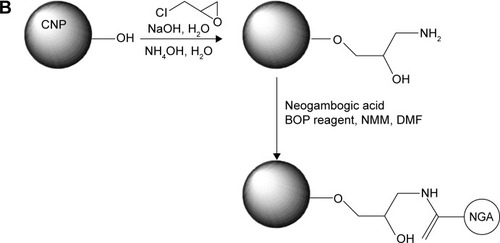
Figure 3 FTIR and XPS analysis of NGA-CNPs.
Notes: (A) FTIR spectrum of CNPs before and after modification with epichlorohydrin. Absorption at 1,100 cm−1 and 1,650 cm−1 corresponding to hydroxide and amine groups, respectively, was observed for amine-functionalized CNPs (NH2-CNP; red line) relative to unmodified CNPs (black line). (B) XPS spectrum of functionalized CNPs. Numbers represent positions of peaks.
Abbreviations: FTIR, Fourier transform infrared spectroscopy; XPS, X-ray photoelectron spectroscopy; NGA-CNP, ceria nanoparticle modified with neogambogic acid; CNP, ceria nanoparticle; au, arbitrary unit.
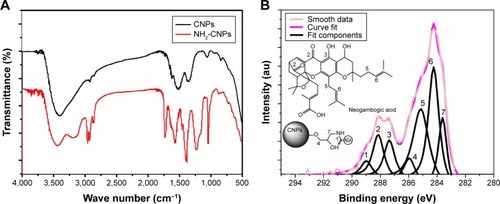
The CNPs had two functional groups available for modification by NGA, which had carboxyl groups that could react with available amine groups of surface-functionalized CNPs. The coupling reaction involved the standard peptide-coupling reagents BOP and NMM, and it was carried out in DMF, a polar solvent that facilitates the reaction. The attachment of NGA to the anime groups of CNPs was confirmed by the C 1s XPS spectrum (), with the various peaks corresponding to different C positions in the NGA-CNPs.
NGA-CNPs enhance radiation-induced growth inhibition
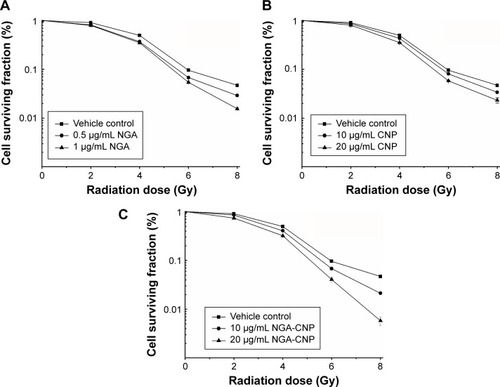
Previous studies have documented the anticancer activities of CNPs and NGA in various tumor models.Citation20,Citation29 To determine whether NGA, CNPs, or NGA-CNPs increase cellular sensitivity to radiation, breast cancer cell growth upon exposure to both was evaluated with the clonogenic assay. MCF-7 cells were exposed to three different concentrations of NGA (0 μg/mL, 0.5 μg/mL, or 1 μg/mL), CNP (0 μg/mL, 10 μg/mL, or 20 μg/mL), or NGA-CNP (0 μg/mL, 10 μg/mL, or 20 μg/mL) for 24 hours prior to irradiation at one of five doses (0 Gy, 2 Gy, 4 Gy, 6 Gy, or 8 Gy). The doses of NGA were the same as those used for the synthesis of NGA-CNP. Exposure to 4 Gy radiation decreased the colony formation rate to 49.03%. However, at 0.5 μg/mL and 1 μg/mL NGA, colony formation was reduced to 37.01% and 34.89%, respectively (P<0.05 vs 4 Gy radiation only); at 10 μg/mL and 20 μg/mL CNP, colony formation was reduced to 46.32% and 35.27%, respectively; and at 10 μg/mL and 20 μg/mL NGA-CNP, colony formation was reduced to 40.69% and 31.97%, respectively (P<0.05 vs 4 Gy radiation only) (). The inhibitory effects of other doses of radiation on colony formation were likewise enhanced by combined treatment with NGA-CNPs, which reduced the colony-forming efficiency of cancer cells to a greater extent than NGA or CNP, suggesting that NGA-CNPs sensitize cells to the effects of radiation.
Figure 4 Enhancement of radiation-induced growth inhibition by NGA, CNP, and NGA-CNPs.
