Abstract
Purpose
We retrospectively analyzed the results of prenatal diagnosis in women with high-risk (HR) serological screening results, and discussed the reasonable application of diagnostic testing.
Patients and Methods
Diagnostic testing was done in 2239 pregnant women who had HR results from serological screening in two prenatal diagnosis centers. According to the HR results, they were divided into simple HR, HR combined with ultrasound abnormalities, and HR combined with other indication groups. After receiving counselling from clinicians, they were allowed to choose either the traditional karyotype analysis and/or chromosomal microarray analysis (CMA).
Results
Those who underwent CMA comprised 49.3%, 97.6%, and 100% of the HR group, HR combined with ultrasound abnormalities, and HR combined with other indication groups, respectively. Among the 100 (4.47%) clinically significant results, 55 (2.46%), 15 (0.67%), and 30 (1.34%) were chromosomal aneuploidies, chromosomal structural abnormalities, and pathogenic copy number variations (CNVs), respectively. The rate of abnormalities was 3.77%, 13.71%, and 19.05% in the simple HR, HR combined with ultrasound abnormalities, and HR combined with other indication groups, respectively. The increasing rate of clinical pathogenic CNVs was 1.34% using CMA in HR pregnant women, 9.52% in the HR combined with other indication group, and 1.24% in the simple HR group. Among the 573 women who chose both diagnostic tests, 45 had abnormal results. Only one case detected using karyotype analysis was missed on CMA. The incidence of chromosomal aneuploidy tended to increase with increase in HR values. However, chromosomal structural abnormalities and pathogenic CNVs did not increase.
Conclusion
CMA should be recommended as the first-line diagnostic testing for women with HR screening results, especially combined with other abnormal indications.
Introduction
It is well known that prenatal screening and diagnosis are the effective ways to avoid severe birth defects. In the past three decades, prenatal screening in the first and/or second trimester has been widely used in pregnant women.Citation1 Gestational age, maternal age and weight, maternal biochemical markers, and ultrasound measurements are the most common methods used.Citation2 According to previous studies, the detection rate of Down syndrome was 50%~75% in the second trimesterCitation3,Citation4 and 75%~85% in the first trimester.Citation5,Citation6 The false-positive rate (FR) is considerableCitation7,Citation8 and women with high-risk (HR) results from serological screening should undergo diagnostic testing. However, due to the availability and cost-effectiveness of serological screening, it is still being used as the first-line screening method in China.
With the development of the high-throughput sequencing technology, noninvasive prenatal screening (NIPS) is widely used as the first-line screening method for fetal trisomy 21, 18, and 13 in some countries, whereas it is still considered as the second-line test in China. NIPS is mainly used for women with intermediate-risk results from prenatal serological screening, and in women with high risk or advanced maternal age who refuse invasive prenatal screening. Nevertheless, NIPS is still an effective method for prenatal screening.Citation9–Citation11 Recently, many pregnant women were willing to undergo NIPS due to its high accuracy. Current diagnostic method mainly includes karyotype analysis, chromosomal microarray analysis (CMA), fluorescence in situ hybridization (FISH), rapid PCR.Citation12 Karyotype analysis is regarded as the traditional method for prenatal diagnosis. It can detect major chromosomal abnormalities, such as aneuploidy, unbalanced rearrangements, translocation, and mosaicism. As a high-resolution genomic technology without the need of cell culture, CMA is becoming widely applied in prenatal diagnosis. It has considerable diagnostic and prognostic values but has not fully replaced karyotype analysis. It offers additional diagnostic benefits by revealing sub-microscopic imbalances or copy number variations (CNVs), which cannot be detected by standard karyotype analysis.Citation13,Citation14 Currently, karyotype analysis and CMA are the most common methods for prenatal diagnosis in China. According to the technical specifications of prenatal diagnosis in China, the appropriate gestational age for prenatal diagnosis in the second trimester is from 18 weeks to 23 weeks 6 days. Within this gestational range, both karyotype analysis and CMA can be used as prenatal diagnostic tests. Women with simple HR results from prenatal serological screening are allowed to choose either karyotype analysis and/or CMA for diagnostic testing. Clinicians usually recommended CMA as the first choice for women at high risk with abnormalities detected on ultrasound or chromosome abnormalities from one of the partners may. Most prenatal women usually choose both methods. However, it is unclear whether to use both diagnostic tests for pregnant women at high risk. In this study, we aimed to discuss this interesting question and provide scientific guidance for clinical diagnosis.
