Abstract
Conventional in vitro culture studies on flat surfaces do not reproduce tissue environments, which have inherent topographical mechanical signals. To understand the impact of these mechanical signals better, we use a cell imprinting technique to replicate cell features onto hard polymer culture surfaces as an alternative platform for investigating biomechanical effects on cells; the high-resolution replication of cells offers the micro- and nanotopography experienced in typical cell–cell interactions. We call this platform a Bioimprint. Cells of an endometrial adenocarcinoma cell line, Ishikawa, were cultured on a bioimprinted substrate, in which Ishikawa cells were replicated on polymethacrylate (pMA) and polystyrene (pST), and compared to cells cultured on flat surfaces. Characteristics of cells, incorporating morphology and cell responses, including expression of adhesion-associated molecules and cell proliferation, were studied. In this project, we fabricated two different topographies for the cells to grow on: a negative imprint that creates cell-shaped hollows and a positive imprint that recreates the raised surface topography of a cell layer. We used two different substrate materials, pMA and pST. We observed that cells on imprinted substrates of both polymers, compared to cells on flat surfaces, exhibited higher expression of β1-integrin, focal adhesion kinase, and cytokeratin-18. Compared to cells on flat surfaces, cells were larger on imprinted pMA and more in number, whereas on pST-imprinted surfaces, cells were smaller and fewer than those on a flat pST surface. This method, which provided substrates in vitro with cell-like features, enabled the study of effects of topographies that are similar to those experienced by cells in vivo. The observations establish that such a physical environment has an effect on cancer cell behavior independent of the characteristics of the substrate. The results support the concept that the physical topography of a cell’s environment may modulate crucial oncological signaling pathways; this suggests the possibility of cancer therapies that target pathways associated with the response to mechanical stimuli.
Introduction
The involvement of physical forces across a range of tissues has been recognized in physiology for some time. For example, mechanical stimulation can influence fracture healing and bone repair, although the mechanisms are still uncertain,Citation1,Citation2 and forces associated with tonic hydrostatic distension and cyclic mechanical deformation are necessary for normal fetal lung development.Citation3 In addition, several cancer-related studies under reduced gravity or aboard a space station have observed a distinct cell behavior compared to that of cells in normal gravity.Citation4 There were, eg, differences in gene expression, cell signaling, and microtubule reorganization of Jurkat human leukemia cells and CaSki cervical carcinoma cells.Citation5,Citation6 With particular relevance to the understanding of cancer proliferation, it has been noted that mechanical forces also exert control during the cell cycle.Citation7
More recent evidence suggests that a deficiency in cancer treatments is the absence of attention to the physical environment of cells.Citation8 The cells attach in vivo to their neighbors and are incorporated into an environment of three dimensions influenced by the extracellular matrix (ECM). There have been studies observing ECM remodeling in wound healing,Citation9 interactions of breast cancer cells with ECM,Citation10 and ECM mediation of the activity of nicotine during lung cancer development.Citation11 However, those discussions include limited acknowledgment of the possible contributions of mechanical forces on the full process. It is becoming an increasingly attractive hypothesis that a physical and mechanical network involving cells and the physical microenvironment operates to regulate cell behavior in parallel to the well-known biochemical processes. In other words, the structure of the neighborhood, as distinct from its composition, can affect cell functioning.Citation12,Citation13 It is already known that tumors are often stiffer than healthy tissues,Citation14 thereby providing a different mechanical environment. Therefore, consideration of this aspectCitation15 is crucial in defining tumor development.
In this study, we explored the biological impact of physical topography on endometrial cancer cells. Previously, we developed a bioimprinting methodology using soft lithography to replicate biological cells on hard polymer.Citation16–Citation18 This technique can produce two different surfaces for the cells to grow on: a negative imprint that creates cell-shaped hollows, or a positive imprint that recreates the raised surface topography of a cell layer. We used the technique (Bioimprint) to form negative-imprinted polymethacrylate (pMA) substrates for cell culture and both negative and positive polystyrene (pST) imprints. The behaviors of the cells cultured on these surfaces were compared to those on nonimprinted, flat surfaces of the respective polymer.
It has been observed that there is dynamic communication between cells and the structured microenvironment,Citation19,Citation20 which in vivo consists of neighboring cells and ECM. Cells on patterned elastomer substrates experience forces that have dependency on the area and intensity of focal adhesions formed between cells and substrate surface.Citation21 These cell–substrate forces are modulated by geometrical alteration of the adhesion site, which acts through distribution of cell stress fibers.Citation19,Citation22 Therefore, in this study, the substrates imprinted with cell-like features will provoke mechanical forces on the cells according to the nature of the adhesion site formation. Thus, we study cancer cells when they are cultured in a physical environment that, importantly, has features similar to themselves, and we compare cell growth and aspects of metabolism with those of cells grown on a conventional flat surface. Here, we report the results of our studies of cells cultured on these surfaces in terms of, first, differences in cell preferential adhesion on either imprinted or flat surface. Next, we assess the expression of β1-integrin because it is a major mediator of the interaction between a cell and its microenvironment;Citation23,Citation24 then, we report the downstream expression of focal adhesion kinase (FAK), an effector enzyme in the complex that is linked to integrin receptor; and next, we describe the characteristics of the cytoskeleton as reflected in the expression and structure of actin and cytokeratin.
Materials and methods
Cell maintenance
Cells of human Ishikawa endometrial cancer cell line (a gift from Dr Mishato Nishida, Ibraki-ken, Japan) and C2C12 murine myoblast cells (a gift from collaborative partners at Plant and Food Research, Hamilton, New Zealand) were cultured at 37°C in 5%/95% CO2/air in Dulbecco’s Modified Eagle’s Medium (DMEM)-F12 supplemented with 10% (v/v) fetal bovine serum (FBS), penicillin (100 units/mL), streptomycin (100 μg/mL), Fungizone™ (1 μg/mL), and Gluta-MAX™ (2 mM) (Gibco Thermo Fisher Scientific, Waltham, MA, USA). All experiments were conducted according to the regulations and guidelines that pertain to biological studies in the University of Otago.
