Abstract
Tumors frequently contain hypoxic regions that result from a shortage of oxygen due to poorly organized tumor vasculature. Cancer cells in these areas are resistant to radiation- and chemotherapy, limiting the treatment efficacy. Macrophages have inherent hypoxia-targeting ability and hold great advantages for targeted delivery of anticancer therapeutics to cancer cells in hypoxic areas. However, most anticancer drugs cannot be directly loaded into macrophages because of their toxicity. In this work, we designed a novel drug delivery vehicle by hybridizing macrophages with nanoparticles through cell surface modification. Nanoparticles immobilized on the cell surface provide numerous new sites for anticancer drug loading, hence potentially minimizing the toxic effect of anticancer drugs on the viability and hypoxia-targeting ability of the macrophage vehicles. In particular, quantum dots and 5-(aminoacetamido) fluorescein-labeled polyamidoamine dendrimer G4.5, both of which were coated with aminederivatized polyethylene glycol, were immobilized to the sodium periodate-treated surface of RAW264.7 macrophages through a transient Schiff base linkage. Further, a reducing agent, sodium cyanoborohydride, was applied to reduce Schiff bases to stable secondary amine linkages. The distribution of nanoparticles on the cell surface was confirmed by fluorescence imaging, and it was found to be dependent on the stability of the linkages coupling nanoparticles to the cell surface.
Introduction
The cell membrane, a semipermeable lipid bilayer, defines the cell boundary and consists of lipids, proteins and carbohydrates that are responsible for selective uptake of molecules, cell–cell interactions, cell–matrix interactions, and many other vital cell activities. Because of the importance of cell surface interactions to cell and tissue function, various cell surface engineering approaches have been explored to modify the cell surface to manipulate cell behavior and function.Citation1,Citation2
Considerable progress has been made in introducing nonnative chemical species to the cell membrane, permitting a wide range of applications in biology, medicine, drug delivery, and tissue engineering.Citation3 A molecule of interest can be attached to the cell surface through a fatty tether including glycosyl-phosphatidylinositol (GPI)-anchored proteinsCitation4 and cholesterol-tethered compounds.Citation5 A viable alternative is to apply enzyme-catalyzed chemical reactions to modify carbohydrates on the cell surface. For example, existing surface glycoforms can be utilized as acceptors for reactions with an exogenously applied glycosyltransferase and appropriate activated sugar donor.Citation1,Citation6 Sialic acids are the most common terminal sugar residue on the cell surfaces and play an important role in cell adhesion and recognition. Unnatural sialic acid precursors can be incorporated into cell surface glycoforms by metabolic engineering.Citation7 Direct covalent reactions can also be applied to enable chemical modification of cell surfaces.Citation8,Citation9 One possible approach is to couple biomolecules of interest to the cell surface via reactive cell surface groups such as aldehydes and ketones.Citation10,Citation11 The generation of these reactive cell surface groups may be achieved by direct chemical or enzymatic treatment, or by the metabolic incorporation of an unnatural molecule that contains the chosen chemical species.
Cell surface engineering has generated tremendous advantages for drug delivery and tissue engineering. For example, a synthetic adenovirus receptor inserted to the cell surface via a metabolic engineering approach facilitates the entrance of adenovirus into cells that are normally resistant to infection by this virus.Citation12 Uptake of exogenous proteins could be enhanced by inserting appropriate synthetic receptors.Citation13 Selectively killing tumor cells could be realized by controlling the targeting of an antibody through tagging tumor cells with a non-natural sugar into the polysialic acid molecules on the cell surface.Citation14 Cell surface modification approaches have also been applied to generate three-dimensional cell aggregates or tissue-engineered constructs through cell cross-linking. Citation15 A recent study reported that polyethylene glycol (PEG) can be attached to the surface of islets to circumvent immune rejection during transplantation of pancreatic islets from donor to a patient.Citation9
In this work, we designed a novel hypoxia-targeted drug delivery vehicle by hybridizing macrophages with nanoparticles through cell surface modification. Tumors frequently contain hypoxic regions that result from a shortage of oxygen due to poorly organized tumor vasculature. Cancer cells in these areas are resistant to radiation- and chemotherapy, limiting the treatment efficacy.Citation16 Macrophages and monocytes have inherent hypoxia-targeting ability and hold great advantages for selective delivery of anticancer therapeutics to cancer cells in hypoxic areas.Citation16–Citation19 However, most anticancer drugs cannot be directly loaded into macrophages or monocytes because of their toxicity. Hybridizing a macrophage or monocyte cellular vehicle with a synthetic carrier, particularly nanoparticles, may represent a novel approach for effective delivery of anticancer drugs to hypoxic regions in solid tumors. Immobilizing nanoparticles on the cell surface provide numerous new sites for anticancer drug loading, hence potentially minimizing the toxic effect of anticancer drugs on the viability and hypoxia-targeting ability of the macrophage or monocyte vehicles.
