Abstract
The transcription factor NF-κB is a critical regulator of immune and inflammatory responses. In mammals, the NF-κB/Rel family comprises five members: p50, p52, p65 (Rel-A), c-Rel, and Rel-B proteins, which form homo- or heterodimers and remain as an inactive complex with the inhibitory molecules called IκB proteins in resting cells. Two distinct NF-κB signaling pathways have been described: 1) the canonical pathway primarily activated by pathogens and inflammatory mediators, and 2) the noncanonical pathway mostly activated by developmental cues. The most abundant form of NF-κB activated by pathologic stimuli via the canonical pathway is the p65:p50 heterodimer. Disproportionate increase in activated p65 and subsequent transactivation of effector molecules is integral to the pathogenesis of many chronic diseases such as the rheumatoid arthritis, inflammatory bowel disease, multiple sclerosis, and even neurodegenerative pathologies. Hence, the NF-κB p65 signaling pathway has been a pivotal point for intense drug discovery and development. This review begins with an overview of p65-mediated signaling followed by discussion of strategies that directly target NF-κB p65 in the context of chronic inflammation.
Keywords:
Introduction
The NF-κB signaling system is a highly dynamic protein interaction network made up of components that regulate each other. The system includes five transcriptional monomers, two precursor proteins, three ankyrin repeat containing inhibitory IκB proteins, three stimulus-responsive inhibitory kinases (IKK complex: IKK-α and IKK-β, and IKK-γ or NF-κB essential modulator/NEMO), and ankyrin repeat containing regulatory proteins.Citation1 While all NF-κB translational monomers that function as homo- or heterodimers share conserved amino terminal dimerizing rel homology domain (RHD), only RelA/p65, C-rel, and RelB possess the transactivation domain (TAD) necessary for transcriptional activity. The TAD lacking monomers p50 or p52 function as trans-repressors as homodimers but can stimulate transcription when heterodimerized with a transactivating Rel subunit.Citation2
In resting state, the NF-κB dimers remain in the cytoplasm as an inactive complex with the IκB inhibitory proteins that mask the nuclear localization signals (NLSs) in the RHD and block their nuclear import.Citation2,Citation3 Activation of NF-κB occurs through canonical or noncanonical pathways and depends on phosphorylation-induced ubiquitination of IκB proteins. The canonical NF-κB pathway is activated within minutes of exposure to proinflammatory signals such as cytokines, pathogens, and danger-associated molecular patterns. It is mediated by activation of the NEMO-containing IKK complex, which in turn designates the classical IκBs (IκBα, IκBβ, or IκBε) for degradation with consequent release of the NF-κB dimers for nuclear translocation.Citation2–Citation4 The noncanonical NF-κB pathway is triggered by developmental cues that signal via a select group of receptors such as a subset of tumor necrosis factor family of receptors (TNFRs), B cell-activating factor receptor, lymphotoxin β receptor, and receptor activator of NF-κB and occurs at slower kinetics.Citation5,Citation6 It is mediated by NF-κB inhibitory kinase (NIK) independent of NEMO. The NIK in turn phosphorylates and activates IKKα, which in turn directly acts on the non-IκB substrates of the NF-κB subunits and modulates the transcriptional responses.Citation6–Citation8 Interestingly suppression of canonical NF-κB activation by mutations or loss of NEMO increases NIK accumulation with aberrant noncanonical NF-κB signaling.Citation9 This suggests that the cross talk between the two pathways and a rate-limiting canonical NF-κB signaling perhaps determine the basal pool of NIK and the constitutive NF-κB activity.Citation2,Citation7,Citation8
Induced activation of NF-κB p65:p50
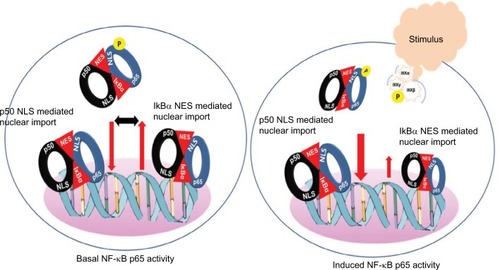
The most abundant form of NF-κB activated by the canonical pathway is the heterodimer of p50 and p65.Citation2 As stated above typically triggered by an NEMO-dependent activation of IKK, phosphorylation of IκB proteins followed by ubiquitination and degradation by proteasomes releases the NF-κB p65:p50 dimers from the inhibitory complex.Citation4,Citation9,Citation10 This exposes the arginine- and lysine-rich NLS of the p65 and the p50 subunits for interaction with the importin α/β heterodimers and nuclear translocationCitation11 (). Mechanistically, most often the importin α proteins initially bind the NLS and then recruit importin β to transport the cargo into the nucleus through the nuclear pore complexes (NPCs). However, importin β has been shown to directly bind the NLS of p65 and promote its nuclear translocation.Citation11,Citation12 Within the nucleus, the Ran-GTP protein binds and dissociates the importin-β from the complex with p65. The free p65 then binds specific nucleotide sequence of the target genes and mediates tightly controlled transcriptional programs that exhibit a wide degree of tissue- and context-specificity.Citation13–Citation16 Sequence-specific DNA-binding of p65 is greatly stimulated by the interaction with various components of the basic transcription machinery such as the TFIIB, TBP-associated factors and several coactivators including p300 and Tip60.Citation15,Citation17,Citation18 Export of p65 out of the nucleus is facilitated by the exportin proteins that bind the nuclear export sequence of p65 and the Ran GTP. The complex then passes through the NPC into the cytoplasm, wherein hydrolysis of Ran GTP to Ran GDP by Ran GTPase-activating protein results in the dissociation of p65.Citation7,Citation8 Structural analyses of the NF-κB:IκB complex showed that the IκB proteins mask the NLS of p65 but not that of the p50 subunit.Citation19–Citation21 Hence, in physiologic conditions the importin α/β dimers can bind the NLS of p50 and facilitate the nuclear import of p50:p65 dimers. Exportin proteins bind the nuclear export sequence of IκBα in complex with the p65 in the nucleus and facilitate export of the complex out of the nucleus (). Continuous shuttling of the NF-κB:importin:IκBα complex between the nucleus and cytoplasm maintains a low level of basal p65 activity.Citation4,Citation19,Citation22
Figure 1 Model for basal and induced NF-κB p65 activity.