Notes: The surviving fraction of MCF-7 cells was decreased by RT in a dose-dependent manner, an effect that was potentiated by the addition of NGA-CNPs. (C), compared with NGA (A), or CNPs (B).
Abbreviations: NGA, neogambogic acid; CNP, ceria nanoparticle; NGA-CNP, ceria nanoparticle modified with neogambogic acid; RT, irradiation.
NGA-CNPs enhance radiation-induced cell death
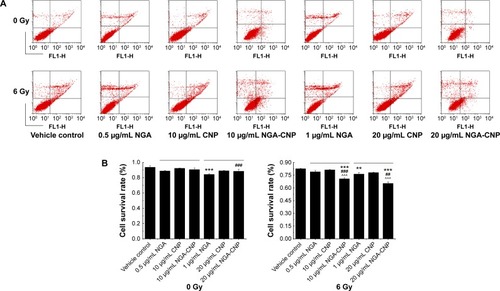
To determine the rate of apoptosis and cell mortality following combined treatment with NGA/CNPs/NGA-CNPs and radiation, cells were analyzed by annexin V/PI staining and flow cytometry. Annexin V–FITC is used as a nonquantitative probe to detect cell surface expression of phosphatidylserine, an early marker for apoptosis.Citation37 PI permeates cells with a damaged cell membrane and is therefore used to identify apoptotic or necrotic cells.Citation37 Pretreatment of cells with NGA-CNPs and radiation inhibited proliferation to a greater degree than radiation alone or combined NGA and radiation treatment, while pretreatment with CNPs and radiation-induced cell death to an extent similar to radiation alone (). Radiation delivered at a dose of 6 Gy or treatment with 0.5 μg/mL NGA, 10 μg/mL CNP, or 10 μg/mL NGA-CNP induced death in 17.40%, 11.14%, 7.63%, and 8.13% of cells, respectively. However, 10 μg/mL NGA-CNPs combined with 6 Gy radiation increased cell death to 29.26% (percent apoptosis: 11.49%) (P<0.001 vs 6 Gy radiation only), while NGA or CNPs combined with radiation increased cell death only to 21.04% (percent apoptosis: 5.02%) and 18.73% (percent apoptosis: 9.09%), respectively. When an NGA-CNP concentration of 20 μg/mL was used in conjunction with 6 Gy radiation, the rate of cell death was 34.65% (percent apoptosis: 8.24%) (P<0.001 vs 6 Gy radiation only), whereas treatment with 20 μg/mL NGA-CNP alone did not increase cell death. These results confirm that NGA-CNP potentiates the radiation-induced death of cancer cells to a greater extent than NGA or CNP alone and is far less toxic than NGA, and can therefore reduce damage to surrounding tissue in radiation therapy. In subsequent experiments, we investigated the mechanism underlying the sensitization of cells to the effects of radiation by NGA-CNP.
Figure 5 Enhancement of radiation-induced apoptosis by NGA, CNPs, and NGA-CNPs.
Notes: (A) MCF-7 cells were exposed to indicated concentrations of NGA/CNPs/NGA-CNPs or 6 Gy radiation or both, and stained with annexin V–FITC/PI before flow cytometry analysis. In each plot, the lower left corner shows annexin V-/PI-negative cells (living cells); the lower right corner shows annexin V-positive cells (apoptotic cells); the top right corner shows PI-positive cells (dead cells with membranes permeable to PI) stained with annexin V; and the top left corner shows dissociated nuclei. (B) Quantitative analysis of dead vs living cells. Data were analyzed by analysis of ANOVA followed by the Bonferroni post hoc test. **P<0.01, and ***P<0.0001, the groups compared with vehicle control group; ##P<0.01 and ###P<0.001, the NGA-CNP group compared with NGA group; ^^^P<0.001, the NGA-CNP group compared with CNP group.
Abbreviations: NGA, neogambogic acid; CNP, ceria nanoparticle; NGA-CNP, ceria nanoparticle modified with neogambogic acid; FITC, fluorescein isothiocyanate; PI, propidium iodide; ANOVA, analysis of variance.