The focus of the current work was to demonstrate the feasibility of immobilizing nanoparticles including polyamidoamine (PAMAM) dendrimers and quantum dots (Qdots) to the macrophage surface through cell surface chemical modification. Dendrimers are highly branched macromolecules with low polydispersity and well-defined surface functionality. Utility of dendrimers in this work allowed us to take advantage of their versatility to explore optimal approaches for cell-nanoparticle hybridization and realize a high drug payload and assembly of multiple functional entities necessary for hybridization and drug delivery. Commercially available quantum dots coated with amine-derivatized PEG was also studied for cell-nanoparticle hybridization. Qdots have been explored for fluorescence imaging of living cells.Citation20 Although the limited number of surface groups of Qdots is not advantageous for drug loading, hybridizing cells with Qdots possessing long-term photostability offers a noninvasive way for cell tracking in vivo and would provide further evidence to demonstrate the feasibility of the cell-nanoparticle hybridization approach explored in this work.
Experimental
Materials
Qdot® 525 ITK™ amino (PEG) quantum dots (simply referred to as QD525) and 5-(aminoacetamido) fluorescein (AAF) were purchased from Invitrogen (Carlsbad, CA). PAMAM dendrimer G4.5 was purchased from Dendritech (Midland, MI). PEG diamine (MW= ~3350 g mol−1), N-hydroxysuccinimide (NHS), 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide hydrochloride (EDC), sodium cyanoborohydride (NaCNBH3), sodium periodate (NaIO4), and sodium phosphate buffer (10×) were purchased from Sigma-Aldrich (St. Louis, MO). 4',6-diamidino-2-phenylindole (DAPI), Dulbecco’s modified Eagle medium (DMEM), sodium hydroxide, paraformaldehyde, phosphate-buffered saline (PBS), fetal bovine serum (FBS), penicillin, and Trypan blue were purchased from Fisher Scientific (Pittsburgh, PA).
Preparation of AAF-labeled PEGylated G4.5 dendrimers (AAF-G4.5-PEG)
Synthesis
The synthesis of G4.5-PEG followed our previous work.Citation21 Upon removal of methanol from G4.5 PAMAM stock solution by rotary evaporation, G4.5 PAMAM dendrimer (5 mg) was dissolved in 2 mL of sodium PBS (0.1 M, pH = 5.5). To this solution were added 5.4 mg of EDC and 3.2 mg of NHS. After a 15 minute reaction at room temperature, 68.5 mg of PEG diamine (MW = ~3350 g mol−1) predissolved in the buffer solution was added to the reaction solution dropwise. Subsequently, 1.4 mg of AAF predissolved in the buffer was slowly added to the reaction solution. The reaction mixture solution was stirred in the dark overnight. The resultant AAF-G4.5-PEG nanoparticles were purified by dialysis and then freeze dried.
Proton nuclear magnetic resonance (1H-NMR) spectroscopy
The 1H-NMR spectra of dendrimer derivatives were recorded on a Varian Mercury-300 MHz NMR spectrometer (Varian, Palo Alto, CA). The solvent used was deuterium water (D2O), which has a chemical shift of 4.8 ppm.
Fluorescence spectroscopy
Fluorescence emission spectra of AAF and AAF-G4.5-PEG in water were recorded on a Varian Cary Eclipse fluorescence spectrophotometer with an excitation wavelength of 488 nm.
Preparation of macrophage–nanoparticle hybrid vehicles
As illustrated in , nanoparticles (QD525 or AAF-G4.5- PEG) were coupled to the surface of macrophages via either a transient Schiff base linkage or a stable secondary amine linkage.
Scheme 1 Hybridization of nanoparticles and macrophage through cell surface modification. Sialic acid residues on the cell surface are modified with sodium periodate to generate aldehydes. Aldehydes react with amine group of PEG conjugated to the nanoparticle surface to form Schiff bases. Schiff bases can be further reduced to stable secondary amine linkages using sodium cyanoborohydride.
Abbreviation: PEG, polyethylene glycol.