Antibodies
Antibodies for Western blotting and immunofluorescence staining, β1-integrin, cytokeratin-18, actin, FAK and phosphorylated FAK (pFAK), and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) were purchased from Santa Cruz Biotechnology Inc. (Dallas, TX, USA). Texas Red®-X Phalloidin was purchased from Invitrogen (Thermo Fisher Scientific, Waltham, MA, USA).
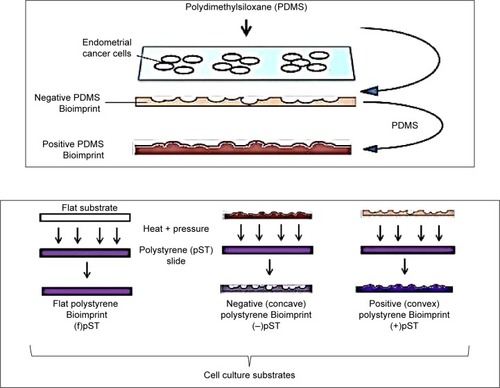
The Bioimprints
Two polymers were used in this study: pMA and pST. To make Bioimprints, a glass microscope slide was attached to a polydimethylsiloxane (PDMS) sheet with three round wells cut in it (diameter: 15 mm) using a cork borer. Cell suspensions were added to the wells and cultured and fixed with IC Fixation Buffer (Invitrogen).
Flat and bioimprinted pMA culture disks
pMA polymer was prepared from a 6:3:1 mixture of ethylene glycol dimethacrylate (EGDMA) (Sigma Aldrich, St Louis, MO, USA): methacrylic acid (Acros Organics, Thermo Fisher Scientific): Irgacure® 2022 (Ciba Specialty Chemicals, Basel, Switzerland). Next, the EGDMA mixture was cured rapidly (240 seconds) under 40% ultraviolet (UV) light (Omnicure S2000; EXFO, Toronto, ON Canada). Next, the EGDMA mixture was pipetted over the fixed cells, which formed an imprinted disk; the surface of the pMA disk captured the impression of the cells. On the other hand, flat pMA was made by curing the same mixture on cell-free glass slides.
Disks of pMA Bioimprint and flat disks were cleaned by sonication in 10% sodium dodecyl sulfate (SDS), followed by shaking with trypsin (0.05%) for at least 40 minutes. The disks were then soaked in double-distilled water overnight, followed by sterilization in absolute ethanol and washing with DMEM-F12 without FBS. Prior to cell seeding, disks were fitted into 24-well plates and cells were grown on their surfaces.
pST Bioimprint
For the pST Bioimprint, the imprint was of two forms. One was “negative” and the features comprised hollows below the horizontal plane. The second was “positive” and the features were of humps above the horizontal plane ().
PDMS master molds
pST Bioimprint was made from PDMS master molds. First, cells were cultured and fixed on a glass slide, followed by washing with phosphate-buffered saline (PBS) and distilled water. Then, PDMS was poured on top and rapidly cured at 34°C for 2 minutes, followed by 2 hours’ curing at 80°C. This PDMS with negative cell impressions was peeled off the glass slide, cleaned with 10% SDS and trypsin, and was ready for use as the mold for positive pST Bioimprint.
Positive pST Bioimprint
pST Bioimprint was made from PDMS master molds. To make the pST Bioimprint, a pST slide (Ted Pella Inc, CA, USA) was pressed against a PDMS mold and the imprint was transferred onto pST slides at 180°C for 15 minutes.
Negative pST Bioimprint
To make positive PDMS molds, a PDMS negative Bioimprint prepared as described in the section on “PDMS master molds” was treated in 0.1% hydroxypropyl methycellulose (HPMC) (Sigma Aldrich, St Louis, MO, USA) for 10 minutes on a shaker, followed by washing thoroughly with distilled water. This treatment prevents sticking between two PDMS layers. A positive PDMS Bioimprint was then made by pouring another layer of PDMS onto this HPMC-treated PDMS and cured at 80°C. This is the positive PDMS mold for the negative pST Bioimprint. To make pST Bioimprint, a pST slide was pressed against the PDMS mold and the imprint was transferred onto pST slides at 180°C.
Preferential attachment of cells
To study the cell attachment and localization on substrates, cells were seeded on low-density Bioimprint so that cells had a chance to deposit either on Bioimprint or flat surface. Then, the “before” pictures of pST Bioimprint and “after” images of cells on the platform were taken; the areas of Bioimprint surface and flat surface were measured. Next, the coverage by cells was determined and the relative percentage coverage was calculated.
RGDS and RGES treatment
Ishikawa endometrial cancer cells were premixed with 100 μM RGDS (Arg-Gly-Asp-Ser) or RGES (Arg-Gly-Glu-Ser) before seeding on flat and Bioimprint pST substrates. After 3 hours’ incubation time, adherent and nonadherent cells were counted and the percentage of adherent cells was calculated.
Western blotting
Cells lysate was prepared using RIPA buffer containing protease inhibitor cocktail tablets (Complete Mini, Hoffman-La Roche Ltd., Basel, Switzerland) and was analyzed for total protein concentration using BCA Protein Assay Kits (BioRad, Hercules, CA, USA). Then, 10 μg of protein was run through SDS polyacrylamide gel electrophoresis under nonreducing conditions and was then transferred onto a polyvinylidene difluoride membrane, blocked, and incubated with primary antibodies overnight at 4°C. On the following day, the membrane was washed with 0.1% Tween-20 Tris-buffered saline before and after incubation with secondary antibody. The membrane was then analyzed using ECL Plus (GE Health-care UK Ltd, Little Chalfont, UK) with Alliance 4.7 imaging system (Unitec, Cambridge, UK). Densitometric analysis of bands of each sample was carried out using ImageJ.Citation25
Some commonly used reference proteins, eg, actin, cytokeratin, and GAPDH, were sensitive to the physical environment on which the cells were grown. Therefore, Western blot data for cells on pMA substrates are presented using the same sample loading, ensured by total protein measurement, and the Western blotting was repeated at least six times.