NGA-CNPs enhance radiation-induced G2/M arrest
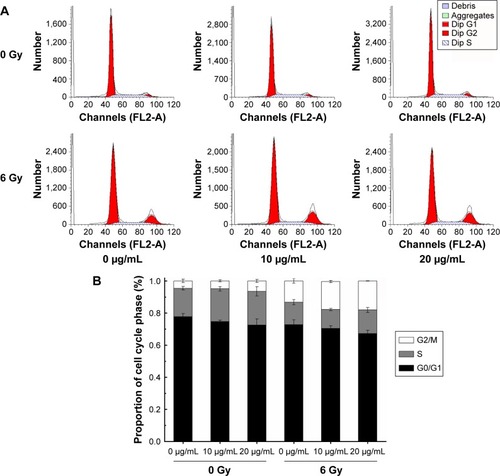
To clarify the mechanism by which NGA-CNPs enhance cancer cell sensitivity to radiation, we examined the effects of combined treatment on cell cycle regulation by flow cytometry. Treatment with NGA-CNPs and radiation decreased the fraction of cells in G0/G1 phase and increased the fraction in G2/M phase as compared to untreated control cells (). For instance, 4.57% of control cells were in G2/M phase; NGA-CNP treatment at concentrations of 10 μg/mL and 20 μg/mL increased the G2/M fraction to 4.89% and 6.44%, respectively; these values increased to 17.39% and 18.03%, respectively (P<0.05 vs 6 Gy radiation only), when NGA-CNPs were combined with 6 Gy radiation. These results indicate that simultaneous exposure of cells to NGA-CNPs and radiation causes acceleration through G1/S and arrest in G2/M phase. Since cells are most sensitive to the effects of radiation in the latter phase and most resistant in G0/G1,Citation33 one potential mechanism by which NGA-CNPs enhance the effects of radiotherapy is by regulating cell cycle progression.
Figure 6 Effect of NGA-CNPs on MCF-7 cell cycle distribution.
Notes: (A) Cells were pretreated with the vehicle DMSO or NGA-CNPs for 24 hours before exposure to 0 Gy or 6 Gy radiation, and the fraction of cells in each phase of the cell cycle was analyzed by flow cytometry. (B) Quantitative analysis of cell cycle distribution.
Abbreviations: NGA-CNP, ceria nanoparticle modified with neogambogic acid; DMSO, dimethyl sulfoxide; G2, second gap phase; M, mitosis phase; S, synthesis phase; G0, zero gap phase; G1, first gap phase.
Combined NGA-CNP and radiation treatment induces autophagy
Recent reports suggest that radiation and some anticancer drugs exert their effects by inducing autophagy, and consequently, tumor cell death.Citation38–Citation48 Autophagic activity is typically low under basal conditions but can be upregulated by endogenous and external signals such as nutrient starvation, energy depletion, radiation, and invasion by pathogens.Citation49,Citation50 Autophagy can be detected by TE microscopy or other methods that allow the visualization of autophagosome accumulation in the cytoplasm.Citation13 To determine whether autophagy was induced by combined NGA-CNP and radiation treatment, we used a commercial autophagy detection kit that employs the green fluorescent probe Cyto-ID to label vacuolar components of the autophagy pathway. It should be noted that unlike lysosomotrophic dyes such as acridine orange, monodansylcadaverine, and LysoTracker Red, which primarily detect lysosomes, the Cyto-ID only weakly stains lysosomes and serves as a selective marker of autolysosomes and earlier autophagic compartments.
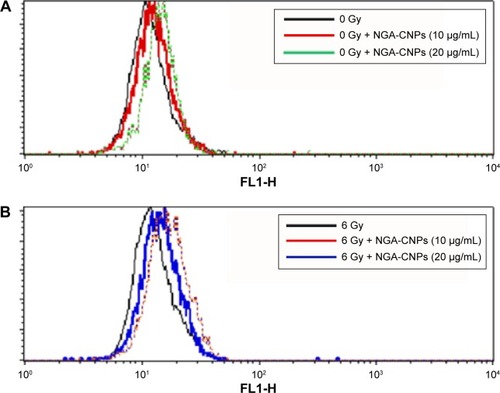
Cells were incubated with NGA-CNPs for 24 hours, exposed to 6 Gy radiation, and then labeled with Cyto-ID as well as Hoechst 33342 to detect nuclei. Quantitative analysis of autophagic cells is shown in . Cells containing at least three green dots or a green cluster corresponding to microtubule-associated light chain protein 3 (LC3) puncta were determined as autophagic cells; at least 150 cells were counted in triplicate samples.Citation15 Exposure to radiation or 10 μg/mL or 20 μg/mL NGA-CNPs alone did not lead to the obvious formation of LC3-positive puncta in confocal images (). However, a combination of NGA-CNP and 6 Gy radiation increased the number of LC3-positive puncta relative to those treated with radiation alone (P<0.001 vs 6 Gy radiation only). These results were confirmed by flow cytometry and suggest that the enhancement of autophagy is a mechanism by which NGA-CNPs sensitize cancer cells to the effects of radiation (). However, additional studies are needed to determine the relationship between the induction of autophagy and the increased rate of cell death.