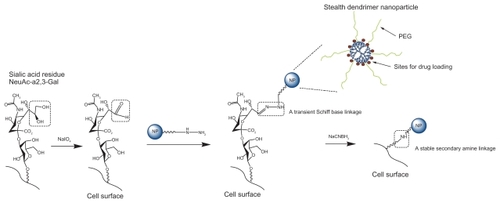
Macrophage–nanoparticle hybrids with a transient linkage (macrophage–T-nanoparticle hybrids)
RAW264.7 macrophages were plated on coverslips at a density of 1.2 × 104 cells/coverslip. At 70% confluence, the cells were treated with 0.1 mM cold NaIO4 in 100 μL of PBS (pH 7.4), incubated with shaking for 15 minutes at 4 °C in the dark, and then rinsed with cold PBS three times to remove NaIO4.Citation15 Following the NaIO4 treatment, the cells were incubated with 12 μg of nanoparticles dissolved in 100 μL of PBS for 0–24 hours. The resultant macrophage–T-nanoparticle hybrids were washed with PBS and incubated in PBS. At predetermined time points, macrophage–T-nanoparticle hybrids were fixed with 300 μL of 4% paraformaldehyde for fluorescence or confocal imaging. Untreated macrophages incubated with nanoparticles were used as a control.
Macrophage–nanoparticle hybrids with a stable linkage (macrophage–S-nanoparticle hybrids)
Macrophage–T-nanoparticle hybrids were further treated with 100 μL of 0.1 mM NaCNBH3 for 1~2 hours to convert the transient Schiff base linkages to stable amide linkages, washed with PBS, and incubated in PBS. At predetermined time points, macrophage–T-nanoparticle hybrids were fixed with 4% paraformaldehyde for imaging. Untreated macrophages incubated with nanoparticles were used as a control.
pH-dependent cell viability assay
Since RAW264.7 macrophages were subjected to surface modification and the rate of the stablizing amide reaction is pH-dependent, the effect of pH of modifying solutions on the RAW264.7 macrophage viability was studied. In brief, RAW264.7 macrophages were initially incubated for two hours in media at different pH values, including DMEM (pH 7.4, control) and PBS (pH 8, pH 9 and pH 10, adjusted with sodium hydroxide), washed with PBS (pH 7.4) three times, and grown in DMEM supplemented with 10% FBS and 5% penicillin for 48 hours. Viable cells were counted by using the Trypan blue assay. In addition, toxicity of 0.1 mM NaCNBH3 in DMEM at pH 7.4 was also evaluated. Cell viability was then determined as follows: Cell viability (%) = total number of viable cells in each group/total number of viable cells in the control ×100.
Fluorescence image analysis
True-color fluorescence images of macrophage–nanoparticle hybrids were taken under a Zeiss Axiovert 200 inverted fluorescence microscope (Carl Zeiss AG, Oberkochen, Germany) or a Leica TCS-SP2 AOBS confocal laser scanning microscope (Lecia, Solms, Germany). DAPI nuclear stain was applied for colocalization of nanoparticles. Images were analyzed with ImageJ software (National Institutes of Health, Bethseda, MD).
Results and discussion
Preparation of macrophage–nanoparticle hybrids
In this study commercial QD525 coated with an amine-derivatized PEG layer and synthesized PEGylated carboxyl terminated PAMAM den-drimer G4.5 were employed for hybridization with macrophages. PEGylated PAMAM dendrimer G4.5 was labeled with AAF to allow confirmation of immobilization of nanoparticles on the cell surface by fluorescence imaging. Polyanionic PAMAM dendrimer G4.5 has a relatively high number of surface groups, negligible toxicity and immunogenicity, and favorable biodistribution.Citation22 The negatively charged dendrimers have negligible cellular uptake due to their low non-specific interaction with the negatively charged cell surface. Inclusion of PEG provides dendrimers with favorable pharmacokinetic and tissue distribution and reduces potential accumulated toxicity and immunogenicity of the nanoparticles. Importantly, PEG helps nanoparticles to escape from phagocytosis of macrophages. This is a critical step for constructing the proposed hybrid delivery system. PEGylated dendrimers of larger molecular weight and with more branches tend to have a lower accumulation in cells as demonstrated previously.Citation23
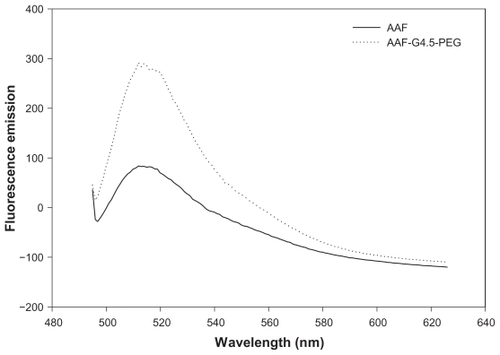
Similarly to the QD525 employed in this study, PEGylated G4.5 was designed to have amine-derivatized PEG chains on the surface, based on which cell surface modification chemistry was explored. PEGylated dendrimers have been well characterized previously.Citation21,Citation24–Citation27 According to the 1H-NMR measurement, an average of eight PEG chains were coupled to G4.5. PEGylated G4.5 was labeled with a moderately water-soluble fluorescence probe, AAF. As shown in , AAF coupled to the dendrimer has an emission wavelength of 515 nm, which is identical to the emission wavelength of free AAF. The unaltered emission wavelength of AAF following this coupling strategy was also found in a previous report.Citation28 Furthermore, the fluorescence emission intensity of AAF coupled to PEGylated G4.5 is significantly higher than that of AAF prepared in water at saturation, suggesting that AAF coupled to PEGylated G4.5 has higher water solubility than its unmodified form. The increase in the water solubility of AAF was attributed to the successful conjugation of AAF to the PEGylated dendrimer.
