Abstract
Background
An early detection of precursor lesions of cervical cancer will help to eliminate the worldwide burden of cervical cancer.
Methods
This exploratory study aimed to identify, by matrix-assisted laser desorption/ionization (MALDI) time-of-flight (TOF) mass spectrometry (MS), serum protein profiles that distinguish cervical intraepithelial neoplasia grades CIN 1 or lower (≤CIN 1) from CIN 2+ among 127 women infected with human papillomavirus (HPV) 16. Of these 127 women, 25 and 23 were diagnosed with CIN 2 or CIN 3, respectively (cases), and 79 were diagnosed with ≤CIN 1 (non-cases). Serum protein profiles were generated by MALDI-TOF-MS. A total of 95 m/z peaks were tested for association with case status by two racial groups, African American (AAs) and Caucasian American (CAs).
Results
Overall, 2 protein peaks identified by our study demonstrated higher specificity for identifying CIN 2+ than previously published studies. An increasing intensity of [m/z 4459] was associated with a higher risk of being a case, regardless of race with a specificity of 58% for CIN 2 and a specificity of 75% for CIN 3. An increasing intensity of [m/z 4154] was not only associated with a higher risk of being a case only among CAs, but also had an opposite effect among AAs.
Conclusion
Identification of specific proteins associated with the peaks detected in serum and development of antibody-based tests such as ELISA should lead to the development of race-specific, non-invasive and cost effective screening tests with higher specificity for identifying HPV 16 associated CIN 2+.
Introduction
Worldwide, cervical cancer (CC), which is caused mainly by 13 high-risk or carcinogenic genotypes of human papillomaviruses (HPVs), is the third most prevalent type of cancer in women.Citation1,Citation2 HPV is the most common sexually transmitted virus.Citation3 In addition, HPV infections and CC risks are compounded by infections with human immunodeficiency virus (HIV) and the resulting acquired immune deficiency syndrome.Citation4,Citation5 Although CC is a priority global health condition affecting millions of women, it is preventable by use of organized screening programs and regular follow-up of at-risk women. A cytology-based screening test for CC (Pap test) has been used in high-income countries for the last 50 years. Compared with cytology, testing for HPVs is more sensitive in detecting cervical intraepithelial neoplasia grades CIN 2+, but with lower specificity.Citation6 For much of the world, screening by the Pap test or by HPV tests may not however, be a viable option because of the need for specialized practitioners or lack of laboratory infrastructure to perform these tests. Therefore, development of improved screening tools that can be applied worldwide, and without the need for such infrastructure, will greatly aid in the prevention and control of CC.
We have previously conducted an exploratory study to identify candidate surface-enhanced laser desorption/ ionization (SELDI) time of flight (TOF) mass spectrometry (MS) protein profiles in plasma that may distinguish cervical intraepithelial neoplasia 3 (CIN 3) from CIN 1 among women infected with high-risk HPVs. The results of this study suggested the possibility of using plasma SELDI protein profiles to identify women who are likely to have CIN 3 lesions.Citation7 The current report describes an exploratory study to identify, by matrix-assisted laser desorption/ionization (MALDI) TOF MS, protein profiles in serum that may distinguish CIN 1 or lower (≤CIN 1) from CIN 2+ among AA and CA women infected with HPV 16, one of the most carcinogenic types of HPVs.
Materials and methods
Patient population
The study is based on the analysis of serum samples from 127 HPV 16 positive women who were enrolled in a prospective follow-up study funded by the National Cancer Institute (R01 CA105448, Prognostic Significance of DNA and Histone Methylation). The study has been described in a previous publication.Citation8 All women were diagnosed with abnormal cervical cells in clinics of the Health Departments in Jefferson County and surrounding counties in Alabama and were referred to the University of Alabama at Birmingham (UAB) for further examination by colposcopy and biopsy. The women were 19–50 years old, had no history of cervical cancer or other cancers of the lower genital tract, no history of hysterectomy or destructive therapy of the cervix, were not pregnant, and were not using antifolate medications such as methotrexate, sulfasalazine, or phenytoin. Of these 127 women, 25 and 23 were diagnosed with CIN 2 or CIN 3, respectively (cases), and 79 were diagnosed with ≤CIN 1 (normal cervical epithelium, n = 3, HPV cytopathic effect, n = 6, reactive nuclear enlargement, n = 15 or CIN 1, n = 55, non-cases). All women tested positive for HPV 16 in exfoliated cervical cells. All women included in this analysis participated in an interview that assessed sociodemographic variables and lifestyle risk factors. Height and weight measurements were obtained by use of standard protocols. The BMI was calculated using the height and weight measurements (weight kg/[height m]Citation2). Pelvic examinations and collection of cervical cells and biopsies were accomplished following the protocols of the colposcopy clinic. Fasting blood samples were collected from all women and processed immediately to isolate serum. Several serum sample aliquots were stored at −80°C. Serum samples which were not subjected to freeze-thaw cycles were used to generate serum protein profiles. The study protocol and procedures were approved by the UAB Institutional Review Board.