Materials and Methods
Patients and Design
A total of 2239 pregnant women with HR results from serological screening from February 2016 to December 2018 underwent prenatal diagnosis at two prenatal diagnosis centers: the Changzhou Maternity and Child Health Care Hospital affiliated to Nanjing Medical University and the Lianyungang Maternal and Child Health Hospital affiliated with Yangzhou University.
Depending on the gestational age at the time of visit, clinicians recommended the pregnant woman to undergo karyotype analysis and/or CMA. For pregnant women whose gestational age was suitable for karyotype analysis (from 18 weeks to 23 weeks 6 days), clinicians would recommend both karyotype analysis and CMA. The pregnant women were allowed to choose which method they want to use. Clinicians usually recommend CMA for pregnant women with a gestational age greater than 23 weeks 6 days.
Samples Collect
Gestational age was calculated using the last menstrual period or ultrasonography. A 3 mL of blood was collected from each pregnant woman by simple needle aspiration. After being placed at room temperature for 0.5 h, the samples were centrifuged at 3000 rpm for 5 minutes and the cells removed. The serum was stored at 4°C in assays for 7 days and at −80°C for long term.
Prenatal Screening in the Second Trimester
As described in our previous studies,Citation9,Citation15 all the subjects received prenatal screening in the second trimester after genetic counseling and obtaining their informed consent. Their blood samples were collected between 15 weeks 0 days and 22 weeks 6 days. The levels of α-fetoprotein (AFP) and free β subunit human chorionic gonadotropin (fβhCG) were quantified by time-resolved fluoroimmunoassay (TRFIA) using Wallac 1235 AutoDELFIA (DELFIA1235: Perkin Elmer, Waltham, MA). Combined with maternal age, gestational age, maternal weight, and insulin-dependent diabetes, the risk values were calculated using the Lifecycle software (4.0), including the risk value of neural tube defects (NTD), T21 and T18. High risk: T21 >1/300, T18 >1/350. The intermediate risk was T21 1/300~1/1000, T18 1/350~1/1000.Citation16–Citation18 Women aged ≥35 were considered at an advanced maternal age.
Women with HR results received genetic counseling. Most of them underwent prenatal diagnosis. The pregnant women voluntarily chose traditional karyotype analysis and/or CMA after receiving genetic counselling from the clinicians. Both centers used the same detection platform, experimental scheme, and quality control standards, and participated in the laboratory quality control evaluation plan.
Karyotype Analysis for Prenatal Diagnosis
As described in our previous studies,Citation19,Citation20 pregnant women received amniocentesis between 18 weeks 0 days and 23 weeks 6 days. All experiments were performed by two individuals using two independent cell culture systems. After cell culture and sample preparation, GSL-120 (Leica Biosystems Richmond, Inc) and software (CytoVision Automated Cytogenetics Platform) were used for chromosome karyotype scanning and analysis. At least five karyotypes were analyzed and 20 karyotypes were counted. Sixty to one hundred karyotypes were counted for the cases with chromosome mosaics.
Chromosomal Microarray Analysis (CMA) for Prenatal Diagnosis
The procedure for prenatal CMA was described in our previous study.Citation21 After the amniotic fluid was collected, DNA was extracted by QIAamp DNA Mini Kit (Qiagen Inc., Valencia, CA). A 250 ng DNA was amplified, labeled, and hybridized to GCS 3000Dx v.2 platform (Affymetrix, USA). Single nucleotide polymorphism (SNP) array test was processed by a commercial 750K microarray chip (Affymetrix CytoScan 750K Array). After hybridization with fragmented DNA, the chip was washed with buffer and scanned by a laser scanner. Data were analyzed using Chromosome Analysis Suite v3.2 (ChAs) software package.
Statistical Analysis
The data were analyzed using EmpowerStats software (X&Y solutions, inc.) and R (http://www.R-project.org).Citation22 The chi-squared test was used to compare the differences in continuous variables between the two groups. In all the analyses, a P < 0.05 was considered as statistically significant.