Immunofluorescence staining
Cells were grown on respective substrates until 70%–80% confluent and were fixed as described in the section “The Bioimprints”. For cytokeratin-18 staining, cells were permeabilized with cold methanol for 10 minutes at −20°C and, for actin, β1-integrin, and pFAK staining, cells were permeabilized with Triton X for 5 minutes at room temperature. After permeabilization, cells were washed four times with cold PBS, each of 5 minutes’ duration. Then, samples were blocked with 5% w/v bovine serum albumin in PBS for 60 minutes at room temperature, followed by washing with PBS (pH 7.4). Then, cells were incubated with mouse anti-cytokeratin-18 (Santa Cruz Biotechnology Inc.) for localization of cytokeratin-18 or Texas Red-X Phalloidin (Life Technologies Thermo Fisher Scientific) for actin filament staining or with both anti-β1 integrin and anti-pFAK (Santa Cruz Biotechnology Inc.) at 4°C overnight. After primary antibody incubation, cells were washed with cold PBS prior to secondary antibody incubation. After washing with cold 0.1% v/v PBS–Tween 20, samples with antifading solution were visualized with Zeiss Axio Imager epifluorescence microscope (Carl Zeiss Meditec AG, Jena, Germany).
Cell morphology and size
Cells were cultured on substrates and were immunofluorescently stained for cytokeratin-18 and were examined under an epifluorescence microscope. The shapes of cells were observed using optical microscopy, and the sizes were measured using ImageJ.
Cell number
The number of cells was estimated using a standard curve that related the level of Coomassie Blue staining to cell number. Cells were grown on different substrates, and then cell culture media were removed and cells were washed with PBS, followed by cell fixing with IC Fixation Buffer for 15 minutes. After washing off the excess fixation buffer, cells were stained with Coomassie Blue for 5 minutes. The stained cells were washed with distilled water three times.
To make a standard curve, a cell suspension was prepared and after cell counting, serial dilutions were made and stained with Coomassie Blue for 5 minutes. The stained samples were washed with distilled water three times and air-dried. The stains of this standard curve, together with the stained experimental samples, were dissolved in 10% SDS in 0.1 N HCl for 10 minutes. Absorbance at 595 nm was obtained and the cell number was calculated from the standard curve.
Results
Substrate resolution
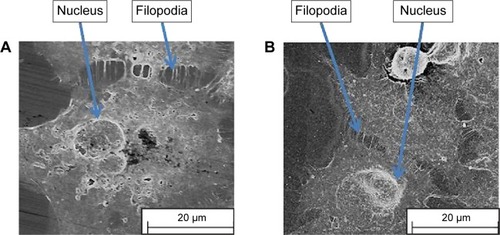
Our previous studies have shown that imprints obtained using UV-curable pMA can replicate cell features of Ishikawa cells derived from endometrial cancer down to the nanoscale.Citation16–Citation18 We chose this polymer as a cell substrate because it satisfied the criteria for a successful bioimprint process, namely, nanoscale resolution, fast curing time, and low temperature process, in addition to biocompatibility. Here, we also produced imprints with a polymer that is commonly used in studies of cell behavior, namely, pST. The results demonstrated that pST imprints were also capable of very high resolution and were observed to capture a cell’s fine features such as cell filopodium (). The fidelity was observed in negative imprints and retained during the later steps to produce a positive imprint.
Preferential substrate for growth
Previously, we observed that there was preferential cell growth on imprinted areas on negative pMA relative to flat areas.Citation26 Here, we undertook quantitative cell population analysis. Using imaging and counting described in the “Materials and methods” section, the fraction of cells that could potentially have adhered to the flat area or to the imprinted area on each slide was calculated. It can be seen that the Ishikawa cells selectively grew on a pST substrate negatively imprinted with Ishikawa cells (). No selective presence was observed on the substrate that was positively imprinted ().
Figure 3 Preferential growth on polystyrene Bioimprint.
Notes: Ishikawa cells were cultured on polystyrene imprints of (A) Ishikawa cells and (B) C2C12 cells. The fractions of cells that could potentially have adhered to the flat area or the imprinted area on each slide were calculated. Data are presented as mean ± SEM of measurements from 25 grid areas on each of 4 incubations per substrate topography (**P<0.01).
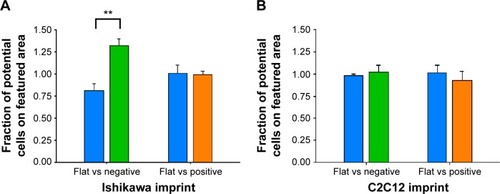
To investigate the possibility that the preference was a result of a nonspecific effect of surface patterning, a different cell line, namely C2C12 mouse myoblast cells, was used to make Bioimprints. The same procedure was performed using an imprint of C2C12 cells as substrate for the endometrial cancer cells (). In contrast to Ishikawa cells cultured on Ishikawa cell imprints, Ishikawa cells cultured on a substrate negatively imprinted with C2C12 cells showed no selective presence. Further, there was again no selection on a substrate positively imprinted with C2C12 cells.
Cell morphology
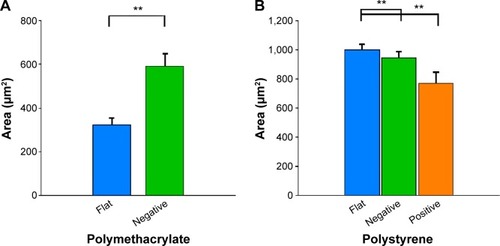
We investigated the area of cells as an indication of cell response to the imprinted surface (). Cells on (−)pMA imprint had bigger areas (590.1±56.7 μm2) than cells on flat surface (f)pMA (323.2±31.0 μm2) (). In contrast, cells on both (−)pST (947.5±39.6 μm2) and (+)pST (772.9±74.2 μm2) imprints were smaller than on (f) pST (1,002.0±37.5 μm2) (). The cells on pMA substrates had smaller surface area than cells on pST surfaces. This was due to the hydrophobicity of pMA substrates, which limited cell spreading. In this context, the contact angle (CA) of pMA is 65°–80°Citation27 and that of pST is 58.89°.Citation26
Figure 4 Effects of Bioimprint substrate on Ishikawa cell size.
