Abstract
The nuclear factor-κB (NF-κB) plays a central role in the activation and survival of lymphocytes. NF-κB, therefore, is pivotal for acquired immunity, but the dysregulation of NF-κB signaling leads to inflammatory diseases and lymphomagenesis. Accumulating evidence has demonstrated that the mucosa-associated lymphoid tissue (MALT) lymphoma-related molecules, B-cell lymphoma 10 (BCL10) and MALT-lymphoma-translocation gene1 (MALT1), are essential signaling components for NF-κB and mitogen-activated protein kinase (MAPK) activation, mediated by the immunoreceptor tyrosine-based activation motif (ITAM)-coupled receptors involved in both innate and adaptive immunity. CARMA1 (also referred to as CARD11 and Bimp3) is a crucial regulator for ITAM-mediated signaling as it forms a complex with BCL10-MALT1 in lymphoid lineage cells such as T, B, natural killer (NK), and natural killer T (NKT) cells, known as the lymphoid CARMA1-BCL10-MALT1 (L-CBM) complex. In this review, recent understanding of the molecular and biological functions and the signal regulation mechanisms of the L-CBM complex are described and its role in disease development and potential as a therapeutic target is further discussed.
CARMA1-BCL10-MALT1 (CBM) complexes
Immunoreceptors such as T cell receptors (TCRs), B cell receptors (BCRs), Fc receptors (FcRs), activating natural killer cell receptors (NKRs), as well as multiple immunoglobulin (Ig)- or C-type lectin-family receptors expressed on myeloid cells, transduce activation signals by associating with signaling chains (eg, CD3s, Igα, Igβ, FcRγ, and DAP12) containing the immunoreceptor tyrosine-based activation motifs (ITAMs), bearing a consensus sequence of YxxL-x6-8-YxxL (where x denotes any amino acids), in their cytoplasmic domains.Citation1 Upon engagement of these ITAM-coupled receptors, an activation signal cascade is initiated with phosphorylation of specific tyrosines in ITAMs by the Src family of kinases.Citation2 The phosphorylated ITAMs recruit the tyrosine kinase Syk or zeta-associated protein, 70 kDa (ZAP-70), which bind via the tandem Src homology domain 2 (SH2) and initiate a downstream signaling cascade leading to activation of transcription factors such as nuclear factor-κB (NF-κB), nuclear factor activated T-cells (NFAT), and activating protein-1 (AP-1).
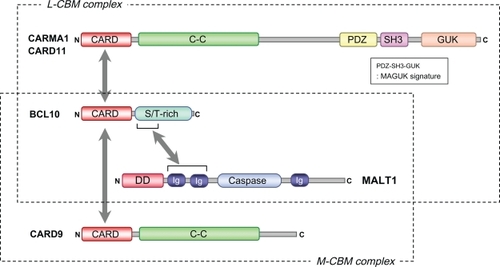
The NF-κB family of transcription factors plays a critical role in immune cell functions such as immune regulation, inflammation, cell survival, and cell cycle progression.Citation3 Accumulating evidence has revealed that the mucosa-associated lymphoid tissue (MALT) lymphoma-related molecules, B-cell lymphoma 10 (BCL10) and MALT-lymphoma-translocation gene1 (MALT1), form a complex and play an essential role in NF-κB activation through all of the ITAM-coupled receptors expressed by immune cells, as well as some G-protein-coupled receptors expressed on nonimmune cells.Citation4 Recent genetic-based studies have identified the two caspase recruitment domain (CARD)-family adaptor molecules, CARMA1 (also known as CARD11 and Bimp3) and CARD9, regulate the BCL10-MALT1-mediated NF-κB, as well as mitogen-activated protein kinase (MAPK) activation signaling in a cell-type-specific, nonredundant manner. CARMA1 acts in lymphoid cells such as T/B lymphocytes and natural killer (NK) cells whereas CARD9 acts in myeloid cells such as macrophages and dendritic cells (DCs),Citation5–Citation7 and thus play a crucial role in both innate and adaptive immune responses. Based on their cell specificity, the CARMA1-BCL10-MALT1 and CARD9-BCL10-MALT1 complexes are referred to as the lymphoid CARMA1-BCL10-MALT1 (L-CBM) complex and the myeloid CARD9-BCL10-MALT1 (M-CBM) complex, respectively ().Citation8 In this article, the role of L-CBM-mediated signaling controlled by CARMA1, with particular emphasis on its regulation for NF-κB activation, in lymphocyte development and activation, as well as in disease, has been reviewed.
Figure 1 L-CBM complex and M-CBM complex.
Abbreviations: L-CBM, lymphoid-CARMA1-BCL10-MALT1; M-CBM, myeloid-CARD9-BCL10-MALT1; CARD, caspase recruitment domain; C-C, coiled-coil domain; PRD, protein-kinase-C-regulated domain; PDZ, PSD95, DLGA and ZO1 homology domain; SH3, Src-homology 3 domain; GUK, guanylate kinase domain; MAGUK, membrane-associated guanylate kinase; S/T-rich, serine/threonine-rich domain; DD, death domain; Ig, immunoglobulin-like domain.
