Abstract
Micro ribonucleic acids (miRNAs) are short noncoding RNAs that inhibit gene expression through the post-transcriptional repression of their target mRNAs. Increasing evidence shows that miRNAs have emerged as key players in diverse biologic processes. Aberrant miRNA expression is also closely related to various human diseases, including kidney diseases. From clinical and experimental animal studies, emerging evidence demonstrates a critical role for miRNAs in renal pathophysiology. Renal fibrosis is the hallmark of various chronic kidney diseases and transforming growth factor beta (TGF-β) is recognized as a vital mediator of renal fibrosis because it can induce production of extracellular matrix proteins resulting in dysfunction of the kidneys. The relationship between TGF-β signaling and miRNAs expression during renal diseases has been recently established. TGF-β positively or negatively regulates expression of several miRNAs, such as miR-21, miR-192, miR-200, and miR-29. Both miR-192 and miR-21 are positively regulated by TGF-β1/Smad3 signaling and play a pathological role in kidney diseases. Conversely, members of both miR-29 and miR-200 families are negatively regulated by TGF-β/Smad3 and play a protective role in renal fibrosis by inhibiting the deposition of extracellular matrix and preventing epithelial-to-mesenchymal transition, respectively. Clinically, levels of miRNAs in circulation and urine may be potential biomarkers for detecting early stages of renal diseases and targeting miRNAs also provides promising therapeutic effects in rodent models of chronic kidney disease. However, mechanisms and roles of miRNAs under disease conditions remain to be explored. Thus, understanding the function of miRNAs in the pathogenesis of kidney diseases may offer an innovative approach for both early diagnosis and treatment of renal diseases.
Introduction
Micro ribonucleic acids (miRNAs) are small, endogenous, noncoding RNAs that bind to their respective target mRNAs and recruit the RNA-induced silencing complex (RISC). The biogenesis of miRNAs includes multiple steps. Firstly, miRNAs are transcribed by RNA polymerase II or RNA polymerase III as long stem-loop primary miRNA (Pri-miR) in the nucleus. The Pri-miR is then cleaved into a double-stranded shorter miRNA precursor (Pre-miR) by RNase III enzyme Drosha and its partner DiGeorge syndrome critical region 8 (DGCR8).Citation1,Citation2 The Pre-miR is subsequently exported into the cytoplasm by the Ran-guanosine triphosphate (GTP) and Exportin-5.Citation3 In the cytoplasm, Pre-miR is further cleaved into the 20–22 base pairs (bp) double-stranded RNAs (mature form) by another RNase III enzyme Dicer and this mature miRNA-miRNA duplex is unwound and the functional strand (“guide strand”) is loaded onto the RISC.Citation4 The mature miRNA guides the RISC complex to bind to the 3′ untranslated region (3′ UTR) of a target messenger RNA (mRNA), resulting in post-transcriptional gene silencing by mRNA degradation or by translation inhibition. Thus, miRNAs can inhibit gene expression via mRNA degradation, translation inhibition, or transcriptional inhibition.
Microarray assays further demonstrate that several miRNAs, including miR-192, -194, -204, -215, and -216, are highly expressed in the kidney, compared to other organs.Citation5,Citation6 These studies also mark the starting point of miRNA research in kidney diseases. To date, over ten miRNAs have been reported in kidney diseases.Citation7–Citation10 Several comprehensive reviews about the biogenesis of miRNAs and the role of miRNAs in normal kidney have been published.Citation3,Citation4,Citation7,Citation10–Citation12 This article highlights recent novel insights into miRNA function and the implications for miRNAs in renal disease.
Role of TGF-β1 in renal pathophysiology
Renal fibrosis and inflammation are common features of chronic kidney disease (CKD) progressing to end-stage renal failure, which is characterized by interstitial extracellular matrix (ECM), myofibroblast accumulation, and infiltration of inflammatory cells accompanied with destruction of renal tubules.Citation13,Citation14 It is well accepted that transforming growth factor-beta (TGF-β) is a master cytokine/growth factor in fibrosis and inflammation ().Citation15–Citation20 TGF-β exerts its biological effects by activating the downstream mediators Smad2 and Smad3.Citation21–Citation27 During fibrosis, TGF-β is known to upregulate many fibrogenic genes, such as ECM proteins, via Smad2, Smad3, or mitogen-activated protein kinases (MAPKs).Citation28–Citation30 Advances in study of TGF-β biology reveal that TGF-β also regulates several miRNAs that are involved in renal fibrosis. TGF-β1 is able to upregulate miR-21, miR-192, miR-491-5p, miR-382, and miR-377, but downregulates the miR-29 and miR-200 families ().Citation31–Citation35 These TGF-β-regulated miRNAs have been shown to modulate renal fibrosis (). In addition, aberrant expression of these miRNAs is observed in mouse models of renal injury,Citation31–Citation33 demonstrating their critical roles in TGF-β-induced fibrosis. In this review, we focus on four groups of TGF-β-regulated miRNAs, including miR-21, miR-192, miR-200, and miR-29 as they have been shown to modulate TGF-β-induced renal fibrosis. Both miR-21 and miR-192 promote fibrosis via amplification of TGF-β signaling while miR-29 and miR-200 families reduce fibrosis by inhibiting the ECM deposition, and preventing epithelial-to-mesenchymal transition (EMT), respectively.
Figure 1 Mechanisms of TGF-β-regulated miRNAs in renal fibrosis.
Abbreviations: AKT, protein kinase B; ECM, extracellular matrix; ERK, extracellular signal-regulated kinase; miRNA, micro ribonucleic acid; MMP, matrix metalloproteinase; PPAR, peroxisome proliferator activated receptor; PTEN, phosphatase and tensin homolog; TGF-β, transforming growth factor beta; Spry, sprouty.