Figure 7 Autophagy in MCF-7 cells treated with NGA-CNPs and radiation.
Notes: (A) Cells were incubated with indicated concentrations of NGA-CNPs for 24 hours followed by exposure to indicated doses of radiation; 24 hours later, cells were stained Cyto-ID (green) for detection of autophagic vacuoles and counterstained with Hoechst 33342 to label nuclei. (B) Quantitative analysis of autophagic cells (ie, with at least three green dots or a green cluster). Data were analyzed by analysis of ANOVA followed by Bonferroni post hoc test. ***P<0.0001, the groups compared with vehicle control groups (0 μg/mL +0 Gy group, or 0 μg/mL +6 Gy group).
Abbreviations: NGA-CNP, ceria nanoparticle modified with neogambogic acid; ANOVA, analysis of variance.
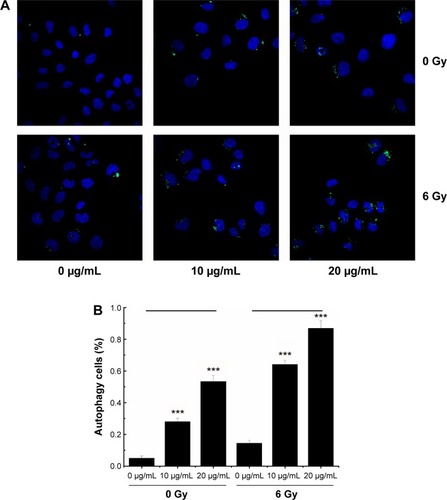
Figure 8 Detection of autophagy in MCF-7 cells by flow cytometry.
Notes: Cells were treated with indicated concentrations of NGA-CNPs without (A) or with (B) subsequent radiation treatment. A 488 nm laser source and the FL1 channel were used to detect the green fluorescence of autophagic cells.
Abbreviation: NGA-CNP, ceria nanoparticle modified with neogambogic acid.
NGA-CNPs suppress the generation of ROS
Oxidative stress, which elicits major changes in cellular function such as the induction of antioxidant enzymes, cell cycle arrest, and apoptosis, is frequently the cause of radiation-induced toxicity.Citation51 Radiation and gold and silver NPs have been shown to cause ROS-mediated cell death.Citation52 Given that ROS can also induce autophagy,Citation53 we investigated the changes in ROS levels resulting from combined NGA-CNP and radiation treatment with a fluorometric assay that detects intracellular oxidation of DCFH-DA by flow cytometry. DCFH-DA is hydrolyzed by esterases to DCFH, which remains trapped within the cell and is oxidized to the fluorescent molecule dichlorofluorescein (DCF) by the action of cellular oxidants; the detection of DCF provides an indication of intracellular ROS level.
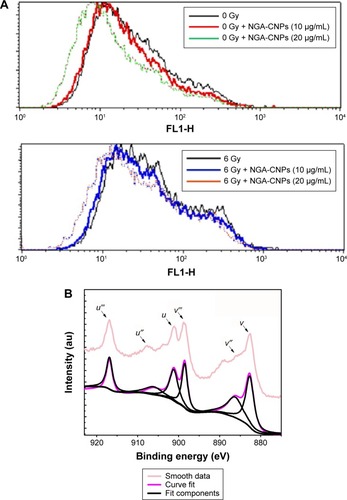
NGA-CNP treatment inhibited endogenous and radiation-induced ROS formation (), in contrast with the reported effects of other nanomaterials.Citation36 This may be explained by the oxygen vacancy sites on the surface of the nanoceria lattice,Citation54 which have cerium (3+) atoms at the center surrounded by cerium (4+) atoms that can absorb ROS.Citation55 Ce 3D features collected for reference powders (NGA-CNPs) were investigated in order to determine the positions of various components and were deconvoluted using a peak-fitting process. The NGA-CNP spectrum was composed of two multiplets (u and v) corresponding to the spin–orbit split 3d3/2 and 3d3/5 core boles shown in .Citation56 The spin–orbit splitting was approximately 18.6 eV, and the intensity ratio I(3d5/2)/I(3d3/2) was fixed at 1.5.Citation57 The highest binding energy peaks, u″′ and v″′ were located at around 916.9 eV and 898.3 eV, respectively, while the satellite peaks u″′ and v″′ associated with Ce 3d3/2 indicated the presence of Ce4+ and Ce3+, respectively, in NGA-CNPs. The lowest binding energy states u, u″, v, and v″ located at 901.3 eV, 907.3 eV, 882.7 eV, and 888.5±0.1 eV, respectively, were the result of Ce 3dCitation94fCitation2 O 2pCitation4 and Ce 3dCitation94fCitation1 O 2pCitation5 final states.Citation58,Citation59 These findings suggest that the NGA-CNP-induced sensitization of cancer cells to the effects of radiation is not achieved via the generation of ROS.