Testing for HPVs
DNA was extracted from cervical cells using the QIAamp MiniElute Media Kit (Qiagen, Inc, Valencia, CA) following the manufacturer’s instruction for HPV genotyping test. HPV genotyping test (Linear Array, Roche Diagnostics, Indianapolis, IN) was performed according to the manufacturer’s instructions by a research associate trained by personnel from Roche Diagnostics. Briefly, target DNA amplified by Polymerase Chain Reaction (PCR) utilized the PGMY09/11 L1 consensus primer system and included co-amplification of a human cellular target, β-globin, as an internal control. Detection and HPV genotyping were achieved using a linear array HPV genotyping test and this test included probes to genotype for 37 anogenital HPV types (6, 11, 16, 18, 26, 31, 33, 35, 39, 40, 42, 45, 51, 52, 53, 54, 55, 56, 58, 59, 61, 62, 64, 66, 67, 68, 69, 70, 71, 72, 73 (MM9), 81, 82 (MM4), 83 (MM7), 84 (MM8), IS39, and CP6108). HPV 16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, 68 were considered to be high-risk (HR) HPV types and all other types were considered to be low-risk (LR) HPVs.
Generation of serum protein profiles
A high-affinity, solid-core lipophilic extraction resin was used to enrich the low-molecular-weight protein fraction of the samples. Bondapak C18 125A, 37–55 μm resin (Waters, Milford, MA, USA) was packed into 96-well, 0.45-μm Unifilter plates (Whatman, Florham Park, NJ, USA) and the packed resins were activated with 80% acetonitrile (aqueous). Serum samples were thawed, diluted (1:50) in distilled water, acidified by adding trifluoroacetic acid (TFA) to a final concentration of 1% v/v (475 μL of sample plus 25 μL of 20% TFA), and mixed with the activated C18 resins. The unbound serum proteins were removed by centrifugation of the 96-well plate for 5 minutes at 1500 g. The resin was washed twice with 200 μL of 1% TFA per well, and the bound peptides and low-molecular-weight proteins were eluted with 100 μl of 70% CH3CN:0.1% TFA (aqueous). Eluants were mixed with an equal volume of matrix consisting of 20 mg/mL sinapinic acid (Fluka, St Louis, MO, USA) in 50:50 CH3CN:0.1% TFA) and spotted onto a MALDI target plate for MALDI-TOF analysis. Profiles of peptides and low-molecular-weight proteins were obtained using a 200 Hz MALDI-TOF/TOF MS (Ultraflex III, Bruker Daltonics). Spectra were acquired in a linear positive ion mode with the mass window set to acquire from 2–20 kDa. Mass calibration was accomplished externally by use of a mixture of standards consisting of insulin, cytochrome c, myoglobin, and ubiquitin (Bruker Daltonics, Bremen, Germany).
Data obtained from the mass spectrometry analysis of the serum samples were exported as text files for preprocessing and further analysis. In-house spectra analysis tools built with MATLAB were used to pre-process the mass spectrometry data. The spectral preprocessing carried out included baseline and noise estimation followed by subtraction of background noise using a local (in m/z) noise estimator, normalization using total ion current, peak detection with a signal-to-noise ratio (S/N) cutoff of 4, and finally peak alignment based on a common set of peaks that appear in at least two-thirds of all spectra. The preprocessing step resulted in 95 peaks in the mass-to-charge (m/z) range of 2–20 kDa on which statistical analysis as described below was carried out.