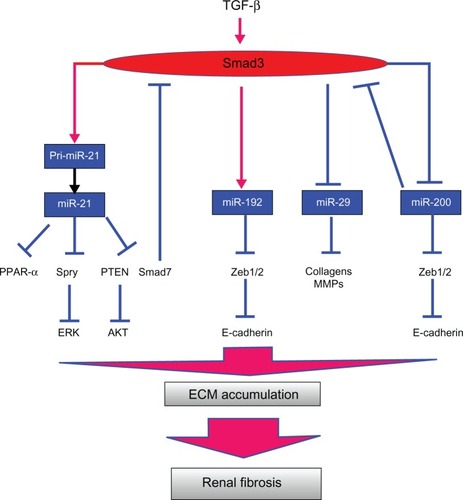
Table 1 Roles of miRNAs in animal models of kidney diseases
Role of miRNAs in TGF-β-mediated kidney diseases
miR-21
miR-21 is a prominent miRNA implicated in the genesis and progression of human cancers.Citation36 As TGF-β1 has been shown to upregulate miR-21 expression,Citation37–Citation39 the role of miR-21 in fibrotic events has been widely studied (). The functional importance of miR-21 in fibrosis was first demonstrated in heart failure.Citation40 Its expression is upregulated in cardiac fibroblasts of failing hearts and treatment with a miR-21 antagomir in a mouse model of cardiac hypertrophy prevents interstitial fibrosis and improves cardiac function.Citation40 Next, miR-21 was found to play a pathological role in lung fibrosis as elevation of miR-21 expression was observed in patients with idiopathic pulmonary fibrosis and in mice with bleomycin-induced lung fibrosis.Citation41 Inhibition of miR-21 by antisense oligonucleotides reduces the severity of experimental lung fibrosis in mice.Citation41
Table 2 Role of miR-21 in different fibrotic diseases
The pathological role of miR-21 in renal diseases has been confirmed by several models. In normal kidney tissue, expression levels of miR-21 are always low. However, its expression levels are highly elevated in both human samples of kidney diseases and animal models of CKD and acute kidney injury (AKI).Citation42–Citation47 In both mouse models of obstructive and diabetic nephropathy, miR-21 expression is elevated in both glomerular and tubulointerstitial area where fibrosis occurs.Citation43,Citation45,Citation46 Expression of renal miR-21 is also significantly increased after ischemia-reperfusion injury.Citation44,Citation47
Results of in vitro studies show that miR-21 actively participates in renal fibrosis because miR-21 positively regulates expression of ECM and α-smooth muscle actin (α-SMA) α-SMA in tubular epithelial cells (TECs) and mesangial cells (MCs) after treatment of TGF-β1 or under diabetic conditions.Citation43,Citation46 In addition, overexpression of miR-21 in kidney cells promotes renal inflammation but knockdown of miR-21 reduces renal inflammation under diabetic conditions. Blocking renal miR-21 expression reduces macrophage infiltration in diseased kidneys.Citation46,Citation48 These results suggest that miR-21 also plays a role in promoting renal inflammation during kidney injury.Citation46,Citation48 The suppression of miR-21 successfully reduces fibrosis in rodent models of heart, lung, and kidney diseases.Citation40,Citation41,Citation43 In a mouse model of diabetic nephropathy, inhibition of miR-21 also improves kidney function and halts the progression of renal injury,Citation46 suggesting that targeting miR-21 should possess a therapeutic potential to ameliorate progressive kidney disease.
miR-21 knockout (KO) mice have been successfully generated and they are phenotypically normal and exhibit normal fertility.Citation42 As expected from the results of unilateral ureteral obstruction (UUO) and renal ischemia reperfusion injury (ISI) models, deletion of the miR-21 gene reduces tubule atrophy, fibrosis, capillary destruction, and P42/P44 MAPK pathway activation in diseased kidneys compared to wild type mice.Citation42 Microarray analyses from miR-21 KO kidneys demonstrate the negative relationship between the presence of miR-21 and genes that are involved in lipid metabolism, fatty acid oxidation, and redox regulation. This study also shows that miR-21 promotes renal fibrosis by silencing metabolic pathways via suppressing peroxisome proliferator-activated receptor-α (PRAR-α; ).Citation42
However, the precise mechanism of how miR-21 regulates fibrosis and inflammation may be relevant for other putative target genes of miR-21. Studies in cardiac fibrosis demonstrate that Sprouty (Spry) and Phosphatase and Tensin Homolog (PTEN) are potential targets of miR-21 ().Citation40,Citation49 Spry is a potent inhibitor of Ras/mitogen-activated protein kinase (MEK)/extracelluar signal-regulated kinase (ERK) and activation of ERK promotes TGF-β-dependent fibrogenic activities.Citation50 In the heart, inhibition of miR-21 decreases ERK-MAPK activity and interstitial fibrosis.Citation40 Suppression of PTEN by miR-21 upregulates phosphatidylinositide 3-kinases (PI3 K) and Akt activity, and subsequently induces matrix metalloproteinase (MMP)-2 expression.Citation49 A study on diabetic kidney injury demonstrates that AKT1 substrate 1 (PRAS40), a negative regulator of Tor complex 1 (TORC1), and Smad7 are targets of miR-21.Citation46,Citation51 Although blocking miR-21 reduces macrophage infiltration in diseased kidneys,Citation46,Citation48 some studies demonstrate anti-inflammatory properties of miR-21 in macrophages by tar-geting proinflammatory programmed cell death 4 (PDCD4).Citation52,Citation53 Negative correlation between miR-21 and PDCD4 has been reported in TECs with induction of ischemia.Citation44 Further studies should be done to clarify whether miR-21 regulates inflammation in a cell type-dependent fashion.