Results
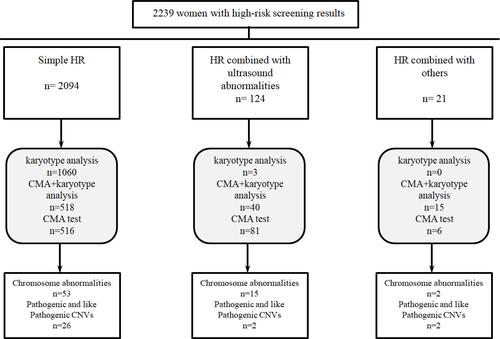
In this study, 2239 pregnant women with HR results from serological screening received a prenatal diagnosis in two centers. According to their HR results, they were divided into three groups: simple HR (2094 cases), HR combined with ultrasound abnormalities (124 cases), and HR combined with other indications (21 cases, mainly including couples with chromosomal abnormalities, history of adverse pregnancy, advanced maternal age, etc.). All the pregnant women were reminded by phone calls, consulted with clinicians, and underwent prenatal diagnosis. They were informed about the two diagnostic tests (karyotype analysis and CMA) and allowed to choose based on their indications and willingness to perform the test. Of the 2094 pregnant women with simple HR, 1060 (50.6%), 516 (24.6%), and 518 (24.7%) requested only traditional karyotype analysis, only CMA, and both diagnostic tests, respectively. Unlike the simple HR group, most women in the HR combined with other abnormal indication groups requested for CMA for prenatal diagnosis. The rates of choosing CMA for prenatal diagnosis were 97.6% (121/124) and 100% (21/21) in the two groups other than the simple HR group, respectively. summarizes the study results.
shows all the abnormal results of prenatal diagnosis and the differences among the three groups. Among the 100 (4.47%) cases with clinically significant results 55 (2.46%), 15 (0.67%), and 30 (1.34%) were chromosome aneuploidy, chromosome structural abnormalities, and pathogenic CNVs. The rate of abnormalities (including chromosome aneuploidy, chromosome structural abnormalities and pathogenic CNVs) was 3.77%, 13.71%, and 19.05% in the simple HR, HR combined with ultrasound abnormalities, and HR combined with other indication groups, respectively. Compared with the simple HR group, the detection rate of chromosomal abnormalities and pathogenic CNVs increased significantly in the other two groups (P<0.05). Among the abnormal results, 30 women were confirmed to have pathogenic CNVs (), all of which were detected by CMA but missed by karyotype analysis. The increasing rate of clinical pathogenic CNVs was 1.34% using CMA in HR pregnant women. The pathogenic CNVs were mainly detected in the HR combined with other indications (9.52%) group. However, even in pregnant women with simple HR, 1.24% of pathogenic CNVs were detected. CMA was recommended for women with simple HR results after a prenatal screening. They received prenatal counseling from a genetic professional. Among the 30 pathogenic CNVs, 16 had reduced and/or variable penetrance and expressivity, combined with the characteristics of pathogenicity, penetrability, familial inheritance, and so on. Eight of the 30 cases chose the follow-up clinical treatment and continued pregnancy. Considering the severity of the pathogenic CNV and the adverse consequences of continuing pregnancy, 22 of the pregnant women decided to terminate the pregnancy. We followed up all the women who decided to continue with the pregnancy for 5 to 16 months. None of them showed significant abnormalities during growth and development. We found 82 cases of variants of uncertain significance (VUS) and two were likely benign (LB). These cases had been followed for approximately 2 years. One case was stillbirth. Among the 72 babies that were born, one had neonatal intestinal obstruction, while the others did not have any abnormalities.
Table 1 The Results of Prenatal Diagnosis in the Women with High Risk After Serological Screening
Table 2 The Results of Pathogenic CNVs in the Women with High Risk After Serological Screening
In this study, 573 women chose both karyotype analysis and CMA for prenatal diagnosis. Forty-five abnormal results were found. Among these abnormalities, 24 cases were detected by both diagnostic tests, 20 only by CMA, and one by only karyotype analysis. Furthermore, 21 chromosome polymorphisms were detected by karyotype analysis, but the results were not clinically significant in CMA. Although severe defects are not usually encountered, long-term follow-up is still recommended.