Statistical methods
Two sets of analyses were conducted in this study. In the first set of analyses, the 95 peaks were tested for association with case status. The case group included the 48 women with CIN 2 or CIN 3 and the non-case group included the 79 women with diagnosis ≤CIN 1. In the second set of analyses, the 95 peaks were tested for association with extreme diagnoses of cervical lesions, ie, cases diagnosed with CIN 3 (n = 23) vs non-cases with diagnosis <CIN 1 (n = 24).
Within each set of analyses, logistic regression models were used to test the association of peak intensities and participant characteristics with case status. Using each individual peak, 5 initial models were fitted: a bivariate model with peak intensity as a predictor of case status, and 4 models for interactions between peak intensity and 4 participant characteristics (age, BMI, race, and infection with multiple HR-HPV types), respectively. Each interaction model included the main effects for peak intensity and participant characteristic, and an interaction term. A model with only the peak intensity as predictor assumed that the effects of intensity on case status are similar for all participants. A model with a peak-by-characteristic interaction allowed separating effects of intensity on case status, according to the levels or categories of the characteristic. The statistical significance was held at the traditional 0.05 level.
The peaks, characteristics, and peak-by-characteristic interactions that individually showed significant association with case status were then used to construct a multivariable logistic model that predicted the case status. The redundant predictors were dropped from the multivariable model using a backward-selection algorithm.
Alternative peak selection procedures used to construct the multivariable models included least absolute shrinkage and selection (LASSO) regression, and least angle regression (LAR).Citation9 In these procedures, the initial set of predictors included all 95 peaks, the 4 participant characteristics, and the 380 peak by characteristic interactions.
Measures of sensitivity and specificity were calculated for the final multivariable models, as well as for the individual predictors used in them. Each logistic regression model estimated the probabilities of case status for the range of values of the predictors. In order to use the model-predicted probabilities to classify individuals into cases or non-cases, a cutoff probability value was required. A natural cutoff point is 0.5, however, this value might not be optimal. For each logistic model, the optimal cutoff probability value was determined using a receiver operating characteristic (ROC) curve. For each predicted probability taken as cutoff value, a ROC curve plots the resulting sensitivity (on the vertical axis) vs 1 – specificity (on the horizontal axis). The optimal cutoff value is that for which sensitivity and 1 – specificity are closest to the ideal values of 1 and 0, respectively. In addition, the area under the ROC curve is a measure of the predictive ability of the model.Citation10 Areas from 0.7 to 0.8 indicate fair predictive ability; areas from 0.8 to 0.9 indicate acceptable predictive ability; and areas greater than 0.9 indicate excellent predictive ability.
Leave-one-out cross-validation was conducted for the final multivariable logistic models (values for each individual were removed from the dataset; then a logistic model was calculated with the remaining individuals and used to predict the status of the removed individual). For the two peaks that showed the strongest association with case status, cross-validated measures of sensitivity and specificity were computed separately by race. All statistical analyses were conducted using SAS v. 9.2 software (SAS Institute, Cary, NC; 2008).
Results
Sample characteristics
Average ages in years for cases and non-cases were 23.6 (SD = 3.7) and 23.4 (SD = 4.8), respectively (difference in mean age P = 0.85). Average BMI measures in kg/m2 for cases and non-cases were 25.9 (SD = 6.8) and 26.9 (SD = 9.2), respectively (difference in mean BMI P = 0.47). The proportions of AAs for cases and non-cases were 39.6% (n = 19) and 49.4% (n = 39), respectively (difference in proportions, P = 0.28). Among cases, the proportion of participants with infections with multiple HPV types was 47.9% (n = 23); among non-cases, this proportion was 44.3% (n = 35, difference in proportions, P = 0.69).
Association of peaks with case status
In the initial set of analyses, which compared 48 cases (CIN 2+) with 79 non-cases (≤CIN 1), significant bivariate associations were detected between case status and the following: 8 peaks, 1 peak by race interaction, and 1 peak by age interaction. Thus, the initial multivariable model included the main effects for 10 peaks, 2 interaction terms, and main effects for age and race. The final multivariable model, however, included only 2 components: (1) a main effect for peak [m/z = 4459], and (2) a race by [m/z = 4154] interaction, which required the inclusion of main effects for race and peak [m/z = 4154].