Figure 9 Detection of ROS in MCF-7 cells by flow cytometry.
Notes: (A) Suppression of ROS generation in MCF-7 cells by NGA-CNPs. Cells were treated with indicated concentrations of NGA-CNPs without (top) or with (bottom) subsequent radiation treatment. A fluorometric assay based on the oxidation of DCFH-DA by intracellular oxidants was used in conjunction with flow cytometry to detect ROS. (B) Ce 3d3/2, 5/2 XPS spectrum for NGA-CNPs.
Abbreviations: ROS, reactive oxygen species; NGA-CNP, ceria nanoparticle modified with neogambogic acid; DCFH-DA, 2′,7′-dichlorofluorescin diacetate; XPS, X-ray photoelectron spectroscopy.
Conclusion
CNPs with sizes of 3–5 nm and modified with NGA were not toxic to breast cancer cells at concentrations of 10 μg/mL or 20 μg/mL. However, when combined with radiation, NGA-CNPs caused G2/M arrest, induced autophagy, and increased the rate of cell death as compared to NGA, CNPs, or radiation treatment alone. These findings suggest that NGA-CNPs can be used as an adjuvant treatment to increase cancer cell sensitivity to the toxic effects of radiation, which would lower the effective doses of radiation that are used and thereby reduce damage to healthy tissue surrounding the tumor site.
Acknowledgments
The study was supported by the National Key Basic Research Program of China (973 Program, grant number: 2013CB933904), the National Nature Science Foundation of China (grant number: 31400721), the Nature Science Foundation of Jiangsu Province (grant number: BK20131355), and the Priority Academic Program Development of Jiangsu Higher Education Institutions. The authors thank the College of Materials Science and Technology, Nanjing University of Aeronautics and Astronautics, and Jiangsu Laboratory for Biomaterials and Devices, Southeast University, for providing technical support for this research.
Disclosure
The authors report no conflicts of interest in this work.
References
- MontazerabadiARSazgarniaABahreyni-ToosiMHAhmadiAAledavoodAThe effects of combined treatment with ionizing radiation and indocyanine green-mediated photodynamic therapy on breast cancer cellsJ Photochem Photobiol B Biol20121094249
- XieQZhouYLanGSensitization of cancer cells to radiation by selenadiazole derivatives by regulation of ROS-mediated DNA damage and ERK and AKT pathwaysBiochem Biophys Res Commun20144491889324813998
- KawasakiESPlayerANanotechnology, nanomedicine, and the development of a new, effective therapies for cancerNanomedicine20051210110917292064
- ShenJSongGAnMThe use of hollow mesoporous silica nanospheres to encapsulate bortezomib and improve efficacy for non-small cell lung cancer therapyBiomaterials201435131632624125776
- EomHJChoiJOxidative stress of CeO2 nanoparticles via p38-Nrf-2 signaling pathway in human bronchial epithelial cell, Beas-2BToxicol Lett20091872778319429248
- XuCQuXGCerium oxide nanoparticle: a remarkably versatile rare earth nanomaterial for biological applicationNPG Asia Mater20146e90
- HussainSAl-NsourFRiceABCerium dioxide nanoparticles induce apoptosis and autophagy in human peripheral blood monocytesACS Nano2012675809582022679913
- CiofaniGGenchiGGLiakosIEffect of cerium oxide nano-particles on PC12 neuronal-like cells: proliferation, differentiation, and dopamine secretionPharm Res20133082133214523661146
- RoccaAMattoliVMazzolaiBCiofaniGCerium oxide nanoparticles inhibit adipogenesis in rat mesenchymal stem cells: potential therapeutic implicationPharm Res201431112952296224805277
- WalkerCDasSSealSCatalytic properties and biomedical applications of cerium oxide nanoparticlesEnviron Sci Nano201521335326207185