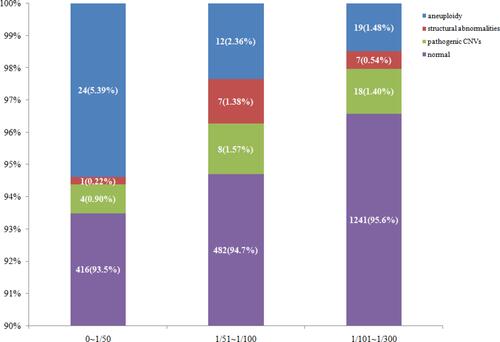
shows the relationship between the HR values and the detected abnormalities. According to the HR values, we divided 2239 women into three groups for comparison: HR values ranged from 0 to 1/50, 1/51 to 1/100 and 1/101 to 1/300. We found that the incidence of chromosomal aneuploidy tended to increase with increase in the HR values. However, chromosomal structural abnormalities and pathogenic CNVs did not show such trend. As shown in and , the rate of aneuploidy was highest in women with HR values ranged 0 to 1/50, reaching 5.39%, while the rates in the other two groups were 2.36% and 1.48% (P<0.05), respectively.
Table 3 Relationship Between the Value of High Risk and Abnormal Rate [n (%)]
Discussion
The procedures for prenatal screening and diagnosis are basically similar, including risk assessment for major aneuploidies first, followed by invasive diagnostic tests for HR women.Citation23 For decades, karyotype analysis from chorionic villus sampling (CVS) and amniocentesis samples has been the most common diagnostic testing for identifying pregnancies affected by chromosomal disorders. Recent developments in prenatal screening and diagnosis include the application of NIPS and CMA. NIPS enables the number of invasive tests for aneuploidies to be reduced considerably.Citation20 CMA can identify a much broader range of abnormalities than karyotyping.Citation24 The diversity of screening and diagnostic methods make prenatal consultation challenging. How to apply the methods reasonably and effectively is an important scientific issue.Citation25 However, most studies only focused on how many abnormal results of prenatal diagnosis were found in HR women, without comparing the different diagnostic methods. In this study, we preliminarily analyzed these problems retrospectively. Our results show that CMA should be recommended as the first-line diagnostic testing for all women with HR screening results, especially when the HR results were combined with other abnormal indications.
Firstly, CMA should be recommended as diagnostic test for women with simple HR results after prenatal screening. Among the 2094 pregnant women with simple HR results, the abnormal rate of prenatal diagnosis was 3.77%, of which 1.24% could only be detected by CMA, accounting for 32.9% of the total abnormalities. In pregnant women with HR results combined with ultrasound abnormalities or other indications, it is necessary to choose CMA as the diagnostic test, which would increase the abnormal rate by 3.6 and 5.1 times, respectively. This increasing trend was manifested not only in chromosomal abnormalities (aneuploidy, structural abnormalities), but also in pathogenic CNVs. There has been no report on the effect of prenatal CMA tests on pregnant women with HR results from serological screening. However, some studies have shown that CMA has beneficial effects on prenatal diagnosis in the general population. Wapner et alCitation24 reported that clinically significant CNVs were observed in 1.7% of fetuses with a normal karyotype in women with advanced maternal age. Levy’s groupCitation13 reported that in prenatal diagnostic samples with a normal karyotype, CMA would diagnose clinically significant CNVs in approximately 1% of structurally normal pregnancies and 6% with ultrasound anomalies. Meanwhile, in 2013, the American Congress of Obstetricians and Gynecologists (ACOG) and the Society for Maternal-Fetal Medicine (SMFM) recommended that CMA should be offered to women with structurally normal fetuses undergoing diagnostic testing for positive aneuploidy screening or maternal anxiety.Citation26 Our findings support this suggestion. However, the application of CMA for routine prenatal diagnosis is still under investigation. After all, prenatal genetic counseling may occasionally be difficult because of the uncertain phenotype with some CNVs.Citation27
Secondly, CMA could replace conventional karyotype analysis in prenatal diagnosis as the first-line test. Among the 573 women who received both karyotype analysis and CMA, the application of CMA did not only detect 96% of the abnormal results in conventional karyotype analysis, but also detected 20 cases of pathogenic CNVs. Only one case was confirmed as 46,XY,inv(1) (p13q21), which was found by karyotype analysis, while missed by CMA. However, it is well known that chromosomal inversion does not lead to an increase or decrease in gene number, but only changes the position of the gene. The clinical phenotypes are mostly normal, although it may increase the risk of stillbirth and abortion. Our finding is also supported by other studies. Many researchers suggested that CMA should be used as the first-line diagnostic test for all pregnant women undergoing invasive prenatal testing, regardless of risk factors.Citation28,Citation29
Thirdly, the incidence of chromosomal aneuploidy tended to increase with increase in the HR values. However, chromosomal structural abnormalities and pathogenic CNVs did not increase. This is also an interesting discovery. It indicates that risk factors are not important referencing index for choosing diagnostic testing. In addition, we found 41 cases of chromosome polymorphism in prenatal diagnosis using karyotype analysis, mainly including pericentric inversion, chromosome constriction, and so on. However, their CMA results did not suggest pathogenic or likely pathogenic CNVs. No serious abnormalities in newborns have been found so far. Therefore, the CMA test can be used during prenatal consultations and can greatly reduce mothers stress.