The alternative LASSO and LAR regression procedures for peak selection retained only [m/z = 4459] as a predictor. Because in the final multivariable logistic model the relationship between case status and the race by [m/z = 4154] interaction was statistically significant in the presence of [m/z = 4459], the larger model with both peaks and interaction was preferred and remained as the final model.
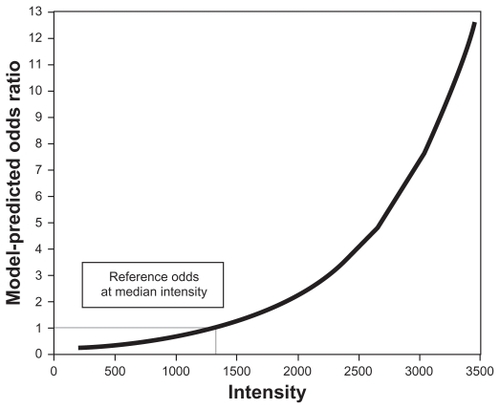
At the observed median [m/z = 4459] intensity, the model-predicted odds of being CIN 2+ was estimated at 0.58 (probability of being a case = 0.37). Using the odds of being a case at the median intensity as reference, shows the model-predicted odds ratios for the observed range of intensity values. According to the model, increasing intensity of [m/z = 4459] was associated with a higher risk of being CIN 2+, regardless of race.
Figure 1 Association between the intensity of peak m/z = 4459 and odds ratio for CIN 2+a.
Abbreviation: CIN, cervical intraepithelial neoplasia.
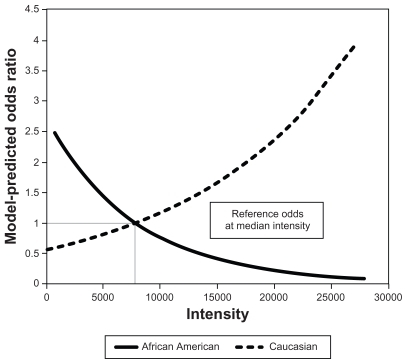
At the observed median [m/z = 4154] intensity, the model-predicted odds of being CIN 2+ among CAs was estimated at 0.75 (probability of being a case = 0.43); among AAs, the model predicted odds of being a case was estimated at 0.56 (probability of being a case = 0.36). Using the odds of being CIN 2+ at the median intensity as reference, shows that the model-predicted odds ratios for the observed [m/z = 4154] range of intensity values differed by race. According to the model, an increasing intensity of [m/z = 4154] was associated with a higher risk of being CIN 2+ only among CAs, but had an opposite effect among AAs. The interaction term was used to model this differential effect by race.
Figure 2 Association between the intensity of peak m/z = 4154 and odds ratio for CIN 2+ for Caucasian Americansa and African Americans.b
Abbreviation: CIN, cervical intraepithelial neoplasia.
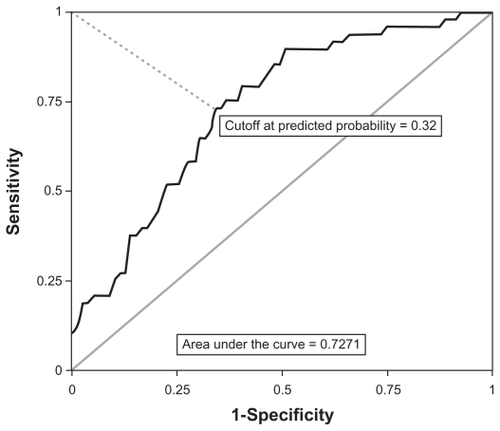
After determining the optimal model-predicted cut-off probability using a ROC curve (), the final multivariate logistic model accurately classified 35 cases and 52 non-cases. However, 13 cases were misclassified as non-cases, and 27 non-cases were misclassified as cases. Concordance with case status was 68.5%; concordance beyond chance, as measured by the Kappa statistic, was estimated at 0.37 (95% CI = 0.21, 0.53). The area under the ROC curve of 0.72 indicated that the final multivariate model had fair predictive ability.
Figure 3 Receiver operating characteristic curve for all cases (CIN 2+) and all controls (≤CIN 1).