Figure 1 Fluorescence emission spectra of AAF and AAF-G4.5-PEG.
Abbreviation: PEG, polyethylene glycol.
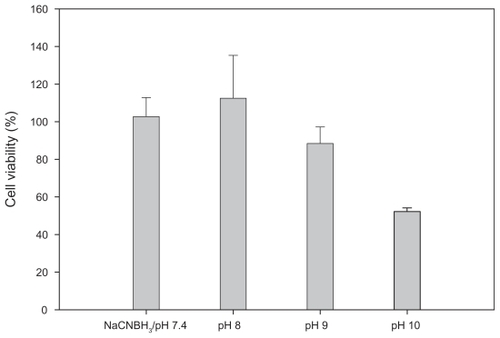
A number of methods have been developed to enable the chemical modification of cell surfaces. In this project, we employed a simple and well-documented methodology to immobilize nanoparticles to the macrophage cell surface. In particular, sialic acid residues embedded on the cell surface were converted to aldehydes with sodium periodate. Our results showed that the toxicity of NaIO4 was negligible at the concentration of 0.1 mM. This was supported by the work of Ong and coworkers.Citation15 Aldehydes reacte with primary amine end groups of PEG on the nanoparticle surface to form Schiff bases. Schiff base linkage is labile and can be cleaved in aqueous solution by hydrolysis. Accordingly, the macrophage– T-nanoparticle hybrids constructed are expected to readily release nanoparticles through the cleavage of the transient Schiff base linkages. We also applied a reducing agent sodium cyanoborohydride to reduce Schiff bases to stable secondary amine linkages.Citation29 It has been reported that sodium cyanoborohydride has high specificity toward the Schiff base. According to our cell viability studies (), sodium cyanoborohydride at 0.1 mM was nontoxic to RAW264.7 macrophages. As the reaction proceeds more efficiently at basic pH,Citation30 the viability of RAW264.7 macrophages at high pH (8–10) was evaluated to determine whether the immobilization chemistry can be performed at high pH. As shown in , pH affects the cell viability. The viability of RAW264.7 macrophages remains intact at pH 8 or pH 7.4 with 0.1 mM sodium cyanoborohydride. The cell viability dropped slightly to 88% at pH 9. By contrast, the viability was reduced drastically to 52% at pH 10. The results suggest that mild basic pH has a minimal impact on the viability of RAW264.7 macrophages and the optimum pH within that range can be explored for increasing the immobilization efficiency.
Figure 2 pH-dependent viability of RAW264.7 macrophages. Cells were incubated for two hours at the indicated pH, and then assessed by the Trypan blue assay 48 hours later. Nontoxicity of 0.1 mM sodium cyanoborohydride in DMEM at pH 7.4 was confirmed.
Note: Bar = SD.
Abbreviations: DMEM, Dulbecco’s modified Eagle’s medium; SD, standard deviation.