miR-192
miR-192 is a highly expressed miRNA in the normal kidney, compared to other organs.Citation5,Citation6 Several studies from rodent models of kidney diseases and cell lines demonstrate a pro-fibrotic role of miR-192 in both MCs and TECs.Citation54–Citation56 Elevated miR-192 expression is found in glomeruli isolated from diabetic mice.Citation56 In cultured MCs and TECs, miR-192 expression is induced by either TGF-β or high glucose conditions.Citation55,Citation56 In vitro, miR-192 also mediates TGF-β-induced collagen expression in MCs by downregulating zinc finger E-box binding homeobox 1/2 (Zeb1/2) expression ().Citation56 Similarly, overexpression of miR-192 promotes and inhibition of miR-192 blocks TGF-β1-induced collagen matrix expression in TECs.Citation55 Recent studies in a mouse model of type I diabetes demonstrate that inhibition of renal miR-192 significantly induces renal expression of Zeb1/2 and suppresses expression of fibrotic markers, such as collagen, TGF-β, and fibronectin.Citation54 More importantly, inhibition of miR-192 in vivo improves renal function by attenuating proteinuria.Citation54 The pathological role of miR-192 in diabetic nephropathy is further confirmed by miR-192 KO mice.Citation57 In these KO type I diabetic mice, albuminuria, proteinuria, renal fibrosis, and hypertrophy are all reduced compared to diabetic wild-type mice.Citation57 Taken together, these studies have established a regulatory role of miR-192 in TGF-β-dependent renal pathologies observed in animal models of diabetic and obstructive nephropathy.
As the results from animal models demonstrate a pro-fibrotic role of miR-192 in diabetic and obstructive nephropathy,Citation54–Citation56 the reverse is true in human nephropathy.Citation58,Citation59 Interestingly, miR-192 expression is reduced in human TECs after TGF-β1 treatment or in human diseased kidneys.Citation58,Citation59 A recent study demonstrates that TGF-β1 suppresses miR-192 expression in human TECs and loss of miR-192 promotes fibrogenesis in diabetic nephropathy.Citation58 Furthermore, renal biopsy samples from diabetic patients show significantly lower miR-192.Citation58 Reduction of miR-192 expression correlates with tubulointerstitial fibrosis and low glomerular filtration rate (GFR) in individual patients. The different findings in expression of miR-192 in human and animal models of diabetic nephropathy renders it necessary to further investigate the potential role of miR-192 and the mechanisms that regulate miR-192 expression during renal fibrosis in different species.
miR-29
The miR-29 family consists of three members that are encoded by two distinct genomic loci in both human and rodent genomes. Citation60 As all members have the same seed binding sequence, they all bind to the same set of target genes.Citation60 In contrast to miR-192 and miR-21, strong evidence has suggested an anti-fibrotic role for miR-29. High expression of miR-29 is found in the kidney, lung, and heart,Citation60 and reduction of miR-29 expression is observed in animal models and human samples of fibrotic diseases in heart, lung, and kidney ().Citation61–Citation63
Table 3 Role of miR-29 in different fibrotic diseases
In a mouse model of obstructive nephropathy, microarrays and real-time polymerase chain reaction (PCR) assays demonstrate that all miR-29 family members (miR-29a, miR-29b, and miR-29c) are substantially reduced in fibrotic kidneys of UUO wild-type mice but significantly increased in Smad3 KO mice in which renal fibrosis was reduced.Citation63 Reduction in expression of miR-29a and miR-29b in renal TECs and lung and cardiac fibroblasts after treatment with TGF-β1 confirms the negative relationship between miR-29 in TGF-β dependent fibrosis.Citation61–Citation63
Similar to the results in the heart in which knockdown of miR-29b relieves the suppression of fibrotic markers,Citation61,Citation64 overexpression of miR-29 suppresses but inhibition of miR-29 promotes expression of fibrotic markers in mouse embryonic fibroblasts and TECs under diabetic conditions, salt-induced hypertensive conditions, or after TGF-β treatment, suggesting an anti-fibrotic effect for miR-29 ().Citation63,Citation65,Citation66 This anti-fibrotic effect of miR-29 is further confirmed by a mouse model of unilateral ureteral obstruction nephropathy.Citation63 Gene delivery of miR-29b either before or after established obstructive nephropathy successfully blocked progressive renal fibrosis. The anti-fibrotic effects of miR-29 may be a result of its ability to inhibit TGF-β-mediated deposition of ECM. More than 20 different ECM-related genes are predicted to be miR-29 targets and some of them are induced by TGF-β signaling.Citation61 In conclusion, miR-29 is a downstream inhibitor of TGF-β/Smad3-mediated fibrosis and may have therapeutic potential for diseases involving fibrosis.
miR-200
The function of miR-200 family is to maintain epithelial differentiation and this family includes miR-200a, miR-200b, miR-200c, miR-429, and miR-141.Citation67 The miR-200 family was firstly discovered by its ability to restore an epithelial phenotype in breast cancer cell lines through the suppression of the E-cadherin transcriptional repressors ZEB1 and ZEB2 ().Citation68–Citation71 Similar to the results in a mouse model of lung fibrosis,Citation72 reduction of expression of miR-200a and miR-141 are observed in fibrotic kidneys during obstructive and diabetic nephropathy ().Citation73,Citation74 In vitro studies also demonstrate that members of the miR-200 family in TECs are downregulated in a Smad signaling dependent manner after TGF-β treatment.Citation73,Citation74 However, another study shows opposite results; that renal expression of miR-200s is increased in mice subjected to UUO.Citation75 Regardless of the discrepancy in miR-200 expression in fibrotic kidneys, the anti-fibrotic role of miR-200 family is confirmed by gene delivery of miR-200b in fibrotic kidneys. The results show that the elevation of collagens and fibronectin in obstructed kidneys can be suppressed by a single injection of a miR-200b precursor.Citation75
Table 4 Role of miR-200 family members in different fibrotic diseases
The maintenance of epithelial differentiation and prevention of EMT may be the mechanism in which miR-200 suppresses fibrosis.Citation71 In epithelial cells, members of the miRNA-200 family inhibit the E-cadherin transcriptional repressors ZEB1 and ZEB2, which were previously implicated in EMT and tumor metastasis.Citation68–Citation71 These miRNAs are also markedly downregulated in cells that have undergone EMT in response to TGF-β,Citation68–Citation71 suggesting that TGF-β regulates the expression of these miRNAs to promote EMT in tumor cells. In response to renal injury, it is generally believed that proximal TECs may undergo EMT and contribute to renal fibrosis.Citation14 However, the notion that EMT contributes to renal fibrosis has recently been challenged. More research is needed to clarify the role of miR-200 in kidney diseases.