There was a small expected decrease in the precision of the prediction after applying the leave-one-out cross-validation algorithm. For the final multivariate model, the cross-validation algorithm resulted in accurate classification of 38 cases and 41 non-cases. Ten cases were misclassified as non-cases, and 38 non-cases were misclassified as cases. Concordance with case status was 62.2%; concordance beyond chance, as measured by the Kappa statistic, was estimated at 0.28 (95% CI = 0.13, 0.43). Cross-validated sensitivity and specificity for the final multivariate model as well as for each of its two components are shown in . As can be seen in this table, the final model resulted in better sensitivity compared with each of its individual components alone; however, the models with individual components resulted in better specificity compared to the final model. This observed variability in the sensitivity and specificity measures, when comparing the final model vs the component-only models, suggests some instability in the prediction, caused by variability in the probability of being CIN 2+ that remained unexplained by the logistic regression models.
Table 1 Cross-validated screening test measures for the final multivariate model and individual models with the two components of the final model predicting cervical intraepithelial neoplasia (CIN) 2+ vs ≤CIN 1 (n = 127)
In the subsample of 24 non-cases with diagnosis <CIN 1 and 23 cases with CIN 3, the average ages in years for cases and non-cases were 23.2 (SD = 3.6) and 22.8 (SD = 3.9), respectively (difference in mean age P = 0.73). Average BMI measures in kg/m2 for cases and non-cases were 25.0 (SD = 5.8) and 29.4 (SD = 9.2), respectively (difference in mean BMI P = 0.06). The proportions of AAs for cases and non-cases were 26% (n = 6) and 70% (n = 17), respectively (difference in proportions, P = 0.0034). Among the cases, the proportion of participants with infections with multiple HR-HPV types was 47.8% (n = 11); among the non-cases, this proportion was 45.8% (n = 11, difference in proportions P = 0.89).
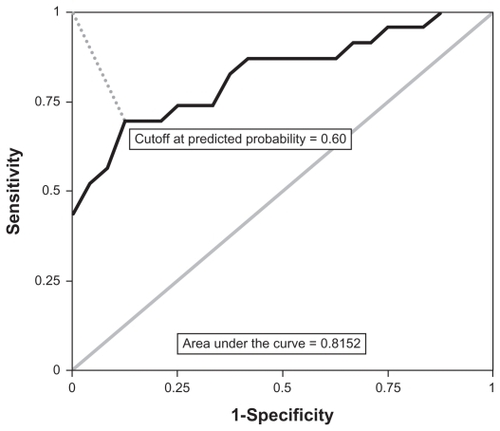
In the second set of analyses, which utilized the sub-sample of 24 non-cases with diagnosis <CIN one and 23 cases diagnosed with CIN 3, significant univariate associations were detected between case status and the following: race, 9 peaks, and 2 peak by BMI interactions. Thus, the initial multivariable model included 11 peaks, 2 interactions, and main effects for BMI and race. The final multivariable model, however, included only 2 components: (1) a main effect for peak [m/z = 4459], and (2) a main effect for race. After determining the optimal cutoff point with a ROC curve (), the final multivariate logistic model accurately classified 21 non-cases and 16 cases. However, 7 cases were misclassified as non-cases, and 3 non-cases were misclassified as cases. Concordance with case status was 78.7%; concordance beyond chance, as measured by the Kappa statistic, was estimated at 0.57 (95% CI = 0.34, 0.80). The area under the ROC curve of 0.8152 indicated that the model had acceptable predictive ability.
Figure 4 Receiver operating characteristic curve for CIN 3 and controls diagnosed with <CIN 1.
The leave-one-out cross-validation algorithm produced similar prediction results compared to those from the original model. Measures of cross-validated sensitivity and specificity for the final multivariate model as well as for each of its two components are shown in . As can be seen in this table, the final model provided more accurate specificity compared with each of its individual components alone, but the sensitivity decreased compared to the race-only model. Again, this observed variability in the sensitivity and specificity measures, when comparing the final model vs the component-only models, suggests some instability in the prediction, caused by variability in the probability of being a case that remained unexplained by the logistic regression models.