L-CBM signaling in T cells
BCL10 is a member of the CARD family of proteins and associates with Ig-like domains of MALT1 through its serine/threonine (S/T)-rich domain (). When overexpressed, this interaction synergistically induces NF-κB activation. CARMA1 is a member of CARD-containing membrane-associated guanylate kinase (MAGUK) family and was found to be a binding partner of BCL10 via CARD-CARD interaction (), similar to CARD9 and the other members of the CARD-MAGUKs: CARMA2 (CARD14/Bimp2), and CARMA3 (CARD10/Bimp1).Citation9,Citation10 It is thought that the BCL10-MALT1 complex binds to CARMA1 following antigen receptor-triggering to form the L-CBM complex. An association of CARMA1 with MALT1 and BCL10 via a coiled-coil (C-C) domain was also reported, which likely contributes to stabilizing the L-CBM complex ().Citation11,Citation12 Genetic studies using knockout mice of CARMA1, BCL10, and MALT1, as well as a CARMA1-deficient Jurkat cell line, have demonstrated the physiological relevance of the L-CBM complex in TCR and BCR signaling. The T cell phenotype of CARMA1-deficent (CARD11−/−) mice closely resembles that of BCL10-deficient (BCL10−/−) and MALT1-deficient (MALT1−/−) mice.Citation13–Citation16
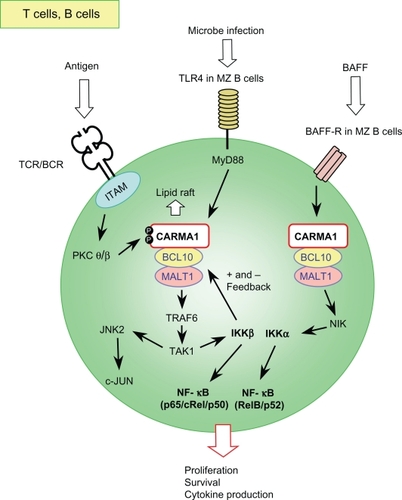
Peripheral mature T cells from these L-CBM-deficient mice show almost complete abrogation of proliferation and cytokine production upon TCR stimulation. As a result, these mice exhibit impaired T cell immunity.Citation13,Citation15 Loss of L-CBM molecules abrogates TCR-induced NF-κB activation owing to defective activation of an inhibitor of κB (IκB) kinase (IKK) (), whereas calcium mobilization and proximal tyrosine phosphorylation are unaffected.Citation13,Citation15 In addition, CARMA1 and MALT1 deficiency affects the activation of the MAPK c-Jun-N-terminal kinases (JNK),Citation14,Citation15 particularly JNK2,Citation17 but not the other MAPKs, Erk and p38 (). The defective JNK2 activation results in reduced expression of c-Jun. The MAP3K, transforming growth factor β kinase 1(TAK1) and the MAPK kinase, MKK7, are likely involved in this pathway as upstream kinases of JNK2 because these molecules have been shown to be associated with BCL10 upon TCR stimulation.Citation17
Figure 2 Schematic signaling pathway mediated by the L-CBM complex in T and B lymphocytes.
L-CBM deficiency did not affect overall development of thymocytes, with normal percentages and numbers of CD4+CD8+, CD4+CD8−, and CD8+CD4− cells, but increased CD4−CD8− double negative (DN) cells.Citation13,Citation15 This increase may be due to defective thymic egression of a T cell subpopulation with a DN phenotype rather than impaired early development of conventional T cell precursors, as these DN cells display a mature T cell phenotype with high levels of CD3 expression. No overt developmental changes in conventional CD4 and CD8 T cells were observed in the peripheral lymphoid organs of mice with L-CBM deficiency; however, the number of naturally occurring regulatory T cells (nTregs) was markedly reduced in the thymus and periphery.Citation18–Citation20 This phenotype could likely be attributed to the defective TCR-mediated NF-κB signaling, as T cells deficient for both PKCθ, the essential upstream kinase of L-CBM, and IKKβ, which controls canonical NF-κB activation pathway through TCRs, also exhibit a similarly significant reduction of nTregs.Citation18 A study using CARD11−/− mice revealed that L-CBM controls an early checkpoint in Treg development by enabling the generation of thymic precursors of Tregs.Citation20 This deficiency is Tregintrinsic, and overexpression of a constitutive active STAT5, an essential mediator of IL-2/IL-15 receptor signaling, as well as of the antiapoptotic protein, BCL2, failed to rescue this deficiency, indicating that L-CBM neither controls IL-2/IL-15 receptor signaling nor mitochondrion-regulated cell survival during Treg development. Thus, L-CBM-mediated signaling may induce undefined factors essential to commit immature thymocytes to the nTreg lineage, although these factors are dispensable to conventional T cell development.
L-CBM signaling in B cells
Reduced follicular (FO) and marginal zone (MZ) B cells in spleen and an almost complete absence of peritoneal B-1 B cells have been consistently observed in CARD11−/−, BCL10−/−, and MALT1−/− mice.Citation14,Citation15,Citation21 This deficiency of L-CBM molecules results in abrogated BCR-induced NF-κB activation and thereby defects in B cell proliferation and survival, although one line of MALT1−/− mice has exhibited only mild defects in B cell activation through a selective defect in c-Rel activation, which controls B cell survival ().Citation14,Citation22 CARMA1, and probably BCL10, control JNK activation through BCRs (),Citation15 whereas MALT1 might be dispensable for it.Citation14,Citation16 CD40-induced activation was also defective in splenic B cells with L-CBM deficiency,Citation13,Citation16,Citation23 possibly owing to defective development of MZ B cells, which are the major cells responding to CD40 stimulation.Citation23
The involvement of L-CBM in Toll-like receptor (TLR) signaling in B lymphocytes has been suggested, although it remains a controversial issue. CARD11−/− whole splenic B cells show impaired proliferation upon TLR4 stimulation, probably due to defective JNK activation.Citation15 A similar deficiency was observed in one of the MALT1−/− mouse models.Citation16 A study that compared FO and MZ splenic B cells revealed that BCL10 deficiency affected only MZ B cells in response to lipopolysaccharide (LPS) due to impaired NF-κB activation ().Citation21 The defective LPS response of whole splenic B cells from L-CBM deficient mice might be due to the reduced number and the impaired response of MZ B cells because MZ B cells have been shown to proliferate much better than FO B cells upon LPS stimulation.Citation24
NF-κB is activated by either a canonical or a noncanonical pathway.Citation25 The canonical pathway, which depends on IKKβ, is activated by engagement of receptors such as TNFR1, IL-1R, TLRs, TCR, and BCR. IKKβ phosphorylates inhibitors of κB molecules bound to NF-κB subunits such as RelA, allowing NF-κB to translocate to the nucleus. The noncanonical pathway, which depends on IKKα, is activated by engagement of receptors for lymphotoxin β (LT-β), RANKL, CD40L, and B-cell activation of the tumor necrosis factor (TNF) family (BAFF). IKKα mediates phosphorylation-dependent processing of the NF-κB2 precursor, p100, resulting in the generation of p52 and its translocation to the nucleus. As discussed so far, L-CBM signaling is crucial for the canonical NF-κB signaling downstream of TCRs and BCRs. However, recent reports have shown that L-CBM also activates the noncanonical pathway and thereby regulates the survival in MZ B cells.Citation26–Citation28 Eμ-driven human BCL10-transgenic (Tg) mice exhibit constitutive activation of both the canonical and noncanonical NF-κB pathway in splenic B cells. The Tg mice show expanded MZ and B1 B cells and reduced FO and B1a cells, with some older mice developing MZ lymphomas. Lack of MALT1 impairs BAFF-induced phosphorylation and degradation of p100 (). The MALT1−/− MZ, but not FO B cells, exhibit reduced survival and antiapoptotic gene induction in response to BAFF in vitro, likely owing to the elevated expression and defective BAFF-induced downregulation of TRAF3, a negative modulator of the BAFF-induced survival signal, particularly in MZ B cells. The phenotypes of BAFF-Tg mice, including increased basal serum Ig, MZ B cells, and B1 B cells, spontaneous germinal center formation, and Ig deposition in the kidney, all disappear in the absence of MALT1 and BCL10. In line with these observations, the B cell phenotype of L-CBM-deficient mice resembles that of the BAFF-R-mutant A/WySnJ mice,Citation29 suggesting a role for L-CBM complex in driving the nonclassical NF-κB pathway in BAFF-mediated survival of B cells.