- ColonJHsiehNFergusonACerium oxide nanoparticles protect gastrointestinal epithelium from radiation-induced damage by reduction of reactive oxygen species and upregulation of superoxide dismutase 2Nanomedicine20106569870520172051
- TarnuzzerRWColonJPatilSSealSVacancy engineered ceria nanostructures for protection from radiation-induced cellular damageNano Lett20055122573257716351218
- ChaachouayHOhneseitPToulanyMKehlbachRMulthoffGRodemannHPAutophagy contributes to resistance of tumor cells to ionizing radiationRadiother Oncol201199328729221722986
- RabinowitzJDWhiteEAutophagy and metabolismScience201033060091344134821127245
- YukJMShinDMSongKSBacillus calmette-guerin cell wall cytoskeleton enhances colon cancer radiosensitivity through autophagyAutophagy201061466019901560
- KimREmiMTanabeKThe role of apoptosis or nonapoptosis cell death in determining cellular response to anticancer treatmentEJSO2006323369377
- MeiWDongCHuiCGambogenic acid kills lung cancer cells through aberrant autophagyPLoS One201491e8360424427275
- ZhouGZZhangSNZhangLSunGCChenXBA synthetic curcumin derivative hydrazinobenzoylcurcumin induces autophagy in A549 lung cancer cellsPharm Biol201452111111624044367
- YanFWangMChenHGambogenic acid mediated apoptosis through the mitochondrial oxidative stress inactivation of Akt signaling pathway in human nasopharyngeal carcinoma CNE-1 cellEur J Pharmacol20116521–3233221118682
- YuXJHanQBWenZSMaLGaoJZhouGBGambogenic acid induces G1 arrest via GSK3β-dependent cyclin D1 degradation and triggers autophagy in lung cancer cellsCancer Lett2012322218519422410463
- ZhouJLuoYHWangJRGambogenic acid induction of apoptosis in a breast cancer cell lineAsian Pac J Cancer Prev201314127601760524460340
- PatilSKuirySCSealSVanfleetRSynthesis of nanocrystalline ceria particles for high temperature oxidation resistant coatingJ Nanopart Res200245433438
- ZhouXDHuebnerWAndersonHURoom-temperature homogeneous nucleation synthesis and thermal stability of nanometer single crystal CeO2Appl Phys Lett2002802038143816
- BumajdadAZakiMIEastoeJPasupuletyLMicroemulsion-based synthesis of CeO2 powders with surface area and high-temperature stabilitiesLangmuir20042025112231123315568879
- XueYLuanQFYangDYaoXZhouKBDirect evidence for hydroxyl radical scavenging activity of cerium oxide nanoparticlesJ Phys Chem C20111151144334438
- BarbaraDASantucciSBenedettiEDi LoretoSPhaniARFaloneSCerium oxide nanoparticles trigger neuronal survival in a human Alzheimer disease model by modulating BDNF pathwayCurr Nanosci200952167176
- CiminiAD’AngeloBDasSAntibody-conjugated PEGylated cerium oxide nanoparticles for specific targeting of A beta aggregates modulate neuronal survival pathwaysActa Biomater2012862056206722343002
- SantosMJQuintanillaRAToroAPeroxisomal proliferation protects from beta-amyloid neurodegenerationJ Biol Chem200528049410574106816204253
- PatilSReshetnikovSHaldarMKSurface-derivatized nanoceria with human carbonic anhydrase II inhibitors and fluorophores: a potential drug delivery deviceJ Phys Chem20071112484378442
- SazgarniaAMontazerabadiARBahreyni-ToosiMHAhmadiAAle-davoodAIn vitro survival of MCF-7 breast cancer cells following combined treatment with ionizing radiation and mitoxantrone-mediated photodynamic therapyPhotodiagnosis Photodyn Ther2013101727823465375
- CuiFBLiRTLiuQEnhancement of radiotherapy efficacy by docetaxel-loaded gelatinase-stimuli PEG-Pep-PCL nanoparticles in gastric cancerCancer Lett20143461536224333735
- ZhangXDWuDShenXSize-dependent radiosensitization of PEG-coated gold nanoparticles for cancer radiation therapyBiomaterials201233276408641922681980
- RoaWZhangXGuoLGold nanoparticles sensitize radiotherapy of prostate cancer cells by regulation of the cell cycleNanotechnology2009203737510119706948