Regulatory mechanisms of TGF-β1 in expression of fibrosis-related miRNAs
The exact mechanism of how TGF-β signaling regulates miRNA expression during kidney diseases is still ongoing. TGF-β signaling promotes synthesis of fibrosis-related miRNAs either by enhancing posttranscriptional processing of the primary miRNA transcript, or by increasing transcription. It has been demonstrated that TGF-β signaling is able to promote the processing of primary transcripts of some miRNAs into their active form by the Drosha complex.Citation37 Receptor-Smads, such as Smad3, physically interact with subunits of the Drosha complex to promote the processing of pri-miR-21 into mature miR-21 (). Davis et al also demonstrate that a consensus sequence (R-SBE) exists within the stem region of the primary transcripts of TGF-β-regulated-miRs (pri-T-miRs).Citation39 Direct binding of Smads to the R-SBE triggers the TGF-β-induced recruitment of Drosha and DGCR8 to pri-T-miRs and promotes the processing of pri-T-miRs.Citation39
Our laboratory also demonstrated that TGF-β/Smad3 signaling regulates the transcription of miR-21, miR-192, and the miR-29 family during renal diseases ().Citation43,Citation55,Citation63 This notion is firstly supported by the results from rodent models of obstructive and remnant kidney diseases induced in mice lacking Smad3, Smad7, or having conditional KO of Smad2 or overexpressing renal Smad7.Citation33,Citation43,Citation55,Citation63 Results from in vitro studies also confirm that TGF-β suppresses miR-29 expression but induces miR-21 and miR-192 expression via the Smad3-dependent mechanism as determined in MCs and TECs overexpressing Smad7, or knocking down Smad2 or Smad3 and in Smad2 or Smad3 KO mouse embryonic fibroblasts.Citation33,Citation43,Citation55,Citation63 In addition, Smad3 regulates the expression of these miRNAs by physically interacting with Smad binding site (SBE) located in their promoters.Citation43,Citation55,Citation63 Binding of Smad3 on SBE in the promoters may either increase transcription and post-transcriptional processing of miRNAs, such as miR-21 and miR-192, or inhibit their transcription, such as for miR-29b.Citation43,Citation55,Citation63 In addition, Smad7, an inhibitory Smad, protects kidneys from fibrosis based on its ability to regulate TGF-β/Smad3-mediated miRNAs via maintaining renal miR-29b but suppressing miR-192 and miR-21.Citation32,Citation33,Citation55
More importantly, miRNAs can also regulate TGF-β/Smad3 signaling (). As TGF-β induces miR-21 expression during renal injury, the elevation of miR-21 suppresses Smad7 expression and, in turn, promotes the TGF-β signaling.Citation41,Citation46 Thus, miR-21 may act as a feed-forward loop to amplify TGF-β signal during renal injury. Activation of ERK/MAP kinase signaling in fibroblasts may be another mechanism by which miR-21 induces renal fibrosis.Citation40 Furthermore, both TGF-β1 and TGF-β2 suppress miR-200a expression in renal cells.Citation74 Since TGF-β2 is one of the target genes for miR-200a, the reduction of miR-200 expression after treatment of TGF-β is associated with an elevation in TGF-β2 expression. On the other hand, overexpression of miR-200a also inhibits both Smad3 activity and TGF-β1-induced fibrosis.Citation74 These results reveal a possible feedback between TGF-β2 and miR-200a. Similar to miR-200, miR-29 is predicted to inhibit TGF-β2.Citation61 Thus, miR-29 may exert its anti-fibrotic effects through inhibition of TGF-β signaling.
In addition, a recent study demonstrates that TGF-β induces a crosstalk circuit between p53 and miR-192 related to the pathogenesis of diabetic nephropathy.Citation57 Expression levels of TGF-β1, p53, and miR-192 are known to be elevated in expanded glomeruli of diabetic mice.Citation54,Citation56,Citation57 Interestingly, inhibition of miR-192 function in vivo is able to suppress p53 expression in the renal cortex of control and streptozotocin injected diabetic mice.Citation57 This positive relationship between miR-192 and renal expression of TGF-β and p53 is confirmed by the miR-192 KO type I diabetic mice.Citation57 Using promoter assay studies in vitro, TGF-β is able to induce activation of miR-192 and p53.Citation57
All the above results suggest the essential roles of miRNAs in TGF-β-induced renal fibrosis and the complexity between TGF-β/Smads signaling and miRNAs under pathophysiological conditions.
Comparison of the role of miRNAs in other fibrotic diseases
In addition to renal fibrosis, miRNAs also participate in fibrotic diseases in other organs. To date, about 40 miRNAs have been reported to be related to fibrosis in various organs, the details of which have been discussed elsewhere.Citation76 In this review, we compare the four groups of miRNAs in fibrotic diseases of heart, lung, kidney, and liver (–). Pro-fibrotic miRNAs, such as miR-21, and anti-fibrotic miRNAs, such as miR-29 and miR-200 families, are dysregulated in these organs as fibrosis occurs.Citation40–Citation47,Citation61–Citation63,Citation66,Citation77–Citation82
These miRNAs directly induce or protect against fibrosis via four different mechanisms. First, regulating the TGF-β pathway is a key mechanism for the fibrotic response of miRNAs during fibrosis ().Citation40–Citation47 In the lung and kidney, miR-21 upregulates TGF-β canonical signaling by suppressing Smad7. However, miR-21 in the heart induces TGF-β non-canonical signaling by targeting Spry1 for derepression of ERK/MAPK. In addition, miR-200 is capable of suppressing expression of TGF-β2 to exert its anti-fibrotic effect in kidney disease ().Citation74
Regulating ECM proteins or enzymes that are involved in ECM remodeling is another vital mechanism for miR-NAs during fibrosis. miR-29 is the best characterized direct regulator of ECM synthesis and has been reported in many major fibrotic diseases in the heart, lung, liver, and kidney ().Citation61–Citation63,Citation66,Citation79–Citation82 Indeed, a large number of ECM and ECM-modulating genes, such as COL1A1, COL3A1, COL4A1, COL5A1, COL5A2, COL5A3, COL7A1, COL8A1, and MMP2, have been validated as direct targets of miR-29 by reporter gene assays.Citation61,Citation66,Citation79 Expression of miR-29 has been reported to be regulated by TGF-β signaling in the heart, lung, liver, and kidney.Citation61–Citation63,Citation66,Citation79–Citation82 In addition, the nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB) pathway is also involved in suppression of hepatic miR-29 expression.Citation80–Citation82 miR-21 is another fibrotic miRNA found in the heart, lung, and kidney although its upregulatory effects may be indirect.Citation40–Citation47,Citation77,Citation78 Thirdly, regulating EMT may be also a mechanism by which miRNAs are involved in the fibrotic process. miR-200 members and miR-192 are closely related to EMT because they negatively regulate the E-cadherin transcriptional repressors ZEB1 and ZEB2 ().Citation56,Citation68–Citation71 Overexpression of miR-200 members or inhibition of miR-192 prevents TGF-β-dependent EMT and reduces expression of ECM proteins.Citation56,Citation68–Citation71 Finally, some miRNAs may exert their regulatory roles in inducing proliferation and resistance to apoptosis during the fibrotic process,Citation76 although no direct evidence was found during the process of fibrosis.