Notes: Effects on (A) (f)pMA and (−)pMA imprint; and (B) (f)pST, (−)pST, and (+)pST imprints. Cells were grown on (A) flat and negative-imprinted pMA substrates; and (B) flat, negative-, and positive-imprinted pST substrates; and were stained for cytokeratin-18; images of cells were taken using epifluorescence microscopy. ImageJ software was used to measure each parameter from the images taken. Data are presented as mean ± SEM of measurements from at least 25 cells from five different images (**P<0.01; paired t-test).
Abbreviations: (−), negative imprint; (+), positive imprint; (f), flat substrate; pMA, polymethacrylate; pST, polystyrene; SEM, standard error of the mean.
Beta-1 integrin expression
The influence of flat and imprinted pMA and pST substrates on β1-integrin expression, an important mediator of signals between the external environment of a cell and its cytoplasmic interior, was examined.
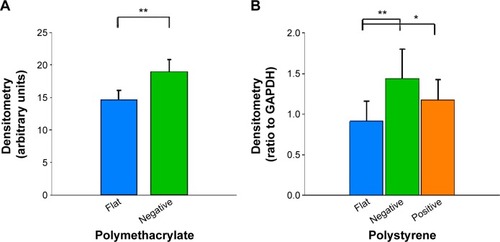
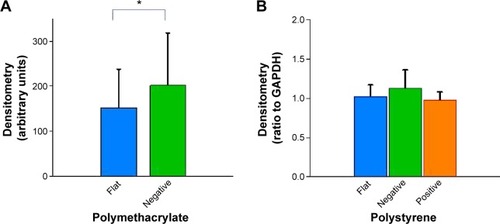
Compared to cells on flat pMA substrate, cells on (−)pMA had higher expression of β1-integrin (). Cells on (−) and (+)pST also had higher expression of β1-integrin than those on (f)pST ().
Figure 5 Effects of Bioimprint on expression of β1-integrin.
Notes: Densitometry results of β1-integrin bands formed after Western blotting of lysates of cells cultured on (A) flat and negative-imprinted pMA substrates and (B) flat, negative-, and positive-imprinted pST substrates. Data are presented as mean ± SEM from at least six tests (*P<0.05; **P<0.01, paired t-test).
Abbreviations: GAPDH, glyceraldehyde 3-phosphate dehydrogenase; SEM, standard error of the mean; pMA, polymethacrylate.
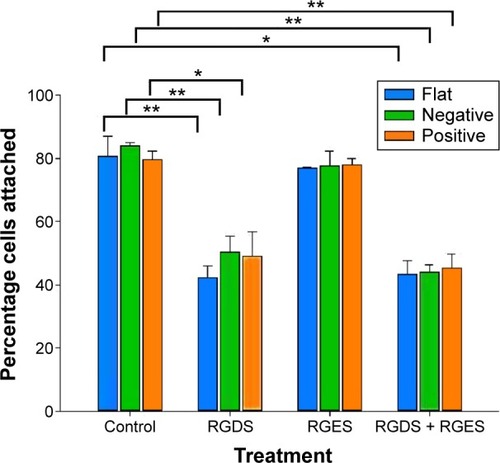
Integrin is known to bind to RGD (Arg-Gly-Asp) moieties in the surrounding ECM. Demonstration that β1-integrin expression was sensitive to the topography of the substrate suggested that there was β1-integrin involvement in the cell’s responses to the physical environment. Therefore, to confirm the role of β1-integrin in cell adhesion on pST, we investigated the effect of RGDS, a tetrapeptide that inhibits binding of β1-integrin to relevant (fibronectin) substrates. The initial binding of cells to pST substrate was indeed blocked by RGDS (). Control peptide (RGES) had no effect on adhesion, which is consistent with the early involvement of β1-integrin in the differential interaction of cells with distinct topographies.
Figure 6 Effects of blocking β1-integrin on initial cell adhesion.
Notes: Cells were grown on respective substrates with either no treatment or with treatments consisting of RGDS alone, RGES alone, or RGDS + RGES for 3 hours and were stained with Coomassie Blue. The number of adherent cells was quantified from a Coomassie Blue staining standard curve. Data are presented as mean ± SEM of measurements from at least three samples, each in triplicate (*P<0.05; **P<0.01; paired t-test).
Abbreviations: RGDS, Arg-Gly-Asp-Ser tetrapeptide; RGES, Arg-Gly-Glu-Ser tetrapeptide; SEM, standard error of the mean.
Focal adhesions
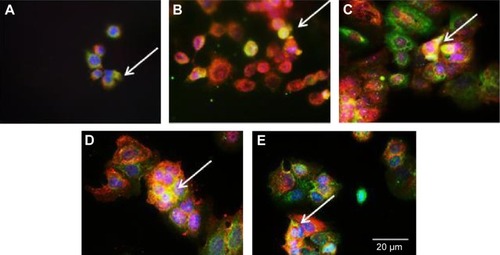
Next, we studied FAK, which is an important component of the focal adhesion complex that forms downstream of β1-integrin as part of the response to biomechanical forces.Citation28 Our immunofluorescence staining results demonstrated that pFAK was expressed with β1-integrin (), indicating functional colocalization.
Figure 7 Colocalization of β1-integrin and pFAK.
Notes: Immunofluorescence staining of β1-integrin of cells cultured on (A) (f)pMAand (B) (−)pMA imprint; and on (C) (f)pST, (D) (−)pST, and (E) (+)pST imprints. β1-integrin was localized using mouse anti-β1-integrin, and pFAK was localized with rabbit anti-pFAK, which were then imaged with AlexaFlour 488 (green) and Atto 594 (red), respectively. Colocalization of β1-integrin and pFAK is indicated by the arrows, where merging of green and red tags to yellow coloration occurs. Cell nuclei were stained with Hoechst 33342 (blue) (original magnifications: ×20).