L-CBM signaling in NK cells
NK cells play as a sentinel in protecting against tumors and intracellular pathogens. NK cell activation is dependant upon multiple germline-encoded activating and inhibitory receptors that recognize specific ligands on target cells.Citation30 These are referred to as activating NK cell receptors (NKRs) and inhibitory NKRs, respectively. When activating NKR signaling dominates inhibitory NKR signaling, NK cells attack targets through two defined effector functions: cytotoxicity; and production of proinflammatory cytokines and chemokines. The activating NKRs are ITAM-coupled receptors and are similar to those in myeloid cells with respect to structure and signaling mechanisms; they belong to either the Ig or C-type lectin family of receptors and use the same ITAM-containing adaptors, DAP12 and FcRγ. The role of the L- or M-CBM complex in activating NKR signaling has been examined by using CARD9−/−, CARD11−/−, BCL10−/−, and MALT1−/− mice.Citation6,Citation7,Citation31 Despite its essential role in myeloid DAP12- and FcRγ-associated receptors, CARD9 is not required for activation through activating NKRs.Citation6,Citation7 Instead, studies have revealed that CARMA1, BCL10, and MALT1 were essential for production of cytokines and chemokines induced by multiple activating NKRs, including FcγRIII (CD16), NK1.1, Ly49H, Ly49D, and NKG2D, demonstrating that L-CBM, not M-CBM, regulates activation of NKR signaling (). However, these deficiencies do not influence either maturation or the repertoire formation of peripheral NK cells. Similar to T and B lymphocytes, the loss of L-CBM impairs NF-κB activation following activation of NKRs (). On the other hand, contribution of L-CBM to MAPK activation, particularly JNK and p38, is unclear. In BCL10−/− and MALT1−/− NK cells, JNK and p38 activation is defective upon stimulation with phorbol myristate acetate plus calcium ionophore (P/I), which bypasses antigen receptor proximal signaling events by directly activating protein kinase Cs (PKCs); while in CARD11−/− NK cells, all MAPK activation following CD16 crosslinking is normal (). Thus, it will be necessary to clarify whether this difference reflects different contributions of CARMA1 and BCL10/MALT1 in NKR-induced MAPK activation or arises from alternative signaling activation by different stimuli. Interestingly, the cytotoxicity of NK cells induced by activating NKRs is not affected by L-CBM deficiency (). This corresponds with normal Vav1 phosphorylation and Ca2+ mobilization, both of which regulate exocytosis of cytotoxic granules, upon activating NKR ligation in CARD11−/− NK cells.Citation6 Together, L-CBM controls a distinct function of NK cells induced by NKR triggering (ie, cytokine and chemokine production, but not cytotoxicity) by selectively regulating the NF-κB activation signal.
Figure 3 Schematic signaling pathway mediated by the L-CBM complex in NK cells.
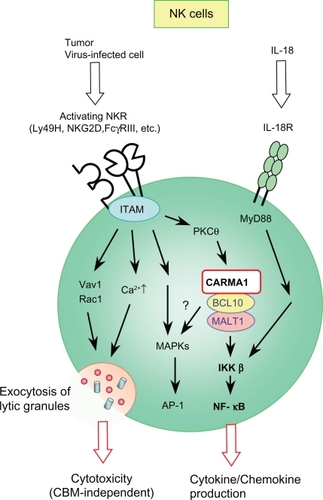
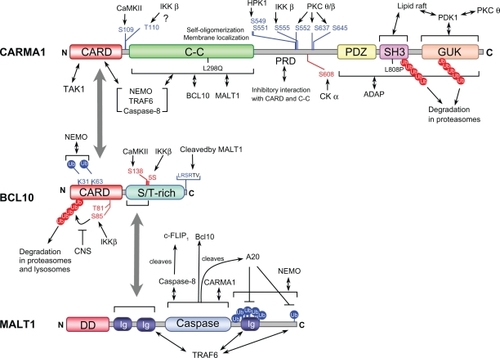
Figure 4 Regulatory phosphorylation sites, polyubiquitylation sites (Ub), proteolytic cleavage sites, and functional domains in L-CBM molecules.
Abbreviations: PRD, PKC-regulated domain; 5S, five serines; CNS, COP9 signalosome.
Signaling regulation of L-CBM complex
In this section, we discuss recent advances in the understanding of the molecular mechanisms that regulate L-CBM signaling. Multiple regulation mechanisms; involving phosphorylation, ubiquitylation, oligomerization, caspase activation, and recruitment to plasma membrane have been proposed to control L-CBM function (). Upon activation of antigen receptors, CARMA1 and BCL10 are phosphorylated by several kinases. Phosphorylation by some kinases represented by PKCs, positively regulates L-CBM signaling by modifying CARMA1 conformation, enabling it to form a signalosome with BCL10/MALT1 and downstream components for the NF-κB activation pathway.Citation32 Phosphorylation by other kinases has been shown to downmodulate L-CBM signaling by interfering with signalosome formation or targeting CARMA1 and BCL10 for degradation. Cascading oligomerization of L-CBM components and downstream molecules is a crucial event for the activation of this pathway. The lysine (K) 63-linked ubiquitination of BCL10 and MALT1 following TCR activation provides docking sites for downstream signaling molecules such as TRAF6 and NEMO, a necessity for the oligomerization cascade leading to IKK activation. By contrast, it has been reported that BCL10 and CARMA1 undergo degradation followed by K48-linked ubiquitination after antigen receptor-induced activation that may work to limit the amplitude of L-CBM signaling. L-CBM complex was shown to interact with caspase-8 and thereby regulate the caspase-8-c-FLIPL-mediated NF-κB activation pathway. Paracaspase activity of MALT1 was recently reported and the proteplytic activity shown to fine tune L-CBM-mediated NF-κB signaling. Various lines of evidence suggest that plasma membrane recruitment of L-CBM molecules is a crucial event for its function in NF-κB activation signaling.