Clinical implications of miRNAs in kidney diseases
Therapeutic potential
As we discussed previously, miRNAs play essential roles in renal injury and miRNAs are always located in the cytoplasm of cells. These two factors support the therapeutic potential of miRNAs in kidney diseases. Furthermore, sequence complementarity provides a possible and specific approach to design a drug that precisely targets a single miRNA. Recent advances in chemical engineering allow the development of chemically modified oligonucleotides that are stable in the circulation and can freely enter cells to bind to specific miRNA and silence it.Citation10 Conventional construction of overexpression or short hairpin (sh)RNA plasmids allow an alternative to restore or suppress miRNA transcription.Citation43,Citation63 In experimental kidney disease models, restoring miR-29 and miR-200 families, or inhibition of miR-21 and miR-192, are able to prevent or inhibit renal fibrosis ().Citation33,Citation43,Citation46,Citation54,Citation63,Citation75 Inhibition of miRNAs by chemical-engineered oligonucleotides has also been applied to other miRNAs.Citation10 Thus, application of miRNAs or its inhibitor in the treatment of kidney disease offers a novel and effective therapeutic approach to combat CKD.
Table 5 Summary of clinical implications of miRNAs
Major obstacles to the therapeutic use of miRNAs are the delivery method and safety concerns. To date, systemic delivery of chemical-engineered oligonucleotides has been widely used for inhibiting miRNA function.Citation10 However, this method may also suppress the function of miRNAs in organs other than the diseased one. Gene delivery systems to regulate miRNA expression in specific organs have also been developed.Citation62,Citation83 For instance, ultrasound-microbubble-mediated gene transfer developed in our laboratory is capable of delivering miRNA overexpression or knockdown plasmids specifically into living kidneys.Citation33,Citation43,Citation46,Citation63,Citation83 Controlling transgene expression at the desired therapeutic levels to minimize side-effects is essential to the success of gene therapy. To avoid any undesirable side-effects caused by overdoses of overexpression or inhibition of miRNA expression, optimal dosages of miRNAs need to clearly be determined and controlled.
The risks of off-target effects and nonspecific immune responses also need to be considered and investigated. For example, the major obstacle to using miR-21 as a therapeutic agent for fibrotic diseases is that downregulation of miR-21 expression induces cell death.Citation43,Citation44,Citation84 Strong pro-apoptotic effects of miR-29b may also hinder the development of miR-29b gene therapy as overexpression of miRNA-29b induces apoptosis in multiple myeloma cells.Citation85 Thus, miRNA therapeutics still require the development of specific and consistent delivery systems that target precise cells and organs.
Biomarkers
Another potential application of miRNAs is as excellent biomarkers of kidney diseases. Recently, circulating nucleic acids have emerged as new biomarkers and many studies are implying that circulating miRNAs are potential biomarkers for cancer growth and organ injury because miRNAs are stable and tissue specific as well as the fact that they can be identified and quantified.Citation86 Several studies confirm the fact that miRNAs are released into urine or blood during kidney disease (). For instance, circulating miR-21 levels have been demonstrated to be significantly higher in patients with severe interstitial fibrosis and tubular atrophy.Citation87 A recent study shows that a total of 27 miRNAs are present at significantly different levels in urine from patients at different stages of diabetic nephropathy.Citation88 More importantly, these miRNAs have previously been shown to play essential roles in signaling pathways of renal fibrosis during diabetic kidney disease. Another study demonstrates that urinary miR-93 levels are related to glomerular scarring while urinary levels of miR-29b and miR-29c are correlated with proteinuria and renal function in immunoglobulin A (IgA) nephropathy.Citation89 These studies demonstrate that miRNAs are not restricted to the kidney only during disease or development. Future studies should focus on the identification of patterns of miRNAs that are released into the urine or blood by damaged kidneys.
Summary and perspectives
Over the past two decades, the progress in discovering miRNAs and characterizing their functions in kidney diseases has been rapid. However, understanding the specific role of miRNAs in renal pathophysiology is still limited. One of the critical issues that hinders the progress of miRNAs in renal research is how to accurately identify miRNA targets because one miRNA is capable of regulating multiple target genes. To date, target prediction programs provide us with a large number of potential miRNA targets. However, the overlap between algorithms is so minimal that only a small portion of these targets can be validated experimentally. Although conservation of the 3′ UTR among species is included, the number of targets predicted is still high for validation. In addition, the power of miRNAs also depends on their ability to target multiple genes that contribute to a pathway or phenotype. This also increases the difficulty in searching for real targets of miRNAs. However, high-throughput validation and proteomic analysis are possible approaches to enable us to identify miRNA targets.