Abbreviations: (−), negative imprint; (+), positive imprint; (f), flat substrate; pFAK, phosphorylated focal adhesion kinase; pMA, polymethacrylate; pST, polystyrene.
Western blotting revealed that cells cultured on (−) pMA imprint exhibited higher mean FAK expression than cells on the flat polymer (, left panel). Moreover, activated pFAK was expressed at similar levels on (−)pMA as on f(pMA) (, left panel). Additionally, on pST substrates, higher expression of FAK was observed in cells cultured on (−)pST and (+)pST imprints relative to those cultured on (f)pST (, right panel).
Figure 8 Effects of Bioimprint on FAK and pFAK expression.
Notes: Densitometry results of (A) FAK and (B) pFAK bands formed after Western blotting of lysates of cells cultured on (left panel) flat and negative-imprinted pMA substrates and (right panel) flat, negative-, and positive-imprinted pST substrates. Data are presented as mean ± SEM from at least six tests (*P<0.05; **P<0.01; paired t-test).
Abbreviations: FAK, focal adhesion kinase; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; pFAK, phosphorylated FAK; pMA, polymethacrylate; pST, polystyrene; SEM, standard error of the mean.
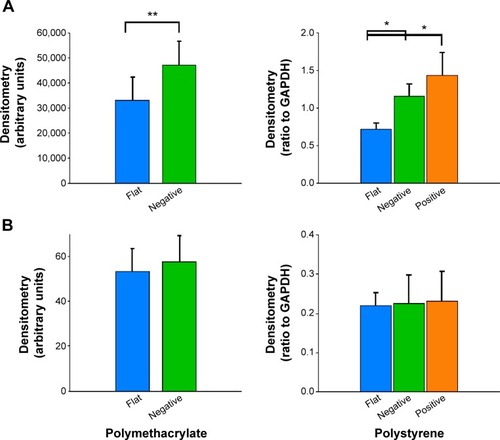
We also found that the ratio of pFAK to total FAK was significantly lower (P<0.01) in cells cultured on bioimprinted pMA than in cells grown on flat substrate. The ratio was also lower in cells grown on (+)pST than on (f)pST (P<0.05).
Actin
Actin is recruited to focal adhesions in response to biomechanical force stimulation,Citation28 and actin has been observed in some cells to modify its structure to subsequently transmit information for relevant gene activation. In cells cultured on pMA Bioimprint (), the actin filaments were more elongated than actin filaments in cells cultured on flat surfaces (cell circularity, determined by ImageJ, on (f)pMA: 0.72; cell circularity on (−)pMA: 0.64; P<0.05). These observations were distinct from those observed in cells cultured on pST. There was no observable difference in actin arrangement between cells cultured on (−)pST and (+)pST surfaces relative to those on (f)pST. Levels of actin expression were similar on the three pST substrates ().
Figure 9 Actin distribution.
Notes: Immunofluorescence staining of actin in cells cultured on (A) (f)pMA, (B) (−)pMA imprint; and on (C) (f)pST, (D) (−)pST, and (E) (+)pST imprints. Cells grown on different surfaces were fixed with 4% paraformaldehyde and permeabilized using Triton X; then actin was probed using Texas Red-X Phalloidin red) (original magnifications: ×20). In cells cultured on pMA Bioimprint the actin filaments were more elongated than in cells cultured on flat pMA. There was no observable difference in the arrangement of actin in cells cultured on (−)pST or (+)pST surfaces relative to actin in cells on (f)pST.
Abbreviations: (−), negative imprint; (+), positive imprint; (f), flat substrate; pMA, polymethacrylate; pST, polystyrene.
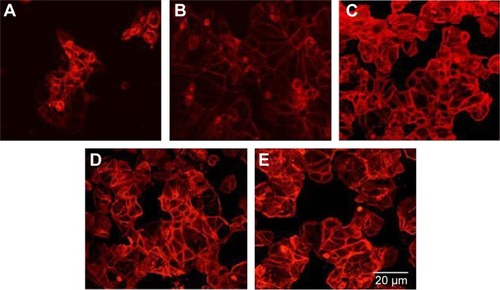
Figure 10 Effects of Bioimprint on expression of actin.
Notes: Densitometry results of actin bands formed after Western blotting of lysates of cells cultured on (A) flat and negative-imprinted pMA substrates and (B) flat, negative-, and positive-imprinted pST substrates. Data are presented as mean ± SEM from at least five tests (*P<0.05; paired t-test).
Abbreviations: GAPDH, glyceraldehyde 3-phosphate dehydrogenase; pMA, polymethacrylate; pST, polystyrene; SEM, standard error of the mean.
Cytokeratin-18
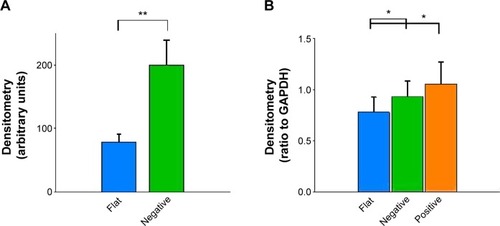
Cytokeratin-18 is a cytoskeleton intermediate filament that functions to maintain the structure of cells and plays a role in adhesion and spreading.Citation29,Citation30 After immunofluorescence staining, cytokeratin-18 was observed to exhibit more elongated distribution in cells grown on (−)pMA imprint compared to the distribution in cells grown on (f)pMA, which was different from the responses to cells on pST, wherein there was no clear difference observed between cytokeratin-18 arrangements (). Cytokeratin-18 has higher expression in cells cultured on (−)pMA imprint than in cells on (f)pMA (). Additionally, the expression on both (−)pST and (+)pST imprints was higher than on (f)pST substrates.
Figure 11 Effects of Bioimprint on expression of cytokeratin-18.