Regulation by PKCs
L-CBM deficiency abrogates NF-κB activation in T, B and NK cells upon stimulation with P/I,Citation6,Citation7,Citation13–Citation15 indicating that L-CBM acts downstream of PKCs. PKC-deficient mice have confirmed this notion; the deficiency of PKCθ in T and NK cells, and of PKCβ in B cells, resulted in profound defects in TCR-, activating NKR- and BCR-induced NF-κB activation, respectively.Citation33,Citation34 This regulation could be explained by findings that CARMA1 is a direct target of these PKCs. Upon antigen receptor stimulation, CARMA1 is phosphorylated by PKCθ and PKCβ in the so called PKC-regulated domain (PRD),Citation32 which is the linker sequence between the C-C and the PDZ domain (). At least three serine residues (S552, S637, and S645 in human CARMA1) have been identified as the target sites.Citation35,Citation36 A model has been proposed where PRD phosphorylation destabilizes and induces conformational changes of CARMA1 from an inhibitory form to an active one that is accessible to BCL10 and other downstream molecules.Citation11,Citation12,Citation32,Citation37 In contrast to this essential PKC-dependent regulation in L-CBM, the fact that CARD9 lacks a PRD suggests that M-CBM-mediated signaling cannot be regulated by PKCs in a similar way to CARMA1. Indeed, lack of CARD9 or BCL10 in bone marrow-derived dendritic cells (BMDCs) did not affect IKK activation and cytokine production in response to P/I.Citation6 In addition, treatment with a PKC inhibitor effectively blocked cytokine production through ITAM-coupled receptors in NK cells, but not in BMDCs. It was so, even when NK cells and BMDCs were activated through the same receptor, CD16. This would indicate that PKC activation is required for L-CBM-, but not M-CBM-mediated signaling. However, loss of CARMA1 abrogates P/I-induced NF-κB activation in BMDCs, suggesting that CARMA1 is essential for PKC-mediated NF-κB activation in both lymphoid and myeloid cells, although the PKC-CARMA1 axis in myeloid cells is not utilized downstream of myeloid ITAM receptors, but might be used in signaling through other receptors. In line with this, the overexpression of CARD9 in CARMA1-deficient cells failed to restore PKC-mediated NF-κB activation. Therefore, these results indicate that L-CBM and M-CBM signaling are regulated by different molecular mechanisms and thus they are functionally not interchangeable.
Regulation by other kinases
It has been suggested that L-CBM signaling is regulated through phosphorylation by kinases other than PKCs. Calmodulin-dependent protein kinase (CaMKII) phosphorylates CARMA1 on Ser109 after TCR engagement and facilitates the interaction between CARMA1 and BCL10 ().Citation38 CaMKII also phosphorylates BCL10 on Ser138 but this phosphorylation is involved in the attenuation of NF-κB activation ().Citation39 Hematopoietic progenitor kinase 1 (HPK1) has been suggested to be involved in the JNK and NF-κB pathways in TCR signaling and regulates activation and survival of T cells.Citation40 A recent report has shown that HPK1 interacts with CARMA1 after TCR stimulation and phosphorylates CARMA1 on Ser549 and Ser551 within the PRD (). Although the precise mechanism is unclear, this phosphorylation is critical for TCR-mediated NF-κB activation and IL-2 production in Jurkat cells.Citation40 IKKβ phosphorylates CARMA1 on Ser578 and probably on Thr119 in chickens (corresponding to Ser555 and Thr110 in humans, respectively) upon BCR stimulation ().Citation41 This phosphorylation is postulated to enhance the assembly of the L-CBM complex, resulting in an increased ability of CARMA1 to activate IKK. Other studies have suggested that IKKβ phosphorylates BCL10 on five serines within the MALT1-interacting, S/T-rich domain and on Thr81 and Ser55 within CARD, upon TCR stimulation ().Citation42,Citation43 The former causes disengagement of BCL10 from MALT1 and interferes with IKK ubiquitination and the latter induces BCL10 degradation via the β-TrCP ubiquitin ligase/proteasome pathway, resulting in negative regulation and attenuation of L-CBM-mediated NF-κB signaling. Casein kinase (CK) 1α has a contrasting role in L-CBM signaling. CK1α associates with the L-CBM complex through the PRD of CARMA1 after TCR stimulation and participates in NF-κB activation, proliferation, and cytokine production. However, CK1α kinase activity subsequently contributes to the negative feedback of NF-κB activation by phosphorylating CARMA1 on Ser608 ().Citation44
Regulation by oligomerization and ubiquitination
Oligomerization and its associated ubiquitination events play an important role in L-CBM signaling. A recent study indicated that CARMA1 constitutively oligomerizes via its C-C domain and this self-oligomerization is required for TCR-induced NF-κB activation ().Citation45 Similarly, a previous study using mice bearing a point mutation (Leu298Gln) in the C-C domain of CARMA1, which is likely to disrupt C-C domain structure, demonstrated that integrity of the C-C domain is crucial for antigen receptor-induced IKK and JNK activation ().Citation46 CARMA1 forms an active conformation after receptor stimulation, which is followed by the recruitment of downstream molecules, triggering a downstream oligomerization and ubiquitination cascade.Citation32,Citation47 BCL10-dependent MALT1 oligomerization induces activation of TNF receptor-associated factor (TRAF) 6, a ubiquitin ligase, which in turn activates the IKK complex through lysine (K) 63-linked ubiquitylation of the regulatory subunit of IKK NEMO (an essential NF-κB modulator). During this process, TRAF6 itself is self-ubiquitinated, enabling recruitment of TAK1 and TAK1-binding protein 2 (TAB2), which then activates IKKα and IKKβ by phosphorylation.Citation48 BCL10 and MALT1 also undergo K63-linked ubiquitination in the CARD domain and the C-terminal region, respectively, upon T cell activation, which is likely to provide a docking surface for the recruitment of NEMO ().Citation49,Citation50 Interestingly, Zhou et al reported that MALT1 itself has an E3-ligase activity at C-terminus that targets both MALT and NEMO for ubiquitination.Citation51