Table 2 Cross-validated screening test measures for the final multivariate model and individual models with the two components of the final model predicting cervical intraepithelial neoplasia (CIN) 3 vs <CIN 1 (n = 47)
Prediction results by race
After determining that peaks [m/z = 4459] and [m/z = 4154] showed the strongest association with case status, the prediction results using these two peaks were compared by race. shows cross-validated sensitivity and specificity measures tabulated by race, for a model predicting CIN 2+ using peak [m/z = 4459] as the only predictor. Because the effect of increasing [m/z = 4154] intensity on CIN 2+ was opposite by race (), two models predicting CIN 2+, using [m/z = 4154] intensity as the only predictor, were fitted for each race, respectively. Cross-validated sensitivity and specificity for these models are shown in . shows cross-validated sensitivity and specificity measures tabulated by race, for a model predicting CIN 3 using peak [m/z = 4459] as the only predictor, among the subsample of 24 non-cases with diagnosis <CIN 1 and 23 cases diagnosed with CIN 3. As can be seen in this table, sensitivity was higher for CAs, but specificity was higher for AAs, suggesting again that the predictive ability of the [m/z = 4459] intensities on extreme diagnoses might vary by race.
Table 3 Cross-validated screening test measures by race for a model predicting cervical intraepithelial neoplasia (CIN) 2+ vs ≤CIN 1, using peak [m/z = 4459] as the only predictorTable Footnotea
Table 4 Cross-validated screening test measures by race for a model predicting cervical intraepithelial neoplasia (CIN) 2+ vs ≤CIN1, using peak [m/z = 4154] as the only predictor
Table 5 Cross-validated screening test measures by race for a model predicting cervical intraepithelial neoplasia (CIN) 3 vs <CIN 1, using peak [m/z = 4459] as the only predictor (n = 47)Table Footnotea
Discussion
An important application of MALDI-TOF MS is the simultaneous analysis of multiple proteins to establish “fingerprint” profiles that discriminate disease from non-disease. This is an important approach, since no single biomarker or protein alone will improve the early detection/diagnosis of diseases, including cancer or pre-cancers. Body fluids, such as serum, are a source of putative protein biomarkers with the potential to elucidate organ-specific carcinogenic events. Because of its high sensitivity for proteins in the low molecular weight range and because of its capability of high throughput screening, MALDI-TOF MS has been used to distinguish healthy controls from patients diagnosed with several cancers including the colon, lung, ovary, breast and esophagus.Citation11–Citation15 Previous studies have also documented differences in serum protein profiles between cervical cancer and healthy controls. Citation16 Three differentially expressed potential biomarkers with relative molecular weights of 3974 Da, 4175 Da and 5906 Da identified in this study demonstrated ~90% sensitivity and specificity in disguising cervical cancer from controls. To our knowledge, the current study is the first to document the usage of MALDI-TOF-MS technology in the analysis of serum protein profiles of patients diagnosed with cervical pre-cancer and evaluated differences in sensitivity and specificity of protein peaks by race.
We focused on women infected with HPV 16 as this virus is the most frequent causative agent for developing cervical cancer world-wide.Citation17 Even though only a fraction of women infected with HPV 16 develops CIN 2+, these lesions have the highest rate of progression to CC.Citation18 Further, the recurrence rate of CIN 2+ after a loop electrosurgical excision procedure was shown to be significantly higher among those who were tested positive for HPV 16 before and after the procedure.Citation19 Therefore, identification of this fraction of women and treatment of their lesions and closer follow-up after treatment are important unmet medical needs in the current management protocols. Currently available tests do not have adequate specificity for identifying women with HPV 16-associated CIN 2+.