Conclusion
CMA should be recommended as the first-line diagnostic test for all women with HR screening results, especially when they are combined with other abnormal indications.
Ethics Approval and Consent to Participate
The study design and protocol were reviewed and approved by the ethics committee of Changzhou Maternity and Child Health Care Hospital affiliated to Nanjing Medical University (No. 2017003). All pregnant women received genetic counseling and signed a written consent before the test. All procedures performed in this study were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Consent for Publication
Informed consent was obtained from all individual participants included in the study.
Acknowledgments
We thank all the project participants for their contributions.
Disclosure
The authors declare that they have no conflicts of interest.
Additional information
Funding
References
- Wald NJ. Prenatal screening for open neural tube defects and Down syndrome: three decades of progress. Prenat Diagn. 2010;30(7):619–621. doi:10.1002/pd.2517
- Yao Y, Liao Y, Han M, Li SL, Luo J, Zhang B. Two kinds of common prenatal screening tests for Down’s syndrome: a systematic review and meta-analysis. Sci Rep. 2016;6:18866. doi:10.1038/srep18866
- Spencer K. Second trimester prenatal screening for Down’s syndrome using alpha-fetoprotein and free beta hCG: a seven year review. BJOG. 1999;106(12):1287–1293. doi:10.1111/j.1471-0528.1999.tb08183.x
- Wang YY, Luo J, Zhu MW, Liu LN, Ma X. Second-trimester double or triple screening for Down syndrome: a comparison of Chinese and Caucasian populations. Int J Gynaecol Obstet. 2006;94(1):67–72. doi:10.1016/j.ijgo.2006.04.030
- Schielen PC, van Leeuwen-spruijt M, Belmouden I, Elvers LH, Jonker M, Loeber JG. Multi-centre first-trimester screening for Down syndrome in the Netherlands in routine clinical practice. Prenat Diagn. 2006;26(8):711–718. doi:10.1002/pd.1486
- Wright D, Spencer K, Nix B. First trimester screening for Down syndrome using free beta hCG, total hCG and PAPP-A: an exploratory study. Prenat Diagn. 2007;27(12):1118–1122. doi:10.1002/pd.1844
- Wright D, Syngelaki A, Bradbury I, Akolekar R, Nicolaides KH. First-trimester screening for trisomies 21, 18 and 13 by ultrasound and biochemical testing. Fetal Diagn Ther. 2014;35(2):118–126. doi:10.1159/000357430
- Spaggiari E, Czerkiewicz I, Sault C, et al. Impact of including or removing nuchal translucency measurement on the detection and false-positive rates of first-trimester down syndrome screening. Fetal Diagn Ther. 2016;40(3):214–218. doi:10.1159/000442198
- Yu B, Lu BY, Zhang B, et al. Overall evaluation of the clinical value of prenatal screening for fetal-free DNA in maternal blood. Medicine (Baltimore). 2017;96(27):e7114. doi:10.1097/MD.0000000000007114
- Zhang B, Lu BY, Yu B, et al. Noninvasive prenatal screening for fetal common sex chromosome aneuploidies from maternal blood. J Int Med Res. 2017;45(2):621–630. doi:10.1177/0300060517695008
- Zhou Q, Zhu ZP, Zhang B, Yu B, Cai ZM, Yuan P. Clinical features and pregnancy outcomes of women with abnormal cell-free fetal DNA test results. Ann Transl Med. 2019;7(14):317. doi:10.21037/atm.2019.06.57
- Cherry AM, Akkari YM, Barr KM, et al. Diagnostic cytogenetic testing following positive noninvasive prenatal screening results: a clinical laboratory practice resource of the American College of Medical Genetics and Genomics (ACMG). Genet Med. 2017;19(8):845–850. doi:10.1038/gim.2017.91
- Levy B, Wapner R. Prenatal diagnosis by chromosomal microarray analysis. Fertil Steril. 2018;109(2):201–212. doi:10.1016/j.fertnstert.2018.01.005
- Ganapathi M, Nahum O, Levy B. Prenatal diagnosis using chromosomal SNP microarrays. Methods Mol Biol. 2019;1885:187–205.