In contrast, signaling inhibition by ubiquitination of L-CBM components has also been reported. CARMA1 is K48-linked-polyubiqutinated in the MAGUK region and degraded by the proteasome after antigen receptor-induced activation in T and B cells ().Citation52 The ubiquitination-dependent degradation of CARMA1 requires the phosphorylation of the PRD by PKC. A region between SH3 and the guanylate kinase domain (GUK), termed the Hook domain, plays a regulatory role in this process. Elimination of the ubiquitination sites from the MAGUK region results in elevated basal and inducible NF-κB and JNK activation, thus this activation-dependent mechanism likely provides an intrinsic feedback control of L-CBM signaling. In vitro experiments have suggested that the cellular inhibitor of apoptosis protein (cIAP) might target this MAGUK ubiquitination, although a cIAP antagonist that depletes the endogenous pool of cIAP did not affect P/I-induced CARMA1 degradation. Another ubiquitination-mediated negative regulation of CARMA1 has been suggested where Cbl-b-promoted monoubiquitination of CARMA1 is involved in the anergy induction in NKT cells.Citation53 It has been shown that BCL10 undergoes degradation following ubiquitination after activation of lymphocytes through antigen receptors,Citation43,Citation54,Citation55 resulting in termination of L-CBM signaling (). NEDD, cIAP, β-TrCP, and Itch have been suggested as ubiquitin ligases (E3) of BCL10. Both the proteasomal pathway via β-TrCP and the lysosomal pathway via NEDD and Itch have been proposed for BCL10 degradation.Citation43,Citation54,Citation55 The proteasomal pathway requires the phosphorylation of CARD by IKKβ,Citation43,Citation54,Citation55 while the lysosomal pathway requires intact CARD and the novel PKC activity.Citation43,Citation54,Citation55 The COP9 signalosome (CNS), a pleiotropic regulator of the ubiquitin/26S proteasome system, controls antigen responses in T cells. The CNS subunit 5 (CNS5) has been shown to interact with L-CBM in activated T cells. The CNS5, as well as CNS2, fine tunes IKK activation by maintaining BCL10 stability, most likely by interfering with the polyubiquitination and degradation of BCL10 ().Citation56 The deubiquitinating enzyme, A20, has been shown to function as a negative regulator for T cell activation.Citation57 Duwel et al have recently reported that A20 catalyzes the removal of the K63-linked ubiquitin chains attached to MALT1 and therefore regulates duration and strength of IKK activity ().Citation58
Regulation by caspases
MALT1 was originally identified as a caspase homolog named paracaspase.Citation59 Recent studies have revealed that the paracaspase domain of MALT1 possesses protease activity, cleaving BCL10,Citation60 and the NF-κB inhibitor, A20Citation61 (). Although the protease activity is required for optimal activation of NF-κB following TCR ligation, the MALT1-dependent BCL10 cleavage is not required for activation of NF-κB and JNK. Instead, it is necessary for TCR-induced cell adhesion to the extracellular matrix protein fibronectin. In contrast to other cells, lymphoid cells, particularly T cells, constitutively express A20. The cleavage of A20 by MALT1 disrupts its inhibitory effect on TCR-induced NF-κB activation and IL-2 production. The proteolytic activity of MALT1, therefore, is likely to act as a fine tuner in L-CBM signaling. This activity of MALT1 might not to be involved in M-CBM signaling because BCL10 cleavage was not detected when THP-1 monocytic cells were stimulated through ITAM-coupled receptors.Citation60
Besides its established role in lymphocyte apoptosis, caspase-8 and its proteolytically inactive homolog, c-FLIPL, are known to be essential for lymphocyte activation in both mice and humans,Citation62–Citation64 probably by interacting with the L-CBM complex and regulating NF-κB activation. A study has shown that MALT1 directly associates with procaspase-8 and induces limited autoprocessing of procaspase-8 (),Citation65 which generates an active form of caspase-8 that lacks the capacity to activate apoptotic substrates including caspase-3, but retains activity toward c-FLIPL to generate p43-FLIP. The cleaved form of c-FLIPL can strongly induce activation of NF-κB signaling pathway through interaction with TRAF2.Citation66 The association of caspase-8 with CARD and the C-C domain of the ‘active’ form of CARMA1 has also been reported ().Citation12
Regulation by membrane recruitment
Membrane recruitment is a crucial event in L-CBM-mediated NF-κB activation. It has been reported that BCL10, PKCθ, PKCβ−, MALT1, procaspase-8, c-FLIPL, and the IKK complex are recruited into lipid rafts after antigen receptor stimulation and that this recruitment is crucial for NF-κB activation.Citation33,Citation67,Citation68 CARMA1 resides in both the cytoplasm and lipid rafts of resting cells, but the amount of CARMA1 in lipid rafts increases after activation. This localization of CARMA1 is of particular importance for L-CBM signaling because mutations such as Leu808Pro mutation (L808P in ) that disrupt membrane localization of CARMA1 impair TCR-induced NF-κB activation.Citation45,Citation67 Genetic studies have demonstrated that CARMA1 is essential for the recruitment of BCL10-MALT1 and IKK complexes to lipid rafts.Citation67,Citation69
CARMA1 was also shown to control PKCθ recruitment in Jurkat cells.Citation67 An SLP-76-binding molecule, adhesion and degranulation-promoting adapter protein (ADAP), which functions in the inside-out signaling for integrin activation, has been shown to be critical for TCR-mediated NF-κB activation by acting as a link between the TCR-ZAP70-SLP76 signaling complex and L-CBM by binding to CARMA1 (). ADAP-deficient T cells revealed impaired L-CBM complex formation in the membrane and defective NF-κB activation.Citation70 It has been shown that 3-phosphoinositide-dependent kinase 1 (PDK1) recruits PKCθ and CARMA1 to lipid rafts upon TCR stimulation ().Citation71 A recent study using mice with a T cell-specific deletion of PKD1 has revealed that CD28 ligation facilitates NF-κB activation by regulating immune synapse localization and phosphorylation of PDK1, which enables the binding of PKCθ and CARMA1.Citation72 In contrast to the essential role of PDK1 in TCR-mediated NF-κB activation, the deletion of PDK1 in a chicken B cell line does not alter BCR-mediated NF-κB activation,Citation41 indicating a possible difference in the regulation mechanism of L-CBM recruitment to the membrane in TCR-versus BCR-mediated signaling pathways for NF-κB activation.