In our population, 40% of women infected with HPV 16 were diagnosed with CIN grades higher than 2 (CIN 2+). Identification, treatment, and closer follow-up of these women would offer a cost-effective strategy to reduce the cervical cancer burden. A meta-analysis showed that detecting any HR-HPV by the Hybrid Capture 2 test among women with abnormal pap demonstrated 97.2% sensitivity for detecting CIN 2+ and 97.1% sensitivity for detecting CIN 3+. This analysis also demonstrated a pooled specificity of 30.6% and 26.1% when the outcome was CIN 2+ and CIN 3+ respectively. Citation20 A recent study demonstrated that the sensitivity of the HPV 16/18 genotyping test for detection of CIN 2+ was >93% while the specificity of the test for detection of CIN 2+ and CIN 3+ was 44.2% and 43%, respectively.Citation21
Two protein peaks identified by our study demonstrated higher specificity for identifying CIN 2+ than these published studies. An increasing intensity of [m/z = 4459] was associated with a higher risk of being a case, regardless of race with a specificity of 58% for CIN 2 and a specificity of 75% for CIN 3. Further, to our knowledge for the first time, we also document interesting racial differences in the associations between peak intensities and higher risk of being diagnosed with CIN 2+. An increasing intensity of [m/z = 4154] was not only associated with a higher risk of being a case only among CAs, but also had an opposite effect among African AAs. With [m/z = 4459], the specificity was higher for AAs, but the sensitivity was higher for CAs suggesting that the predictive ability of the [m/z = 4459] peak intensities varies by race. With [m/z = 4154], on the other hand, the sensitivity was similar for the two races, but the specificity was higher for AAs, indicating that the peak had slightly better predictive ability among AA women.
This report suggests a capacity of serum protein profiles to differentiate between HPV 16 positive women free of true pre-neoplastic lesions (≤CIN 1) and women diagnosed with higher grades of CIN, especially CIN 3, in our population of women infected with HPV 16. In the statistical analyses, infection with other types of HR-HPVs was used as a predictor of case status, by itself and as an interaction with the m/z peaks, but no significant association was found. Therefore, these results suggest that it is unlikely that co-infections with other HPVs interfere with identifying CIN lesions in women infected with HPV 16. Further, the results also suggested that specific serum profiles might be useful for differentiating CIN cases from non-cases in AAs and CAs.
Identification of specific proteins associated with peaks that are significantly different between ≤CIN 1 and CIN 2+ by race may potentially lead to the development of new screening tests which are suitable in different racial groups. Identification of these serum proteins and development of antibody-based tests, such as ELISA, may lead to the development of cost-effective, non-invasive, sensitive, and simple to use tests. Further, serum based screening tests are likely to be more acceptable than cervical cell based tests in populations where the prevalence of obesity is high, as studies indicate lower rates of cervical cancer screening among obese compared with non-obese women due to embarrassment and perceived weight stigma.Citation22 Also, lack of appropriately sized equipment for examination of obese women in some clinical settings may lead to poor quality cervical cell samples that result in unreliable or invalid test results. Therefore, serum based screening biomarkers are likely to be extremely useful in this group of women. Collectively, these results suggest the need for developing race-specific markers to maximize the usefulness of serum protein based tests as effective screening tools. Despite intense screening in the past decades, higher rates of cervical cancer still persist among some sub-groups of women, including AA women.Citation23 Race-specific screening tests are likely to reduce these disparities.
Although these prediction results using serum biomarkers are promising, there was still a considerable amount of variability in the probabilities of case status that remained unexplained by the statistical models used in this study. A classical screening approach used in this study permitted the detection of the two individual peaks associated with case status. Only interactions between individual peaks and 4 participant characteristics (age, BMI, race, and infection with multiple HR-HPV types) were considered. However, peak by peak interactions (or peak by peak profiles) were not considered in either the classical screening approach or in the alternative LASSO and LAR peak selection procedures, due to the overwhelming number of possibilities. With 95 peaks, restricting the interactions or profiles to only a maximum of four peaks at a time, there are 4465 possible 2-way interactions, 138,415 possible 3-way interactions, and 3,183,545 possible 4-way interactions. Because interactions (or peak by peak profiles) cannot be ruled out from being associated with case status, further examination of these peak by peak profiles using larger sample sizes and data mining techniques is warranted in future studies.
Acknowledgment
This publication was supported by Grant Number (U54 CA118948-01) from the National Cancer Institute.
Disclosure
The authors have no conflicts of interest that are directly relevant to the content of this study.