- KlionskyDJAbeliovichHAgostinisPGuidelines for use and interpretation of assays for monitoring autophagy in higher eukaryotesAutophagy20084215117518188003
- MizushimaNYoshimoriTLevineBMethods in mammalian autophagy researchCell2010140331332620144757
- KimSChoiJEChoiJOxidative stress-dependent toxicity of silver nanoparticles in human hepatoma cellsToxicol In Vitro20092361076108419508889
- HarhajiLIsakovicARaicevicNMultiple mechanisms underlying the anticancer action of nanocrystallineJ Appl Polymer Sci20075681–38998
- KondoYKanzawaTSawayaRKondoSThe role of autophagy in cancer development and response to therapyNat Rev Cancer20055972673416148885
- WhiterEDiPaolaRSThe double-edged sword of autophagy modulation in cancerClin Cancer Res200915175308531619706824
- AmaravadiRKLippincott-SchwartzJYinXMPrinciples and current strategies for targeting autophagy for cancer treatmentClin Cancer Res201117465466621325294
- DalbyKNTekedereliILopez-BeresteinGOzpolatBTargeting the prodeath and prosurvival functions of autophagy as a novel therapeutic strategies in cancerAutophagy20106332232920224296
- YangZJCheeCEHuangSSinicropeFAThe role of autophagy in cancer: therapeutic implicationsMol Cancer Ther20111091533154121878654
- OuyangLShiZZhaoSProgrammed cell death pathways in cancer: a review of apoptosis, autophagy and programmed necrosisCell Prolif201245648749823030059
- WhiteEKarpCStroheckerAMGuoYMathewRRole of autophagy in suppression of inflammation and cancerCurr Opin Cell Biol201022221221720056400
- MilaniMRzymskiTMellorHRThe role of ATF4 stabilization and autophagy in resistance of breast cancer cells treated with BortezomibCancer Res200969104415442319417138
- ChenNKarantzaVAutophagy as a therapeutic target in cancerCancer Biol Ther201111215716821228626
- KimRHCoatesJMBowlesTLArginine deiminase as a novel therapy for prostate cancer induces autophagy and caspase-independent apoptosisCancer Res200969270070819147587
- DikicIJohansenTKirkinVSelective autophagy in cancer development and therapyCancer Res20107093431343420424122
- ShintaniTKlionskyDJAutophagy in health and disease: a double-edged swordScience2004306569899099515528435
- ZhangYYuCHuangGWangCWenLNano rare-earth oxides induced size-dependent vacuolization: an independent pathway from autophagyInt J Nanomedicine2010560160920856835
- GaoZSarsourEHKalenALLiLKumarMGGoswamiPCLate ROS accumulation and radiosensitivity in SOD1-overexpressing human glioma cellsNanomed Nanotechnol Biol Med201394558569
- ShiJMBaiLLZhangDMSaxifragifolin D induces the interplay between apoptosis and autophagy in breast cancer cells through ROD-dependent endoplasmic reticulum stressBiochem Pharmacol201385791392623348250
- AzadMBChenYQGibsonSBRegulation of autophagy by reactive oxygen species (ROS): implication for cancer progression and treatmentAntioxid Redox Signal200911477779018828708
- SahuTBishtSMDasRKKiritRNanoceria: synthesis and biomedical applicationsCurr Nanosci201395588593
- BriggsACordeSOktariaSCerium oxide nanoparticles: influence of the high-Z component revealed on radioresistant 9L cell survival under X-ray irradiationNanomedicine2013971098110523473745
- BecheECharvinPPerarnauDCe 3d XPS investigation of cerium oxides and mixed cerium (CexTiyOz)Surf Interface Anal2008403–4264267
- Escamilla-PereaLNavaRPawelecBSBA-15-supported gold nanoparticles decorated by CeO2: structural characteristics and CO oxidation activityAppl Catal A20103811–24253
- TullerHLNowickASDefect structure and electrical properties of nonstoichiometric CeO2 single crystalsJ Electrochem Soc19791262209217
- BoroninAISlavinskayaEMDanilovaIGInvestigation of palladium interaction with cerium oxide and its state in catalysts for low-temperature CO oxidationCatal Today20091443–4201211