L-CBM signaling in disease
Three chromosomal translocations of L-CBM genes, t(11;18) (q21;q21), t(1;14)(p22;q32), and t(14;18)(q32;q21), have been reported and well characterized in MALT lymphoma (MALTL).Citation73 The MALT1 gene was originally identified in a break point of t(11;18)(q21;q21). This translocation generates API2-MALT1 fusion products that comprise the N-terminus of API2 with three intact baculovirus IAP repeat (BIR) domains and the C-terminus of the MALT1 with intact caspase-like domains. The fusion product, but neither API2 nor MALT1 alone, is capable of activating NF-κB. The translocations, t(1;14)(p22;q32) and t(14;18)(q32;q21), bring the BCL10 and MALT1 genes under the regulatory control of the Ig heavy chain (IgH) enhancer, respectively, thereby deregulating its expression and culminating in aberrant NF-κB activation. In addition to chromosomal translocations, BCL10 gene amplification has been reported in pancreatic cancer and nodal diffuse large B cell lymphoma (DLBCL).Citation74 Similarly, MALT1 gene amplification was found in cell lines of marginal zone B cell lymphoma and DLBCL.Citation75,Citation76
In contrast to normal B cells that express BCL10 in the cytoplasm, MALT lymphoma cells bearing t(11;18) (q21;q21) and t(1;14)(p22;q32) express the protein predominantly in the nucleus. Shen et al reported that 70% (28/40) of nasal NK/T cell lymphoma (NL) cases demonstrated aberrant nuclear staining of BCL10 and concomitant nuclear localization of NF-κB, although t(11;18)(q21;q21) and t(1;14)(p22;q32) are not present in NLs, indicating a possible relationship between aberrant BCL10 nuclear localization and tumorigenesis.Citation77
Among the subtypes of DLBCL, the least curable activated-B-cell-like (ABC) subtype DLBCLs, but not the germinal center B cell-like (GCB) subtype, relies on constitutive NF-κB signaling for survival. A loss-of-function RNA interference screen for genes required for survival of ABC DLBCL revealed that CARMA1 is a key upstream signaling component responsible for the constitutive IKK activity in ABC DLBCL.Citation78 In line with this, oncogenic missense mutations of the CARMA1 gene, all within exons encoding the C-C, have been found in ABC DLBCL.Citation79 These mutations constitutively activate the NF-κB pathway and enhance antigen receptor signaling to NF-κB, possibly owing to aggregate formation of the mutant proteins. MALT1 is constitutively active in ABC DLBCL, but not GCB DLBCL lines.Citation80 The oncogenic forms of CARMA1 are more potent than wild type CARMA1 in inducing proteolytic activity of MALT1. Inhibition of MALT1 activity, with the inhibitor z-VRPR-fmk, specifically affected the growth and survival of ABC DLBCLs. Thus, the MALT1 proteolytic activity might be a promising target for DLBCL therapy.
BCL10-transgenic mice elevate BAFF expression and specifically promote survival of MZ B cells and some mice develop splenic MZ lymphomas (MZL).Citation26 It has been reported that BAFF overexpression, with concomitant nuclear expression of BCL10 and NF-κB activation, is associated with Helicobacter pylori-independent growth of gastric DLBCL with histologic evidence of MALT lymphoma.Citation81 Thus, L-CBM-regulated BAFF signaling and vice versa might contribute to the development of MALTL, MZL, and DLBCL.
L-CBM, therefore, is an attractive therapeutic target for B-cell lymphomas and diseases associated with aberrant lymphocyte activation. However, further elucidation of L-CBM signaling is required for such applications, especially in approaches utilizing suppression of L-CBM activity. Mice with the Leu298Gln (L298Q in ) point mutation in CARMA1 exhibit profound defects in humoral immune response, whereas, paradoxically, the mice develop a hyper-IgE syndrome with atopic manifestations as they age.Citation46 This indicates that the amplitude of L-CBM signaling balances immunity versus tolerance and is thereby involved in the maintenance of immune homeostasis.
Disclosures
The authors report no conflicts of interest in this work.