Fluorescence image analysis of macrophage–Qdot hybrids
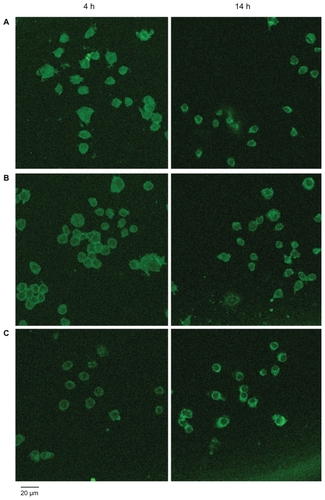
Fluorescence microcopy and confocal microscopy were applied to conf irm the hybridization of nanoparticles with macrophages. As shown in , both macrophage–T-nanoparticle hybrids and macrophage– S-nanoparticle hybrids prepared with QD525 exhibit strong fluorescence intensity at the edge of the cells, clearly outlining the cell surface. Qualitatively, more QD525 were taken up in macrophage–T-nanoparticle hybrids as opposed to macrophage–S-nanoparticle hybrids, reflecting the stability of the linkage between the nanoparticles and the cell. Confocal microscopy images in z-sections were taken to further examine the distribution of QD525 over the course of time. Without cell surface modification, QD525 were phagocytosed quickly and evenly distributed in the cytoplasm (). Although QD525 were still internalized, a significant amount of QD525 accumulated at the edge of the cells treated with sodium periodate () or a combination of sodium periodate and sodium cyanoborohydride (). QD525 immobilized on the cell surface through a stable secondary amine bond displayed good stability overnight (). In contrast, QD525 linked to the cell surface via Schiff base linkages were found to be re-distributed in the cell overnight ().
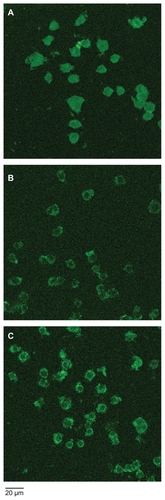
Figure 3 Fluorescence microscopy images of macrophage–Qdot hybrids. A) Macrophage–T-Qdot: macrophages treated with sodium periodate, incubated with QD525 for four hours, and fixed; B) Macrophage–S-Qdot: macrophages treated with sodium periodate, incubated with QD525 for four hours, treated with sodium cyanoborohydride, and fixed.
Notes: Original magnification, ×400; scale bar, 10 μm.
Abbreviation: Qdot, quantum dot.
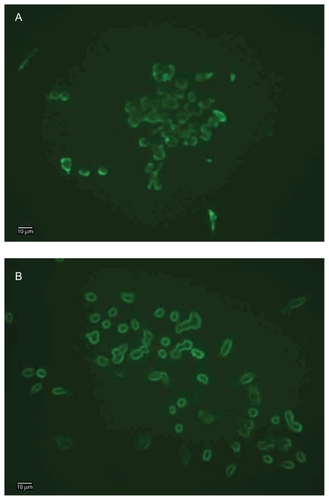
Figure 4 Confocal microscopy images of macrophage–Qdot hybrids at four hours (left panel) or 14 hours (right panel) post-treatment. A) QD525 incubated with untreated macrophages (control); B) Macrophage–T-Qdot hybrids; C) Macrophage–S-Qdot hybrids.
Notes: Original magnification, ×630.
Abbreviation: Qdot, quantum dot.
Fluorescence image analysis of macrophage–dendrimer hybrids
Macrophages hybridized with AAF-labeled PEGylated PAMAM dendrimer G4.5 were also studied. As shown in , the level of the internalization of AAF-G4.5-PEG by untreated macrophages is low. This confirmed that cationic surface charges and PEGylation indeed helped to reduce nonspecific internalization by macrophages. Further, an apparently biased distribution of AAF-G4.5-PEG nanoparticles was observed on the surface of macrophages that were treated with sodium perioate () or the combination of sodium periodate and sodium cyanoborohydride (). A significant increase in fluorescence intensity of macrophage–T-dendrimer hybrids and macrophage–S-dendrimer hybrids as compared to the control suggests that more dendrimer nanoparticles have been immobilized to the cell surface and/or internalized by the cells. The amine linkages connecting dendrimers to the cell enabled macrophage–S-dendrimer hybrids to carry dendrimers stably on the surface for an extended period of time. We also observed that when the surface-treated macrophages were incubated with the same amount of AAF-G4.5-PEG (12 μg), a condition of a high concentration (12 μg/100 μL) and a short incubation (one minute) was relatively equivalent to a condition of a low concentration (12 μg/1.5 mL) and a long incubation (10 minutes) in terms of hybridization efficiency as determined by confocal image analysis. Cell viability remained good during the surface treatment.
Figure 5 Confocal microscopy images of macrophage–dendrimer hybrids. A) Untreated macrophages incubated with AAF-G4.5-PEG for 10 minutes and fixed immediately (control); B) Macrophage–T-dendrimer hybrids fixed immediately following 10-minute incubation with AAF-G4.5-PEG; C) Macrophage–S-dendrimer hybrids fixed at 14 hours following treatment of sodium cyanoborohydride. Original magnification, ×630.