- Jiang T, Ding J, Zhang XQ, et al. Analysis of Down syndrome failed to be diagnosed after prenatal screening: a multicenter study. Medicine (Baltimore). 2017;96(24):e7166. doi:10.1097/MD.0000000000007166
- Wald NJ, Hackshaw AK, Walters J, et al. First and second trimester antenatal screening for down’s syndrome: the results of the Serum, Urine and Ultrasound Screening Study (SURUSS). J Med Screen. 2003;10(2):56–104.
- Wald NJ, Rodeck C, Hackshaw AK, et al. SURUSS in perspective. BJOG. 2004;111:521–531. doi:10.1111/j.1471-0528.2004.00193.x
- Malone FD, Canick JA, Ball RH, et al. First-trimester or second-trimester screening, or both, for Down’s syndrome. N Engl J Med. 2005;353(19):2001. doi:10.1056/NEJMoa043693
- Chen YP, He ZQ, Shi Y, et al. Not all chromosome aberrations can be detected by NIPT in women at advanced maternal age: a multicenter retrospective study. Clin Chim Acta. 2018;486:232–236. doi:10.1016/j.cca.2018.08.018
- Yu B, Li H, Chen YP, et al. Clinical evaluation of NIPS for women at advanced maternal age: a multicenter retrospective study. J Matern Fetal Neonatal Med. 2018;32:1–6.
- Shi Y, Ma J, Xue Y, Wang J, Yu B, Wang T. The assessment of combined karyotype analysis and chromosomal microarray in pregnant women of advanced maternal age: a multicenter study. Ann Transl Med. 2019;7(14):318. doi:10.21037/atm.2019.06.63
- Yu B, Long W, Yang Y, et al. Newborn screening and molecular profile of congenital hypothyroidism in a Chinese population. Front Genet. 2018;9:509. doi:10.3389/fgene.2018.00509
- de Jong A, Maya I, van Lith JM. Prenatal screening: current practice, new developments, ethical challenges. Bioethics. 2015;29:1–8. doi:10.1111/bioe.12123
- Wapner RJ, Martin CL, Levy B, et al. Chromosomal microarray versus karyotyping for prenatal diagnosis. N Engl J Med. 2012;367(23):2175–2184. doi:10.1056/NEJMoa1203382
- Carlson LM, Vora NL. Prenatal diagnosis: screening and diagnostic tools. Obstet Gynecol Clin North Am. 2017;44(2):245–256. doi:10.1016/j.ogc.2017.02.004
- Wou K, Levy B, Wapner RJ. Chromosomal microarrays for the prenatal detection of microdeletions and microduplications. Clin Lab Med. 2016;36(2):261–276. doi:10.1016/j.cll.2016.01.017
- Rotshenker-Olshinka K, Moshe NS, Weiss O, et al. Preimplantation genetic testing (PGT) for copy number variants of uncertain significance (CNV- VUS) in the genomic era: to do or not to do? J Assist Reprod Genet. 2021;38:719–725. doi:10.1007/s10815-020-02055-3
- Sagi-Dain L, Cohen Vig L, Kahana S, et al. Chromosomal microarray vs. NIPS: analysis of 5541 low-risk pregnancies. Genet Med. 2019;21(11):2462–2467. doi:10.1038/s41436-019-0550-x
- Wu X, An G, Xie X, et al. Chromosomal microarray analysis for pregnancies with or without ultrasound abnormalities in women of advanced maternal age. J Clin Lab Anal. 2019;34(25):e23117.