References
- IsakovNRole of immunoreceptor tyrosine-based activation motif in signal transduction from antigen and Fc receptorsAdv Immunol1998691832479646845
- SamelsonLESignal transduction mediated by the T cell antigen receptor: the role of adapter proteinsAnnu Rev Immunol20022037139411861607
- LiQVermaIMNF-κB regulation in the immune systemNat Rev Immunol2002272573412360211
- WegenerEKrappmannDCARD-Bcl10-Malt1 signalosomes: missing link to NF-κBSci STKE20072007pe2117473310
- HaraHIshiharaCTakeuchiAThe adaptor protein CARD9 is essential for the activation of myeloid cells through ITAM-associated and Toll-like receptorsNat Immunol2007861962917486093
- HaraHIshiharaCTakeuchiACell type-specific regulation of ITAM-mediated NF-κB activation by the adaptors, CARMA1 and CARD9J Immunol200818191893018606643
- GrossOGruppCSteinbergCMultiple ITAM-coupled NK-cell receptors engage the Bcl10/Malt1 complex via Carma1 for NF-κB and MAPK activation to selectively control cytokine productionBlood20081122421242818192506
- HaraHSaitoTCARD9 versus CARMA1 in innate and adaptive immunityTrends Immunol20093023424219359218
- ThomeMCARMA1, BCL-10 and MALT1 in lymphocyte development and activationNat Rev Immunol2004434835915122200
- BertinJGuoYWangLCARD9 is a novel caspase recruitment domain-containing protein that interacts with BCL10/CLAP and activates NF-κBJ Biol Chem2000275410824108611053425
- CheTYouYWangDTannerMJDixitVMLinXMALT1/paracaspase is a signaling component downstream of CARMA1 and mediates T cell receptor-induced NF-κB activationJ Biol Chem2004279158701587614754896
- McCullyRRPomerantzJLThe protein kinase C-responsive inhibitory domain of CARD11 functions in NF-κB activation to regulate the association of multiple signaling cofactors that differentially depend on Bcl10 and MALT1 for associationMol Cell Biol2008285668568618625728
- RulandJDuncanGSEliaABcl10 is a positive regulator of antigen receptor-induced activation of NF-κB and neural tube closureCell2001104334211163238
- RulandJDuncanGSWakehamAMakTWDifferential requirement for Malt1 in T and B cell antigen receptor signalingImmunity20031974975814614861
- HaraHWadaTBakalCThe MAGUK family protein CARD11 is essential for lymphocyte activationImmunity20031876377512818158
- Ruefli-BrasseAAFrenchDMDixitVMRegulation of NF-κB-dependent lymphocyte activation and development by paracaspaseScience20033021581158414576442
- BlonskaMPappuBPMatsumotoRThe CARMA1-Bcl10 signaling complex selectively regulates JNK2 kinase in the T cell receptor-signaling pathwayImmunity200726556617189706
- Schmidt-SupprianMTianJGrantEPDifferential dependence of CD4+CD25+ regulatory and natural killer-like T cells on signals leading to NF-κB activationProc Natl Acad Sci U S A20041014566457115070758
- MedoffBDSandallBPLandryADifferential requirement for CARMA1 in agonist-selected T-cell developmentEur J Immunol200939788419130560
- MolineroLLYangJGajewskiTAbrahamCFarrarMAAlegreMLCARMA1 controls an early checkpoint in the thymic development of FoxP3+ regulatory T cellsJ Immunol20091826736674319454668
- XueLMorrisSWOrihuelaCDefective development and function of Bcl10-deficient follicular, marginal zone and B1 B cellsNat Immunol2003485786512910267
- FerchUzum BuschenfeldeCMGewiesAMALT1 directs B cell receptor-induced canonical nuclear factor-κB signaling selectively to the c-Rel subunitNat Immunol2007898499117660823
- PappuBPLinXPotential role of CARMA1 in CD40-induced splenic B cell proliferation and marginal zone B cell maturationEur J Immunol2006363033304317048267
- OliverAMMartinFGartlandGLCarterRHKearneyJFMarginal zone B cells exhibit unique activation, proliferative and immunoglobulin secretory responsesEur J Immunol199727236623749341782
- HaydenMSGhoshSSignaling to NF-κBGenes Dev2004182195222415371334
- LiZWangHXueLEmu-BCL10 mice exhibit constitutive activation of both canonical and noncanonical NF-κB pathways generating marginal zone (MZ) B-cell expansion as a precursor to splenic MZ lymphomaBlood20091144158416819696203
- TuscheMWWardLAVuFDifferential requirement of MALT1 for BAFF-induced outcomes in B cell subsetsJ Exp Med20092062671268319917778
- BhattacharyyaSBorthakurATyagiSBCL10 is required for NF-κB nuclear translocation by both canonical and non-canonical pathways and for NF-κB-inducing kinase (NIK) phosphorylationJ Biol Chem200910.1974/jbc.M109.050815
- AmannaIJDingwallJPHayesCEEnforced bcl-xL gene expression restored splenic B lymphocyte development in BAFF-R mutant miceJ Immunol20031704593460012707337
- LanierLLNK cell recognitionAnnu Rev Immunol20052322527415771571
- MalarkannanSRegunathanJChuHBcl10 plays a divergent role in NK cell-mediated cytotoxicity and cytokine generationJ Immunol20071793752376217785812
- RawlingsDJSommerKMoreno-GarciaMEThe CARMA1 signalosome links the signalling machinery of adaptive and innate immunity in lymphocytesNat Rev Immunol2006679981217063183
- Schulze-LuehrmannJGhoshSAntigen-receptor signaling to nuclear factor κBImmunity20062570171517098202
- TassiICellaMPrestiRNK cell-activating receptors require PKC-theta for sustained signaling, transcriptional activation, and IFN-gamma secretionBlood20081124109411618784374
- SommerKGuoBPomerantzJLPhosphorylation of the CARMA1 linker controls NF-κB activationImmunity20052356157416356855
- MatsumotoRWangDBlonskaMPhosphorylation of CARMA1 plays a critical role in T Cell receptor-mediated NF-κB activationImmunity20052357558516356856
- StiloRLiguoroDDi JesoBPhysical and functional interaction of CARMA1 and CARMA3 with Iκ kinase gamma-NF-κB essential modulatorJ Biol Chem2004279343233433115184390
- IshiguroKGreenTRapleyJCa2+/calmodulin-dependent protein kinase II is a modulator of CARMA1-mediated NF-κB activationMol Cell Biol2006265497550816809782
- IshiguroKAndoTGotoHXavierRBcl10 is phosphorylated on Ser138 by Ca2+/calmodulin-dependent protein kinase IIMol Immunol2007442095210017052756
- BrennerDBrechmannMRohlingSPhosphorylation of CARMA1 by HPK1 is critical for NF-κB activation in T cellsProc Natl Acad Sci U S A2009106145081451319706536
- ShinoharaHMaedaSWataraiHKurosakiTIκB kinase beta-induced phosphorylation of CARMA1 contributes to CARMA1 Bcl10 MALT1 complex formation in B cellsJ Exp Med20072043285329318086859