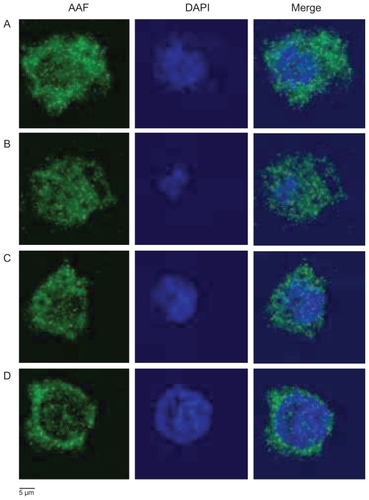
To further evaluate the intracellular localization of nanoparticles, we performed a colocalization assay on AAF-G4.5-PEG with nuclear DAPI staining. Clearly shown in , a significant amount of AAF-G4.5-PEG nanoparticles were internalized within one-minute incubation and accumulated evenly in the cytoplasma with no selectivity towards the cell surface after overnight culture. Entry of the nanoparticles into nuclei was also observed in the macrophages. This became more evident when the cells were incubated with AAF-G4.5-PEG for an extended period of time (overnight incubation, ). In addition, more nanoparticles were internalized by the cells following the overnight incubation as indicated by the increase in the fluorescence intensity. Macrophage–S-dendrimer hybrids () show a stronger fluorescence intensity at the cell surface and less in nuclei as compared to macrophage– T-dendrimer hybrids (), confirming that the stability of immobilized dendrimers in macrophage–S-dendrimer hybrids was higher than macrophage–T-dendrimer hybrids.
Figure 6 Colocalization assay of AAF-G4.5-PEG (green) with nuclei (blue) by confocal microscopy. A) Control 1: untreated macrophages incubated with AAF-G4.5-PEG for one minute, washed and cultured overnight (24 hours), then fixed and counterstained with DAPI; B) Control 2: untreated macrophages incubated with AAF-G4.5-PEG overnight (24 hours), then fixed and counterstained with DAPI; C) Macrophage–T-dendrimer hybrids: sodium periodate-treated macrophages incubated with AAF-G4.5-PEG for one minute, cultured overnight (24 hours), then fixed and counterstained with DAPI; D) Macrophage–S-dendrimer hybrids: sodium periodate-treated macrophages incubated with AAF-G4.5-PEG for one minute, treated with sodium cyanoborohydride, cultured overnight (24 hours), then fixed and counterstained with DAPI.
Notes: Original magnification, ×630.
It is apparent that fluorescently labeled nanoparticles were taken into the macrophages after each treatment. Qualitatively, there is a uniform distribution of fluorescence throughout the untreated control groups, suggesting cellular uptake pathways are responsible for this occurrence. Following surface modification, a pronounced ring of fluorescence is observed towards the cell surface. Quantitative analysis of the distribution of nanoparticle fluorescence was attempted with the intensity profile generated by ImageJ software (). Since all confocal images were taken under identical image acquisition settings, the fluorescence intensity profiles generated allowed us to quantitatively analyze fluorescence distribution in individual cells based on their relative fluorescence intensity. For each group, three representative cells were chosen and analyzed. Individual cells were measured three times from different orientations with each orientation approximately bisecting midlines. From each profile, intensity values were recorded from fluorescence peaks at the two cell boundaries and from a fluorescence peak near the midline of each trace. The results are summarized in . The results from the intensity traces show that for a nanoparticle incubation of one minute, there is 20.8% more fluorescence at the cell boundaries versus the cell center. For an overnight incubation, there was an even distribution of fluorescent intensity where the cell walls exhibited a fluorescence value that was 99.8% of the fluorescence of the midpoint of the slice. Surface modification drastically increased the ratio of fluorescence between the cell walls versus the cell interior. The macrophage–T-nanoparticle hybrids showed an 85.2% increase in fluorescence near the cell exterior, whereas the macrophage–S-nanoparticle hybrids showed a 94.4% increase in cell wall fluorescence. Based on these measurements we show that there is markedly more fluorescence near the cell surface for surface-treated groups, whereas there is an even distribution of nanoparticles in the control groups.
Figure 7 Quantitative analysis of the distribution of fluorescence intensity in representative cells. A) Control 1; B) Control 2; C) Macrophage–T-dendrimer hybrid; D) Macrophage–S-dendrimer hybrid. The treatment conditions are detailed in .