Notes: Immunofluorescence staining of cytokeratin-18 in cells cultured on (A) (f)pMA and (B) (−)pMA imprint; and on (C) (f)pST, (D) (−)pST, and (E) (+)pST imprints. Cells grown on different surfaces were fixed with 4% paraformaldehyde and permeabilized using cold methanol; then cytokeratin-18 was probed using mouse anti-cytokeratin-18 that was labeled with AlexaFluor 488 (green). Cell nuclei were co-stained with Hoechst 33342 (blue) (original magnifications: ×20).
Abbreviations: (−), negative imprint; (+), positive imprint; (f), flat substrate; pMA, polymethacrylate; pST, polystyrene.
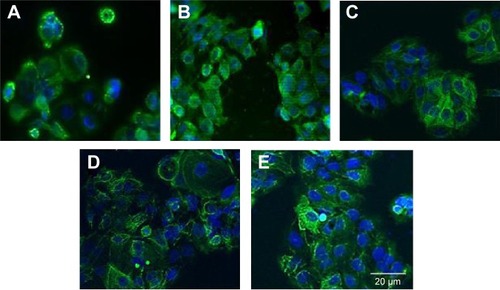
Figure 12 Effects of Bioimprint on cytokeratin-18 expression.
Notes: Densitometry results of cytokeratin-18 bands formed after Western blotting of lysates of cells cultured on (A) flat and negative-imprinted pMA substrates and (B) flat, negative-, and positive-imprinted pST substrates. Data are presented as mean ± SEM from at least 12 tests (*P<0.05; **P<0.01; paired t-test).
Abbreviations: GAPDH, glyceraldehyde 3-phosphate dehydrogenase; pMA, polymethacrylate; pST, polystyrene; SEM, standard error of the mean.
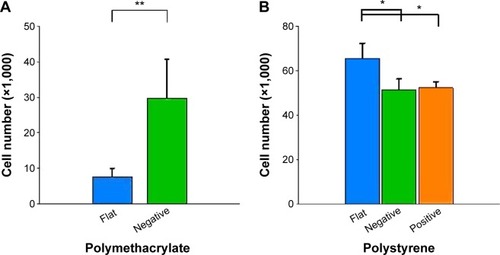
Cell number
The parameters examined, including cell size,Citation31–Citation33 have been observed to be associated with cell growth and we looked at cell number after culture (). Cultures on (−) pMA imprint had a larger population of cells than those on (f)pMA. On the other hand, there were fewer cells on (+)pST than on (f)pST and (−)pST.
Figure 13 Effects of Bioimprint on cell number after 60-hour culture.
Notes: Cells were grown on (A) flat and negative-imprinted pMA substrates and (B) flat, negative-, and positive-imprinted pST substrates for 60 hours and were stained with Coomassie Blue. The number of cells was quantified from the standard curve of cell staining with Coomassie Blue. Data are presented as mean ± SEM of measurements from at least three samples, each as triplicate cultures (**P<0.01; *P<0.05; paired t-test).
Abbreviations: pMA, polymethacrylate; pST, polystyrene; SEM, standard error of the mean.
Discussion
We report the influence of cell culture platform topography on cancer cells’ biological characteristics. A method was developed for providing a substrate in vitro with cell-like features, thereby enabling the study of the effects of a topographic surface that is similar to that experienced by cells in vivo. The observations we report in this project establish that the physical environment has an effect on part of cancer cell behavior independent of the inherent characteristics of the substrate.
We investigated the effect of incubation of cells on two different polymers, pMA and pST. Both pST and pMA were biocompatible and cells were able to grow and proliferate, although there was a detectable difference in percentage initial adherence on pMA relative to that on conventional pST culture surfaces (approximately 89% on (f)pMA compared to approximately 95% on (f)pST; P,0.01). The differences in cell morphology and growth rates that occurred between polymers were likely to be partly associated with the biophysical properties of cell membrane interactions with the substrate surface as reflected by, eg, the water CA (for pST, the angle is 59°, whereas for pMA, the CA is 73°). The observation that cells cultured on pMA surfaces form smaller populations is consistent with the sparing use of pMA in in vitro culture studies and the correspondingly wider use of pST, for which cell growth and maintenance are well characterized.
The behavior of cells on pMA was modified if the MA polymer was imprinted with shapes resembling the cells themselves.Citation16,Citation17 Cells on the imprint expressed higher levels of β1-integrin, FAK, and cytokeratin-18 compared to cells on (f)pMA. In addition, cultures on pMA Bioimprint had a bigger area (600 μm2, compared to 300 μm2) and had higher cell numbers. These results using pMA indicated that behavior and characteristics of cancer cells may be altered by the topography of the microenvironment alone. The observations point to the substantial potential for these substrates to assist in studies using a cell-like topography aimed at, for instance, cell regulation of signaling molecules and molecular target drug specificity.Citation15,Citation34 Because expression of signaling molecules is modified by the physical nature of a cell’s environment, it is important that we reach a greater understanding of the effects of these interactions on the development of cancers.Citation35–Citation37 In addition, we also noted, by using a similar topography with a different polymer, pST, that the topographical environment combines with polymer characteristics to have an important role in influencing the cell behavior through the physical characteristics. Further, cells cultured on imprinted pST had smaller area and slower rate of proliferation compared to cells on (f)pST – the converse to the relationship of cells on pMA. The results with pMA compared to pST confirm that topography itself is not the only factor in the microenvironment that affects behavior and indicate that polymer characteristics (ie, non-ECM-related factors) are also influential. Where the surface is not favorable for growth (ie, the pMA condition in which rounded cells suggest low adherence), cells cultured on an imprinted surface responded by inducing enhanced activity of signaling pathways and increased proliferation. On the other hand, pST platforms with imprints had fewer cells than those that were flat, which indicated that certain modifications of substrate characteristics may alter cell signaling in a manner that suppresses tumor development.