References
- ShantaVKrishnamurthiSGajalakshmiCKSwaminathanRRavichandranKEpidemiology of cancer of the cervix: global and national perspectiveJ Indian Med Assoc200098495211016150
- MuirCSEpidemiology, basic science, and the prevention of cancer: implications for the futureCancer Res199050644164482208102
- ForhanSEGottliebSLSternbergMRPrevalence of sexually transmitted infections among female adolescents aged 14 to 19 in the United StatesPediatrics20091241505151219933728
- SunXWKuhnLEllerbrockTVChiassonMABushTJWrightTCJrHuman papillomavirus infection in women infected with the human immunodeficiency virusN Engl J Med1997337134313499358128
- FerenczyACoutleeFFrancoEHankinsCHuman papillomavirus and HIV co-infection and the risk of neoplasias of the lower genital tract: a review of recent developmentsCMAJ200316943143412952805
- SroczynskiGSchnell-InderstPMühlbergerNDecision-analytic modeling to evaluate the long-term effectiveness and cost-effectiveness of HPV-DNA testing in primary cervical cancer screening in GermanyGMS Health Technol Assess27 Apr20106 Doc05
- PiyathilakeCJOelschlagerDKMelethMSPartridgeEEGrizzleWEPlasma protein profiles differ between women diagnosed with cervical intraepithelial neoplasia (CIN) 1 and 3Cancer Informatics2007234534919458776
- PiyathilakeCJMacalusoMAlvarezRDBellWCHeimburgerDCPartridgeEELower risk of cervical intraepithelial neoplasia in women with high plasma folate and sufficient vitamin B12 in the post-folic acid fortification eraCancer Prev Res (Phila Pa)20092658664
- HastieTTibshiraniRFriedmanJThe Elements of Statistical Learning2nd edNew YorkSpringer-Verlag2009
- HosmerDWLemeshowSApplied Logistic Regression2nd edJohn Wiley & Sons, IncNew York, NY2000
- LiuCPanCShenJWangHYongLMALDI-TOF MS combined with magnetic beads for detecting serum protein biomarkers and establishment of boosting decision tree model for diagnosis of colorectal cancerInt J Med Sci20118394721234268
- LinXLYangSYDuJDetection of lung adenocarcinoma using magnetic beads based matrix-assisted laser desorption/ionization time-of-flight mass spectrometry serum protein profilingChin Med J (Engl)2010123343920137572
- QiuFLiuHYDongZNFengYJZhangXJTianYPSearching for potential ovarian cancer biomarkers with matrix-assisted laser desorption/ ionization time-of-flight mass spectrometryAm J Biomed Sci20091809020664751
- PietrowskaMMarczakLPolanskaJMass spectrometry-based serum proteome pattern analysis in molecular diagnostics of early stage breast cancerJ Transl Med200976019594898
- GuoRPanCShenJLiuCNew serum biomarkers for detection of esophageal carcinoma using Matrix-assisted laser desorption/ionization time-of-flight mass spectrometryJ Cancer Res Clin Oncol201113751351920490864
- LiuCPanCShenJWangHYongLZhangRDiscrimination analysis of mass spectrometry proteomics for cervical cancer detectionMed Oncol2010Nov 16 [Epub ahead of print]
- BoschFXde SanjoseSChapter 1: human papillomavirus and cervical cancer--burden and assessment of causalityJ Natl Cancer Inst Monogr200331312807939
- MatsumotoKOkiAFurutaRJapan HPV And Cervical Cancer (JHACC) Study GroupPredicting the progression of cervical precursor lesions by human papillomavirus genotyping: a prospective cohort studyInt J Cancer20101282898291020734388
- KangWDOhMJKimSMNamJHParkCSChoiHSSignificance of human papillomavirus genotyping with high-grade cervical intraepithelial neoplasia treated by a loop electrosurgical excision procedureAm J Obstet Gynecol201020372.e1e620417477
- CuzickJArbynMSankaranarayananROverview of human papillomavirus-based and other novel options for cervical cancer screening in developed and developing countriesVaccine200826Suppl 10K29K4118847555
- EinsteinMHMartensMGGarciaFAClinical validation of the Cervista HPV HR and 16/18 genotyping tests for use in women with ASC-US cytologyGynecol Oncol201011811612220488510
- AldrichTHackleyBThe impact of obesity on gynecologic cancer screening: an integrative literature reviewJ Midwifery Womens Health20105534435620630361
- SaraiyaMAhmedFKrishnanSRichardsTBUngerERLawsonHWCervical cancer incidence in a prevaccine era in the United States, 1998–2002Obstet Gynecol200710936037017267837