Understanding the regulation of miRNA expression will be another challenge for miRNA research because one miRNA expression can be regulated by many different mediators or pathways. Firstly, miRNAs in a given miRNA cluster may show the same expression pattern. However, some cluster members do not follow the same pattern and exhibit different expression patterns.Citation90 Furthermore, some miRNAs may be encoded from two or more distinct genomic loci, such as miR-29b;Citation60 it is still not known how to control miR-29b expression from two genomic loci. Intronic miRNAs do not always have the same expression pattern as their host gene.Citation91 Furthermore, sometimes both strands of the miRNA are coexpressed and they usually target different genes.Citation92 Further studies are required to understand the mechanism regarding how to regulate miRNA expression. Recent technology advances in deep sequencing may provide us a comprehensive view of gene expression patterns and quantify transcript levels. This information may allow us to correlate expression of miRNAs and target transcripts.
Understanding the pathophysiological role of a specific miRNA in the kidney is not as easy as previous thought because the kidney is composed of several types of cells and these cell types may respond differentially to miRNAs in various renal diseases. Identifying the cell type(s) that expresses the miRNA in a particular disease condition may enable us to understand the role of miRNAs during disease progression. To overexpress or inhibit the function of a specific miRNA in a particular cell type will be beneficial to our understanding of disease mechanisms. Thus, transgenic animal models with conditional overexpression or knockdown of a particular miRNA may provide the best model to study the function and regulation of miRNAs in renal diseases. This approach will improve our knowledge of the role of miRNAs in renal pathology and enable us to find new treatments for halting renal fibrosis and dysfunction. However, identifying the functional significance of a change in miRNA regulation and its target gene(s) may be difficult because a single miRNA can regulate several target proteins and a single protein can be regulated by several miRNAs.
Finally, miRNAs act as important downstream effectors of TGF-β/Smad signaling during renal fibrosis. Advances in our understanding of the specific role of miRNAs involved in TGF-β/Smad signaling during renal fibrosis should provide us an effective, alternative strategy to ameliorate disease progression.
Acknowledgments
This work was supported by grants from Major State Basic Research Development Program of China (973 program, No 2012CB517700 to HYL) and National Natural Science Foundation of China (General Program 81170681 to ACC and Major Program 81130012 to XY); the Research Grant Council of Hong Kong (RGC GRF 468711, 469110, CUHK5/CRF/09, and CUHK3/CRF/12R to HYL; GRF 463612, 464010, 763908, and 764109 to ACC); the Focused Investment Scheme A and B and direct grants from the Chinese University of Hong Kong (2011.1.076, 2012.1.021 to ACC); 2011 Research Grant from Hong Kong Society of Nephrology (6903213 to ACC); Municipal Science and Technology R&D funding of basic research, Shenzhen (Major Program JC201104220290 A to HYL and General Program C.02.12.00501 to ACC).
Disclosure
The authors report no conflicts of interest in this work.
References
- GregoryRIYanKPAmuthanGThe Microprocessor complex mediates the genesis of microRNAsNature2004432701423524015531877
- LeeYAhnCHanJThe nuclear RNase III Drosha initiates microRNA processingNature2003425695641541914508493
- DuTZamorePDmicroPrimer: the biogenesis and function of microRNADevelopment2005132214645465216224044
- FilipowiczWRNAi: the nuts and bolts of the RISC machineCell20051221172016009129
- TianZGreeneASPietruszJLMatusIRLiangMMicroRNA-target pairs in the rat kidney identified by microRNA microarray, proteomic, and bioinformatic analysisGenome Res200818340441118230805
- SunYKooSWhiteNDevelopment of a micro-array to detect human and mouse microRNAs and characterization of expression in human organsNucleic Acids Res20043222e18815616155
- BhattKMiQSDongZmicroRNAs in kidneys: biogenesis, regulation, and pathophysiological rolesAm J Physiol Renal Physiol20113003F602F61021228106
- LiJYYongTYMichaelMZGleadleJMReview: the role of micro-RNAs in kidney diseaseNephrology (Carlton)201015659960820883280
- KatoMArceLNatarajanRMicroRNAs and their role in progressive kidney diseasesClin J Am Soc Nephrol2009471255126619581401
- LorenzenJMHallerHThumTMicroRNAs as mediators and therapeutic targets in chronic kidney diseaseNat Rev Nephrol20117528629421423249
- WesselyOAgrawalRTranUMicroRNAs in kidney development: lessons from the frogRNA Biol20107329629920458188
- SaalSHarveySJMicroRNAs and the kidney: coming of ageCurr Opin Nephrol Hypertens200918431732319424061
- BöttingerEPTGF-beta in renal injury and diseaseSemin Nephrol200727330932017533008
- LiuYCellular and molecular mechanisms of renal fibrosisNat Rev Nephrol201171268469622009250
- RobertsABMolecular and cell biology of TGF-betaMiner Electrolyte Metab1998242–31111199525693
- WangWHuangXRLiAGSignaling mechanism of TGF-beta1 in prevention of renal inflammation: role of Smad7J Am Soc Nephrol20051651371138315788474
- LanHYChungACTransforming growth factor-β and SmadsContrib Nephrol2011170758221659760
- MengXMHuangXRXiaoJDisruption of Smad4 impairs TGF-β/Smad3 and Smad7 transcriptional regulation during renal inflammation and fibrosis in vivo and in vitroKidney Int201281326627922048127