Notes: Original magnification, ×630.
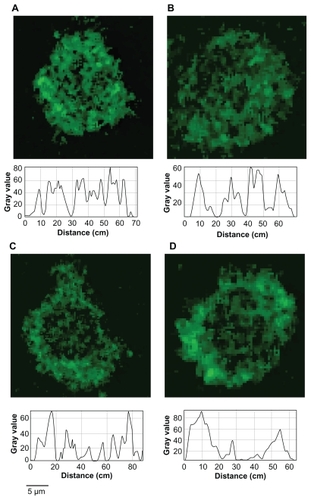
Table 1 Summary of the distribution of AAF fluorescence intensity in macrophages subjected to various treatments as indicated below
From a chemistry perspective, this work demonstrated the proof-of-principle of chemically hybridizing macrophages with nanoparticles through cell surface modification. The reaction conditions explored in this study were mild to the cells. It should be noted that internalization of nanoparticles by macrophages seems to be an inevitable process because of their innate phagocytic capability. Nonetheless, our studies disclosed that cell surface modification provides a means to retard the internalization progress and alter the intracellular distribution of nanoparticles. A comprehensive understanding of the trafficking and dynamic distribution of nanoparticles is needed in order for us to optimize the hybridization process. Reducing nonspecific phagocytic internalization of nanoparticles will be pursued in our laboratory. The sizes of the Qdots and PAMAM dendrimers as an important factor affecting nanoparticle internalization by cells will be studied. PAMAM dendrimers have a versatile structure ideal for construction of drug delivery systems and have been extensively studied by many groups including us.Citation31–Citation34 Future work will include hybridization of drugcarrying dendrimers and/or QD525 with macrophages and studying the delivery efficiency of such a new drug vehicle in terms of drug distribution in hypoxic areas using in vitro spheroid models and animal models.
Conclusions
QD525 and dendrimers were immobilized to the macrophage cell surface through either a transient Schiff base linkage or a stable amine linkage. The distribution of nanoparticles on the cell surface was confirmed by fluorescence imaging and was found to be dependent on the stability of the linkages connecting nanoparticles to the cell surface. Achieving homogeneous distribution of anticancer drugs within tumors remains one of the major challenges in cancer chemotherapy and is critical for treatment effectiveness. The current study has explored an innovative way of utilizing nanoparticles and cellular vehicles for anticancer drug delivery. Development of a cell–nanoparticle hybrid vehicle through cell surface modification would utilize the best aspects of both cellular carriers and nanoparticles and may help to improve anticancer drug distribution and penetration in tumors.
Acknowledgments
This research was supported in part by The Jeffress Memorial Trust (J-873) and the National Institutes of Health (R21NS063200). RAW264.7 macrophages were provided by Dr Xianjun Fang (Department of Biochemistry and Molecular Biology, Virginia Commonwealth University). Confocal microscopy was performed at the VCU Department of Neurobiology and Anatomy Microscopy Facility, supported, in part, with funding from NIH-NINDS Center core grant (5P30NS047463). The authors report no conflicts of interest in this work.
References
- GahmbergCGTolvanenMNonmetabolic radiolabeling and tagging of glycoconjugatesMethods Enzymol199423032448139505
- MahalLKYaremaKJBertozziCREngineering chemical reactivity on cell surfaces through oligosaccharide biosynthesisScience19972765315112511289173543
- KellamBDe BankPAShakesheffKMChemical modification of mammalian cell surfacesChem Soc Rev200332632733714671788
- PremkumarDRFukuokaYSevleverDProperties of exogenously added GPI-anchored proteins following their incorporation into cellsJ Cell Biochem200182223424511527149
- HusseySLHeEPetersonBRA synthetic membrane-anchored antigen efficiently promotes uptake of antifluorescein antibodies and associated protein a by mammalian cellsJ Am Chem Soc200112350127121271311741450
- GuptaSKAgarwalRGalpalliNDComparative efficacy of pilocarpine, timolol and latanoprost in experimental models of glaucomaMethods Find Exp Clin Pharmacol2007291066567118200329
- KepplerOTStehlingPHerrmannMBiosynthetic modulation of sialic acid-dependent virus-receptor interactions of two primate polyoma virusesJ Biol Chem19952703130813147836396