- WegenerEOeckinghausAPapadopoulouNEssential role for IκB kinase beta in remodeling Carma1-Bcl10-Malt1 complexes upon T cell activationMol Cell200623132316818229
- LobryCLopezTIsraelAWeilRNegative feedback loop in T cell activation through IκB kinase-induced phosphorylation and degradation of Bcl10Proc Natl Acad Sci U S A200710490891317213322
- BidereNNgoVNLeeJCasein kinase 1alpha governs antigen-receptor-induced NF-κB activation and human lymphoma cell survivalNature2008
- TannerMJHanelWGaffenSLLinXCARMA1 coiled-coil domain is involved in the oligomerization and subcellular localization of CARMA1 and is required for T cell receptor-induced NF-κB activationJ Biol Chem2007282171411714717428801
- JunJEWilsonLEVinuesaCGIdentifying the MAGUK protein Carma-1 as a central regulator of humoral immune responses and atopy by genome-wide mouse mutagenesisImmunity20031875176212818157
- ThomeMMultifunctional roles for MALT1 in T-cell activationNat Rev Immunol2008849550018575460
- SunLDengLEaCKXiaZPChenZJThe TRAF6 ubiquitin ligase and TAK1 kinase mediate IKK activation by BCL10 and MALT1 in T lymphocytesMol Cell20041428930115125833
- WuCJAshwellJDNEMO recognition of ubiquitinated Bcl10 is required for T cell receptor-mediated NF-κB activationProc Natl Acad Sci U S A20081053023302818287044
- OeckinghausAWegenerEWeltekeVMalt1 ubiquitination triggers NF-κB signaling upon T-cell activationEmbo J2007264634464517948050
- ZhouHWertzIO’RourkeKBcl10 activates the NF-κB pathway through ubiquitination of NEMONature200442716717114695475
- Moreno-GarciaMESommerKShinoharaHBandaranayakeADKurosakiTRawlingsDJMAGUK-controlled ubiquitination of CARMA1 modulates lymphocyte NF-κB activityMol Cell Biol2009
- KojoSEllyCHaradaYLangdonWYKronenbergMLiuYCMechanisms of NKT cell anergy induction involve Cbl-b-promoted monoubiquitination of CARMA1Proc Natl Acad Sci U S A2009106178471785119815501
- ScharschmidtEWegenerEHeissmeyerVRaoAKrappmannDDegradation of Bcl10 induced by T-cell activation negatively regulates NF-κB signalingMol Cell Biol2004243860387315082780
- HuSAlcivarAQuLTangJYangXCIAP2 inhibits anigen receptor signaling by targeting Bcl10 for degredationCell Cycle200651438144216775419
- WeltekeVEitelhuberADuwelMSchweitzerKNaumannMKrappmannDCOP9 signalosome controls the Carma1-Bcl10-Malt1 complex upon T-cell stimulationEMBO Rep20091064264819444310
- SunSCDeubiquitylation and regulation of the immune responseNat Rev Immunol2008850151118535581
- DuwelMWeltekeVOeckinghausAA20 negatively regulates T cell receptor signaling to NF-κB by cleaving Malt1 ubiquitin chainsJ Immunol20091827718772819494296
- UrenAGO’RourkeKAravindLAIdentification of paracaspases and metacaspases: two ancient families of caspase-like proteins, one of which plays a key role in MALT lymphomaMol Cell2000696196711090634
- RebeaudFHailfingerSPosevitz-FejfarAThe proteolytic activity of the paracaspase MALT1 is key in T cell activationNat Immunol2008927228118264101
- CoornaertBBaensMHeyninckKT cell antigen receptor stimulation induces MALT1 paracaspase-mediated cleavage of the NF-κB inhibitor A20Nat Immunol2008926327118223652
- SalmenaLLemmersBHakemAEssential role for caspase 8 in T-cell homeostasis and T-cell-mediated immunityGenes Dev20031788389512654726
- SuHBidereNZhengLRequirement for caspase-8 in NF-κB activation by antigen receptorScience20053071465146815746428
- ChauHWongVChenNJCellular FLICE-inhibitory protein is required for T cell survival and cyclingJ Exp Med200520240541316043518
- KawadlerHGantzMARileyJLYangXThe paracaspase MALT1 controls caspase-8 activation during lymphocyte proliferationMol Cell20083141542118691973
- KataokaTTschoppJN-terminal fragment of c-FLIP(L) processed by caspase 8 specifically interacts with TRAF2 and induces activation of the NF-κB signaling pathwayMol Cell Biol2004242627263615024054
- WangDMatsumotoRYouYCD3/CD28 costimulation-induced NF-κB activation is mediated by recruitment of protein kinase C-theta, Bcl10, and IκB kinase beta to the immunological synapse through CARMA1Mol Cell Biol20042416417114673152
- MisraRSRussellJQKoenigACaspase-8 and c-FLIPL associate in lipid rafts with NF-κB adaptors during T cell activationJ Biol Chem2007282193651937417462996
- HaraHBakalCWadaTThe molecular adapter Carma1 controls entry of IκB kinase into the central immune synapseJ Exp Med20042001167117715520247
- MedeirosRBBurbachBJMuellerKLRegulation of NF-κB activation in T cells via association of the adapter proteins ADAP and CARMA1Science200731675475817478723
- LeeKYD’AcquistoFHaydenMSShimJHGhoshSPDK1 nucleates T cell receptor-induced signaling complex for NF-κB activationScience200530811411815802604
- ParkSGSchulze-LuehrmanJHaydenMSThe kinase PDK1 integrates T cell antigen receptor and CD28 coreceptor signaling to induce NF-κB and activate T cellsNat Immunol20091015816619122654
- DuMQMALT lymphoma: recent advances in aetiology and molecular geneticsJ Clin Exp Hematop200747314218040143
- HolzmannKKohlhammerHSchwaenenCGenomic DNA-chip hybridization reveals a higher incidence of genomic amplifications in pancreatic cancer than conventional comparative genomic hybridization and leads to the identification of novel candidate genesCancer Res2004644428443315231651
- Sanchez-IzquierdoDBuchonnetGSiebertRMALT1 is deregulated by both chromosomal translocation and amplification in B-cell non-Hodgkin lymphomaBlood20031014539454612560219
- DijkmanRTensenCPJordanovaESArray-based comparative genomic hybridization analysis reveals recurrent chromosomal alterations and prognostic parameters in primary cutaneous large B-cell lymphomaJ Clin Oncol20062429630516330669
- ShenLLiangACLuLAberrant BCL10 nuclear expression in nasal NK/T-cell lymphomaBlood20031021553155412900354
- NgoVNDavisRELamyLA loss-of-function RNA interference screen for molecular targets in cancerNature200644110611016572121
- LenzGDavisRENgoVNOncogenic CARD11 mutations in human diffuse large B cell lymphomaScience20083191676167918323416
- HailfingerSLenzGNgoVEssential role of MALT1 protease activity in activated B cell-like diffuse large B-cell lymphomaProc Natl Acad Sci U S A2009106199461995119897720
- KuoSHYehPYChenLTOverexpression of B cell-activating factor of TNF family (BAFF) is associated with Helicobacter pylori-independent growth of gastric diffuse large B-cell lymphoma with histologic evidence of MALT lymphomaBlood20081122927293418628489