- MengXMHuangXRXiaoJDiverse roles of TGF-β receptor II in renal fibrosis and inflammation in vivo and in vitroJ Pathol2012227217518822190171
- MengXMHuangXRChungACSmad2 protects against TGF-beta/Smad3-mediated renal fibrosisJ Am Soc Nephrol20102191477148720595680
- MassaguéJChenYGControlling TGF-beta signalingGenes Dev200014662764410733523
- MiyazonoKPositive and negative regulation of TGF-beta signalingJ Cell Sci2000113Pt 71101110910704361
- MengXMChungACLanHYRole of the TGF-β/BMP-7/Smad pathways in renal diseasesClin Sci2013124424325423126427
- ChungACZhangHKongYZAdvanced glycation end-products induce tubular CTGF via TGF-beta-independent Smad3 signalingJ Am Soc Nephrol201021224926019959709
- ChenHYHuangXRWangWThe protective role of Smad7 in diabetic kidney disease: mechanism and therapeutic potentialDiabetes201160259060120980457
- ZhouLFuPHuangXRMechanism of chronic aristolochic acid nephropathy: role of Smad3Am J Physiol Renal Physiol20102984F1006F101720089673
- LiRLanHYChungACDistinct roles of Smads and microRNAs in TGF-β signaling during kidney diseasesHong Kong Journal of Nephrology20131511421
- SchnaperHWHayashidaTHubchakSCPonceletACTGF-beta signal transduction and mesangial cell fibrogenesisAm J Physiol Renal Physiol20032842F243F25212529270
- HoffmanBBSharmaKZiyadehFNPotential role of TGF-beta in diabetic nephropathyMiner Electrolyte Metab1998242–31901969525704
- ChungACHuangXRZhouLHeuchelRLaiKNLanHYDisruption of the Smad7 gene promotes renal fibrosis and inflammation in unilateral ureteral obstruction (UUO) in miceNephrol Dial Transplant20092451443145419096081
- KantharidisPWangBCarewRMLanHYDiabetes complications: the microRNA perspectiveDiabetes20116071832183721709278
- LanHYChungACTGF-β/Smad signaling in kidney diseaseSemin Nephrol201232323624322835454
- ChungACDongYYangWZhongXLiRLanHYSmad7 suppresses renal fibrosis via altering expression of TGF-β/Smad3-regulated microRNAsMol Ther201321238839823207693
- KriegelAJLiuYCohenBUsaKLiuYLiangMMiR-382 targeting of kallikrein 5 contributes to renal inner medullary interstitial fibrosisPhysiol Genomics201244425926722202692
- KriegelAJFangYLiuYMicroRNA-target pairs in human renal epithelial cells treated with transforming growth factor beta 1: a novel role of miR-382Nucleic Acids Res201038228338834720716515
- JazbutyteVThumTMicroRNA-21: from cancer to cardiovascular diseaseCurr Drug Targets201011892693520415649
- DavisBNHilyardACLagnaGHataASMAD proteins control DROSHA-mediated microRNA maturationNature20084547200566118548003
- ZavadilJNarasimhanMBlumenbergMSchneiderRJTransforming growth factor-beta and microRNA: mRNA regulatory networks in epithelial plasticityCells Tissues Organs (Print)20071851–315716117587821
- DavisBNHilyardACNguyenPHLagnaGHataASmad proteins bind a conserved RNA sequence to promote microRNA maturation by DroshaMol Cell201039337338420705240
- ThumTGrossCFiedlerJMicroRNA-21 contributes to myocardial disease by stimulating MAP kinase signalling in fibroblastsNature2008456722498098419043405
- LiuGFriggeriAYangYmiR-21 mediates fibrogenic activation of pulmonary fibroblasts and lung fibrosisJ Exp Med201020781589159720643828
- ChauBNXinCHartnerJMicroRNA-21 promotes fibrosis of the kidney by silencing metabolic pathwaysSci Transl Med20124121121r a18
- ZhongXChungACChenHYMengXMLanHYSmad3-mediated upregulation of miR-21 promotes renal fibrosisJ Am Soc Nephrol20112291668168121852586
- GodwinJGGeXStephanKJurischATulliusSGIacominiJIdentification of a microRNA signature of renal ischemia reperfusion injuryProc Natl Acad Sci U S A201010732143391434420651252
- WangJGaoYMaMEffect of miR-21 on renal fibrosis by regulating MMP-9 and TIMP1 in kk-ay diabetic nephropathy miceCell Biochem Biophys2013
- ZhongXChungACChenHYmiR-21 is a key therapeutic target for renal injury in a mouse model of type 2 diabetesDiabetologia201356366367423292313
- XuXKriegelAJLiuYDelayed ischemic preconditioning contributes to renal protection by upregulation of miR-21Kidney Int201282111167117522785173
- ZarjouAYangSAbrahamEAgarwalALiuGIdentification of a microRNA signature in renal fibrosis: role of miR-21Am J Physiol Renal Physiol20113014F793F80121775484
- RoySKhannaSHussainSRMicroRNA expression in response to murine myocardial infarction: miR-21 regulates fibroblast metalloprotease-2 via phosphatase and tensin homologueCardiovasc Res2009821212919147652
- DingQGladsonCLWuHHayasakaHOlmanMAFocal adhesion kinase (FAK)-related non-kinase inhibits myofibroblast differentiation through differential MAPK activation in a FAK-dependent mannerJ Biol Chem200828340268392684918669633
- DeyNDasFMariappanMMMicroRNA-21 orchestrates high glucose-induced signals to TOR complex 1, resulting in renal cell pathology in diabetesJ Biol Chem201128629255862560321613227
- SheedyFJPalsson-McDermottEHennessyEJNegative regulation of TLR4 via targeting of the proinflammatory tumor suppressor PDCD4 by the microRNA miR-21Nat Immunol201011214114719946272
- MerlineRMorethKBeckmannJSignaling by the matrix proteoglycan decorin controls inflammation and cancer through PDCD4 and MicroRNA-21Sci Signal20114199ra7522087031
- PuttaSLantingLSunGLawsonGKatoMNatarajanRInhibiting microRNA-192 ameliorates renal fibrosis in diabetic nephropathyJ Am Soc Nephrol201223345846922223877
- ChungACHuangXRMengXLanHYmiR-192 mediates TGF-beta/Smad3-driven renal fibrosisJ Am Soc Nephrol20102181317132520488955
- KatoMZhangJWangMMicroRNA-192 in diabetic kidney glomeruli and its function in TGF-beta-induced collagen expression via inhibition of E-box repressorsProc Natl Acad Sci U S A200710493432343717360662