- BurchenalJEDeibleCRDeglauTEPolyethylene glycol diisocyanate decreases platelet deposition after balloon injury of rabbit femoral arteriesJ Thromb Thrombolysis2002131273311994557
- PanzaJLWagnerWRRiloHLTreatment of rat pancreatic islets with reactive PEGBiomaterials200021111155116410817268
- De BankPAKellamBKendallDAShakesheffKMSurface engineering of living myoblasts via selective periodate oxidationBiotechnol Bioeng200381780080812557313
- LemieuxGABertozziCRChemoselective ligation reactions with proteins, oligosaccharides and cellsTrends Biotechnol199816125065139881482
- ChoiYSHongSRLeeYMStudies on gelatin-containing artificial skin: II. Preparation and characterization of crosslinked gelatin-hyaluronate spongeJ Biomed Mater Res199948563163910490676
- MartinSEPetersonBRNon-natural cell surface receptors: synthetic peptides capped with N-cholesterylglycine efficiently deliver proteins into mammalian cellsBioconjug Chem2003141677412526694
- LiuTGuoZYangQSadSJenningsHJBiochemical engineering of surface alpha 2–8 polysialic acid for immunotargeting tumor cellsJ Biol Chem200027542328323283610976100
- OngSMHeLThuy LinhNTTransient inter-cellular polymeric linkerBiomaterials200728253656366717512584
- BrownJMWilsonWRExploiting tumour hypoxia in cancer treatmentNat Rev Cancer20044643744715170446
- GriffithsLBinleyKIqballSThe macrophage – a novel system to deliver gene therapy to pathological hypoxiaGene Ther20007325526210694803
- PaulSSnaryDHoebekeJTargeted macrophage cytotoxicity using a nonreplicative live vector expressing a tumor-specif ic single-chain variable region fragmentHum Gene Ther200011101417142810910139
- ChoiMRStanton-MaxeyKJStanleyJKA cellular Trojan horse for delivery of therapeutic nanoparticles into tumorsNano Lett20077123759376517979310
- HowarthMTakaoKHayashiYTingAYTargeting quantum dots to surface proteins in living cells with biotin ligaseProc Natl Acad Sci U S A2005102217583758815897449
- YangHLopinaSTPenicillin V-conjugated PEG-PAMAM star polymersJ Biomater Sci Polym Ed200314101043105614661878
- RobertsJCBhalgatMKZeraRTPreliminary biological evaluation of polyamidoamine (PAMAM) starburst dendrimersJ Biomed Mater Res199630153658788106
- XyloyannisMPadilla De JesusOLFrechetJMJDuncanRPEG-dendron architecture influences endocytic capture and intercellular traffickingProc Int Symp Control Rel Bioact Mater200330149
- KailasanAYuanQYangHSynthesis and characterization of thermoresponsive polyamidoamine-polyethylene glycol-poly (D, L-lactide) (PAMAM-PEG-PDLLA) core-shell nanoparticlesActa Biomaterialia200910.1016/j.actbio.2009.08.036
- YangHLopinaSTIn vitro enzymatic stability of dendritic peptidesJ Biomed Mater Res Part A200676A2398407
- YangHLopinaSTDiPersioLPSchmidtSPStealth dendrimers for drug delivery: correlation between PEGylation, cytocompatibility, and drug payloadJ Mater Sci Mater Med20081951991199717952565
- YangHMorrisJJLopinaSTPolyethylene glycol-polyamidoamine dendritic micelle as solubility enhancer and the effect of the length of polyethylene glycol arms on the solubility of pyrene in waterJ Colloid Interface Sci2004273114815415051444
- TempletonACCliffelDEMurrayRWRedox and fluorophore functionalization of water-soluble, tiopronin-protected gold clustersJ Am Chem Soc19991213070817089
- PengLCaltonGJBurnettJWEffect of borohydride reduction on antibodiesAppl Biochem Biotechnol198714291993619438
- HornseyVSProwseCVPepperDSReductive amination for solid-phase coupling of protein. A practical alternative to cyanogen bromideJ Immunol Methods198693183883021857
- YangHKaoJWSynthesis and characterization of nanoscale dendritic RGD clusters for potential applications in tissue engineering and drug deliveryInt J Nanomedicine200721899917722516
- YangHLopinaSTStealth dendrimers for antiarrhythmic quinidine deliveryJ Mater Sci Mater Med200718102061206517558476
- YangHLopinaSTExtended release of a novel antidepressant, venla-faxine, based on anionic polyamidoamine dendrimers and poly(ethylene glycol)-containing semi-interpenetrating networksJ Biomed Mater Res200572A1107114
- SarkarKYangHEncapsulation and extended release of anti-cancer anastrozole by stealth nanoparticlesDrug Deliv200815534334618763165