In a number of studies, a link between integrins and cell signaling pathways has been observed.Citation28 Here, we hypothesized that the physical environment would be detected by the integrin receptor. Indeed, cells on (−)pMA imprint and both (−) and (+)pST imprint exhibited a higher expression of integrin than cells on flat surfaces. The interaction with RGD (Arg-Gly-Asp) moiety during adhesion was demonstrated. In this study, the effect of blocking on phosphorylation of integrin was not evaluated. Secreted ECM components from nonstromal (Ishikawa) cellsCitation38–Citation42 or soluble factorsCitation3,Citation43 may be partly responsible for the interface effect. Physical signaling is complex, and a number of relevant signaling molecules and transcription factors have been identified,Citation43,Citation44 but their roles are not fully defined.Citation45 Here, eg, we observed that a reduction in the ratio of pFAK/FAK accompanied an increase in β1-integrin expression, suggesting activation by the cell topography interaction of a pathway that inhibited phosphorylation.
The connections between topography and cytoskeleton were also investigated and quantitative polymer-dependent differences in actin and cytokeratin-18 were observed. Cells on all imprints showed a higher expression of cytokeratin-18, but not actin, than the cells on flat surfaces. This may indicate that the signaling in the context of these studies was more prominently mediated by cytokeratin-18 than actin. Alternatively or additionally, changes in actin might have been occurring in parallel but were not detected.
An effect of substrate topography on cell behavior has been previously reportedCitation46,Citation47 but with only sparse attention to cancer cells.Citation48,Citation49 Our study is a major advance in that features similar to cells themselves are used in the substrates. Cells on both (−) and (+)pST indeed showed lower growth rate than on flat pST, in parallel with occurrences in cells cultured in 3D models.Citation50,Citation51 However, biophysical properties favored preferential adherence to (−)pST but not to (+)pST substrate, revealing that the details of topography were also important. This study is the first to develop a method in which the imprinted substrate has a cell’s physical shapes but without differential cell secretion and surface chemistry complexities that are present in cell cluster models.
The results have implications for enhancing conventional cell culture practice because a positively imprinted pST provides a more tissue-like topography than does flat pST. It is clear that flat surface is not physiological but has been used widely to investigate cancer cell behavior and is now suggested to often induce nonrelevant behavior. There is an implication in our observations that there is an association between physiological-like topography and control over cancer development. The results together therefore indicate that cancer cells respond to the physical microenvironment, and the response is a result of an interaction between topography and substrate surface characteristics. The potential may be significant both for designing and for more relevant testing of drugs in development.
We investigated whether topography has a role in regulating the behavior of Ishikawa endometrial cancer cells. We demonstrated a link between structure of the microenvironment and activities of the cancer cells. The results are consistent with the concept that the mechanical environment is a vital influence on gene expression.Citation45,Citation52,Citation53 Our observations indicate that increased understanding of the interactions may signal potential for treatment aimed directly at altering the physical environment or, alternatively, at components of the associated signaling pathways.
Acknowledgments
The authors are very grateful for funding support from the Marsden Fund, the MacDiarmid Institute for Advanced Materials and Nanotechnology, and the New Zealand Cancer Society.
Disclosure
The authors report no conflicts of interest in this work.
References
- ChaoEYInoueNBiophysical stimulation of bone fracture repair, regeneration and remodellingEur Cell Mater20036728414722904
- WaldorffEIChristensonKBCooneyLAGoldsteinSAMicrodamage repair and remodeling requires mechanical loadingJ Bone Miner Res20102573474519821772
- Sanchez-EstebanJMechanical forces in fetal lung development: opportunities for translational researchFront Pediatr201315124400295
- BeckerJLSouzaGRUsing space-based investigations to inform cancer research on EarthNat Rev Cancer20131331532723584334
- GuoFLiYLiuYIdentification of genes associated with tumor development in CaSki cells in the cosmic spaceMol Biol Rep2012396923693122302396
- LewisMLCubanoLAZhaoBcDNA microarray reveals altered cytoskeletal gene expression in space-flown leukemic T lymphocytes (Jurkat)FASEB J2001151783178511481229
- ClarkAGPaluchEMechanics and regulation of cell shape during the cell cycleResults Probl Cell Differ201153317321630140
- LeeJMMhawech-FaucegliaPLeeNA three-dimensional microenvironment alters protein expression and chemosensitivity of epithelial ovarian cancer cells in vitroLab Invest20139352854223459371
- KlassBRGrobbelaarAORolfeKJTransforming growth factor beta1 signalling, wound healing and repair: a multifunctional cytokine with clinical implications for wound repair, a delicate balancePostgrad Med J20098591419240282
- MoustakasAHeldinPTGFbeta and matrix-regulated epithelial to mesenchymal transitionBiochim Biophys Acta2014184082621263424561266
- ZhengYRitzenthalerJDRomanJHanSNicotine stimulates human lung cancer cell growth by inducing fibronectin expressionAm J Respir Cell Mol Biol20073768169017600315
- MaedaTSakabeTSunagaAConversion of mechanical force into TGF-beta-mediated biochemical signalsCurr Biol20112193394121600772
- PotierENoaillyJItoKDirecting bone marrow-derived stromal cell function with mechanicsJ Biomech20104380781719962149
- Iglesias-GarciaJDominguez-MunozJEEndoscopic ultrasound image enhancement elastographyGastrointest Endosc Clin N Am20122233334822632955