- DeshpandeSDPuttaSWangMTransforming growth factor-β induced cross talk between p53 and a microRNA in the pathogenesis of diabetic nephropathyDiabetes2013
- KrupaAJenkinsRLuoDDLewisAPhillipsAFraserDLoss of MicroRNA-192 promotes fibrogenesis in diabetic nephropathyJ Am Soc Nephrol201021343844720056746
- WangBHerman-EdelsteinMKohPE-cadherin expression is regulated by miR-192/215 by a mechanism that is independent of the profibrotic effects of transforming growth factor-betaDiabetes20105971794180220393144
- KriegelAJLiuYFangYDingXLiangMThe miR-29 family: genomics, cell biology, and relevance to renal and cardiovascular injuryPhysiol Genomics201244423724422214600
- van RooijESutherlandLBThatcherJEDysregulation of microRNAs after myocardial infarction reveals a role of miR-29 in cardiac fibrosisProc Natl Acad Sci U S A200810535130271303218723672
- XiaoJMengXMHuangXRmiR-29 inhibits bleomycin-induced pulmonary fibrosis in miceMol Ther20122061251126022395530
- QinWChungACHuangXRTGF-β/Smad3 signaling promotes renal fibrosis by inhibiting miR-29J Am Soc Nephrol20112281462147421784902
- YeYHuZLinYZhangCPerez-PoloJRDownregulation of microRNA-29 by antisense inhibitors and a PPAR-gamma agonist protects against myocardial ischaemia-reperfusion injuryCardiovasc Res201087353554420164119
- DuBMaLMHuangMBHigh glucose down-regulates miR-29a to increase collagen IV production in HK-2 cellsFEBS Lett2010584481181620067797
- LiuYTaylorNELuLRenal medullary microRNAs in Dahl salt-sensitive rats: miR-29b regulates several collagens and related genesHypertension201055497498220194304
- HoweENCochraneDRRicherJKThe miR-200 and miR-221/222 microRNA families: opposing effects on epithelial identityJ Mammary Gland Biol Neoplasia2012171657722350980
- GregoryPABertAGPatersonELThe miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1Nat Cell Biol200810559360118376396
- ParkSMGaurABLengyelEPeterMEThe miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2Genes Dev200822789490718381893
- BurkUSchubertJWellnerUA reciprocal repression between ZEB1 and members of the miR-200 family promotes EMT and invasion in cancer cellsEMBO Rep20089658258918483486
- KorpalMLeeESHuGKangYThe miR-200 family inhibits epithelial-mesenchymal transition and cancer cell migration by direct targeting of E-cadherin transcriptional repressors ZEB1 and ZEB2J Biol Chem200828322149101491418411277
- YangSBanerjeeSde FreitasAParticipation of miR-200 in pulmonary fibrosisAm J Pathol2012180248449322189082
- XiongMJiangLZhouYThe miR-200 family regulates TGF-β1-induced renal tubular epithelial to mesenchymal transition through Smad pathway by targeting ZEB1 and ZEB2 expressionAm J Physiol Renal Physiol20123023F369F37922012804
- WangBKohPWinbanksCmiR-200a prevents renal fibrogenesis through repression of TGF-β2 expressionDiabetes201160128028720952520
- ObaSKumanoSSuzukiEmiR-200b precursor can ameliorate renal tubulointerstitial fibrosisPLoS ONE2010510e1361421049046
- VettoriSGaySDistlerORole of microRNAs in fibrosisOpen Rheumatol J2012613013922802911
- van RooijESutherlandLBQiXRichardsonJAHillJOlsonENControl of stress-dependent cardiac growth and gene expression by a microRNAScience2007316582457557917379774
- WeiJFengLLiZXuGFanXMicroRNA-21 activates hepatic stellate cells via PTEN/Akt signalingBiomed Pharmacother201367538739223643356
- CushingLKuangPPQianJmiR-29 is a major regulator of genes associated with pulmonary fibrosisAm J Respir Cell Mol Biol201145228729420971881
- SekiyaYOgawaTYoshizatoKIkedaKKawadaNSuppression of hepatic stellate cell activation by microRNA-29bBiochem Biophys Res Commun20114121747921798245
- RoderburgCUrbanGWBettermannKMicro-RNA profiling reveals a role for miR-29 in human and murine liver fibrosisHepatology201153120921820890893
- PogribnyIPStarlard-DavenportATryndyakVPDifference in expression of hepatic microRNAs miR-29c, miR-34a, miR-155, and miR-200b is associated with strain-specific susceptibility to dietary nonalcoholic steatohepatitis in miceLab Invest201090101437144620548288
- LanHYMuWTomitaNInhibition of renal fibrosis by gene transfer of inducible Smad7 using ultrasound-microbubble system in rat UUO modelJ Am Soc Nephrol20031461535154812761254
- LiTLiDShaJSunPHuangYMicroRNA-21 directly targets MARCKS and promotes apoptosis resistance and invasion in prostate cancer cellsBiochem Biophys Res Commun2009383328028519302977
- ZhangYKWangHLengYOverexpression of microRNA-29b induces apoptosis of multiple myeloma cells through down regulating Mcl-1Biochem Biophys Res Commun2011414123323921951844
- VeluVKRameshRSrinivasanARCirculating microRNAs as biomarkers in health and diseaseJ Clin Diagn Res20126101791179523373057
- GlowackiFSavaryGGnemmiVIncreased circulating miR-21 levels are associated with kidney fibrosisPLoS ONE201382e5801423469132
- ArgyropoulosCWangKMcClartySUrinary microRNA profiling in the nephropathy of type 1 diabetesPLoS ONE201381e5466223358711
- WangGKwanBCLaiFMChowKMLiPKSzetoCCUrinary miR-21, miR-29, and miR-93: novel biomarkers of fibrosisAm J Nephrol201236541241823108026
- KhellaHWWhiteNMFaragallaHExploring the role of miRNAs in renal cell carcinoma progression and metastasis through bioinformatic and experimental analysesTumour Biol201233113114022086373
- BaskervilleSBartelDPMicroarray profiling of microRNAs reveals frequent coexpression with neighboring miRNAs and host genesRNA200511324124715701730
- KhellaHWBakhetMLichnerZRomaschinADJewettMAYousefGMMicroRNAs in kidney disease: an emerging understandingAm J Kidney Dis201361579880823219107