- VinciMGowanSBoxallFAdvances in establishment and analysis of three-dimensional tumor spheroid-based functional assays for target validation and drug evaluationBMC Biol2012102922439642
- MuysJJAlkaisiMMMelvilleDOCellular transfer and AFM imaging of cancer cells using bioimprintJ Nanobiotechnology20064116426461
- SamsuriFAlkaisiMMMitchellJEvansJJReplication of cancer cells using soft lithography bioimprint techniqueMicroelectron Eng201087699703
- MuysJJAlkaisiMMEvansJJBioimprint: nanoscale analysis by replication of cellular topography using soft lithographyJ Biomed Nanotechnol2006215
- AlbertPJSchwarzUSDynamics of cell shape and forces on micropatterned substrates predicted by a cellular Potts modelBiophys J20141062340235224896113
- GilchristCLRuchDSLittleDGuilakFMicro-scale and meso-scale architectural cues cooperate and compete to direct aligned tissue formationBiomaterials201435100151002425263687
- BalabanNQSchwarzUSRivelineDForce and focal adhesion assembly: a close relationship studied using elastic micropatterned substratesNat Cell Biol2001346647211331874
- TheryMPepinADressaireEChenYBornensMCell distribution of stress fibres in response to the geometry of the adhesive environmentCell Motil Cytoskeleton20066334135516550544
- BouvardDPouwelsJDe FranceschiNIvaskaJIntegrin inactivators: balancing cellular functions in vitro and in vivoNat Rev Mol Cell Biol20131443044223719537
- GehlerSPonikSMRichingKMKeelyPJBi-directional signaling: extracellular matrix and integrin regulation of breast tumor progressionCrit Rev Eukaryot Gene Expr20132313915723582036
- SchneiderCARasbandWSEliceiriKWNIH image to imageJ: 25 years of image analysisNat Methods2012967167522930834
- MurrayLMNockVEvansJJAlkaisiMMBioimprinted polymer platforms for cell culture using soft lithographyJ Nanobiotechnology201412289
- SaltzmanWMKyriakidesTRCell interactions with polymersLanzaRLangerRVacantiJPrinciples of Tissue EngineeringSan DiegoAcademic Press2007279296
- Roca-CusachsPIskratschTSheetzMPFinding the weakest link: exploring integrin-mediated mechanical molecular pathwaysJ Cell Sci20121253025303822797926
- BordeleauFGalarneauLGilbertSLorangerAMarceauNKeratin 8/18 modulation of protein kinase C-mediated integrin-dependent adhesion and migration of liver epithelial cellsMol Biol Cell2010211698171320357007
- BordeleauFMyrand LapierreMEShengYMarceauNKeratin 8/18 regulation of cell stiffness-extracellular matrix interplay through modulation of Rho-mediated actin cytoskeleton dynamicsPLoS One20127e3878022685604
- TzurAKafriRLeBleuVSLahavGKirschnerMWCell growth and size homeostasis in proliferating animal cellsScience200932516717119589995
- KafriRLevyJGinzbergMBOhSLahavGKirschnerMWDynamics extracted from fixed cells reveal feedback linking cell growth to cell cycleNature201349448048323446419
- ChenCSMrksichMHuangSWhitesidesGMIngberDEGeometric control of cell life and deathScience1997276142514289162012
- GirardYKWangCRaviSA 3D fibrous scaffold inducing tumoroids: a platform for anticancer drug developmentPLoS One20138e7534524146752
- Agudelo-GarciaPADe JesusJKWilliamsSPGlioma cell migration on three-dimensional nanofiber scaffolds is regulated by substrate topography and abolished by inhibition of STAT3 signalingNeoplasia20111383184021969816
- KriparamananRAswathPZhouATangLNguyenKTNanotopography: cellular responses to nanostructured materialsJ Nanosci Nanotechnol200661905191917025103
- WeigeltBLoATParkCCGrayJWBissellMJHER2 signaling pathway activation and response of breast cancer cells to HER2-targeting agents is dependent strongly on the 3D microenvironmentBreast Cancer Res Treat2010122354319701706
- NedermanTNorlingBGlimeliusBCarlssonJBrunkUDemonstration of an extracellular matrix in multicellular tumor spheroidsCancer Res198444309030976373002
- Niculescu-DuvazIPhanishMKColville-NashPDockrellMEThe TGFbeta1-induced fibronectin in human renal proximal tubular epithelial cells is p38 MAP kinase dependent and Smad independentNephron Exp Nephrol2007105e108e11617347580
- SmithBDNilesRCharacterization of collagen synthesized by normal and chemically transformed rat liver epithelial cell linesBiochemistry198019182018257378375
- CreelyJJDiMariSJHoweAMHaralsonMAEffects of transforming growth factor-beta on collagen synthesis by normal rat kidney epithelial cellsAm J Pathol199214045551731530
- QuaroniATrelstadRLBiochemical characterization of collagens synthesized by intestinal epithelial cell culturesJ Biol Chem1980255835183617410366
- JanmeyPAWellsRGAssoianRKMcCullochCAFrom tissue mechanics to transcription factorsDifferentiation20138611212023969122
- ChiquetMGelmanLLutzRMaierSFrom mechanotransduction to extracellular matrix gene expression in fibroblastsBiochim Biophys Acta2009179391192019339214
- BissellMJHinesWCWhy don’t we get more cancer? A proposed role of the microenvironment in restraining cancer progressionNat Med20111732032921383745
- BiggsMJRichardsRGDalbyMJNanotopographical modification: a regulator of cellular function through focal adhesionsNanomedicine2010661963320138244
- NikkhahMEdalatFManoucheriSKhademhosseiniAEngineering microscale topographies to control the cell-substrate interfaceBiomaterials2012335230524622521491
- ZhangLChunYWWebsterTJDecreased lung carcinoma cell density on select polymer nanometer surface features for lung replacement therapiesInt J Nanomedicine2010526927520517474
- KwonKWChoiJCSuhKYDohJMultiscale fabrication of multiple proteins and topographical structures by combining capillary force lithography and microscope projection photolithographyLangmuir2011273238324321348500
- GruberHEHanleyENJrHuman disc cells in monolayer vs 3D culture: cell shape, division and matrix formationBMC Musculoskelet Disord20001111231882
- SkrobanskaREvangelatovAStefanovaNTopouzova-HristovaTMomchilovaAPankovRCell proliferation in in vivo-like three-dimensional cell culture is regulated by sequestration of ERK1/2 to lipid raftsCell Prolif20144733634624862604
- WangCCBajikarSSJamalLAtkinsKAJanesKAA time- and matrix-dependent TGFBR3-JUND-KRT5 regulatory circuit in single breast epithelial cells and basal-like premalignanciesNat Cell Biol20141634535624658685
- MichorFWeaverVMUnderstanding tissue context influences on intratumour heterogeneityNat Cell Biol20141630130224691256