Notes: Basal NF-κB activity is potentially maintained by a balanced shuttling of the NF-κB p65:IκB complex between the cytoplasm and nucleus. In a steady-state condition, IκBα masks entirely the nuclear localization signal of p65 but not that of p50. This allows shuttling of the NF-κB:IκB complex by passive nuclear import to support basal activity and reciprocal export by the nuclear export signals in the terminal ankyrin repeat of IκBα. Stimulus induced phosphorylation of IKKβ followed by protein kinase-A-mediated phosphorylation of IκBα and rapid release and import of p65:p50 dimers. Postinduction, the newly synthesized IκBα not only sequesters DNA bound p65 but also shuttles to the cytoplasm to quench the free p50:p65 dimers.
Mechanisms and modulations of the p65-mediated transactivation
Induced activation of p65 in response to a variety of stimuli is typically transient but sufficient to upregulate transactivation of target genes of diverse activities such as the cellular proliferation, inflammatory cytokines, chemokines, and mediators of apoptosis.Citation1,Citation23,Citation24 It is tightly controlled by postinduction termination via negative feedback loops. Different importins including the importin α3, α4, α5, and β1 exhibit variable affinities for binding the p65 NLS and hence compete with or substitute for each other in facilitating its nuclear import.Citation11,Citation25 Furthermore, the expression levels of importin α and importin β have been shown to vary widely in different cells and tissues.Citation26 Hence, the combined effects of the expression levels of the importin proteins and the differential p65 affinities determine the extent of nuclear translocation of p65 and subsequent transactivation. Efficient and controlled transactivation of the target genes is facilitated by cross talk with other transcription factors with either synergistic (eg, AP-1) or antagonistic (eg, cEBP/β) effects.Citation2,Citation27 In addition, a variety of posttranslational modifications including degradation and regulatory ubiquitination, acetylation, methylation, and phosphorylation play key roles in the fine-tuning and specification of the NF-κB p65 activation outcomes. These modifications often occur in a hierarchical manner with phosphorylation or acetylation as typical starting points and degradative ubiquitination as characteristic irreversible endpoints.Citation4,Citation28
p65 phosphorylation
Inducible p65 phosphorylation can occur both in the cytoplasm and in the nucleus in response to a variety of stimuli.Citation4,Citation28 Phosphorylation is mediated by protein kinases that transfer the γ-phosphate of adenosine triphosphate to the hydroxyl group of a serine (Ser), threonine (Thr), or tyrosine (Tyr) residue of the target protein.Citation29 The NF-κB p65 possesses many Ser and Thr residues distributed in the RHD and the TAD. Phosphorylation of individual Ser/Thr/Tyr by multiple kinases generates a heterogeneous pool of modified NF-κB p65 that transactivates genes and mediates context-dependent functional responses.Citation4,Citation28
Ser-276 in p65 RHD is predominantly phosphorylated by protein kinase A (PKAc). In resting cells, the catalytic subunit of PKAc is masked by the cytosolic IκB:p65 complex. Activation of the IKK complex releases the PKAc, which in turn phosphorylates Ser-276 inducing a structural change in p65 that facilitates interaction with the cofactors p300/CBP (cAMP response element binding [CREB] protein) and enhances transcriptional activity.Citation30 The mitogen and stress-activated protein kinase-1 (MSK-1)/MSK-2 phosphorylates Ser-276 in the nucleus and enhances inflammatory gene expression.Citation4,Citation9,Citation31 This MSK1/2-mediated phosphorylation depends on the nature of stimuli and may require additional kinases such as casein kinase 2 and ribosomal s6 kinase (RSK1).Citation9,Citation28
Phosphorylation of p65 at Thr 254 leads to a phosphorylated Thr–proline motif that is recognized by the peptidyl-prolyl isomerase Pin1. Pin1-mediated prolyl isomerization of p65 results in reduced IκBα binding, increased p65 stability, and nuclear accumulation mediating enhanced transcription.Citation4,Citation32 Mutation of Thr 254 dramatically reduced the half-life of p65 protein and inhibited p65 transcriptional activity.Citation32
Phosphorylation of Ser-468 in p65 T mediated by IKKε or glycogen synthase kinase 3β in unstimulated cells has been shown to negatively regulate basal NF-κB activity.Citation33,Citation34 Phosphorylation of Thr-505 by checkpoint kinase 1 has been identified as a means of cross talk between DNA damage and the activation of NF-κB.Citation4,Citation35
Ser-536 in the p65-TAD is also phosphorylated by multiple kinases including IKK kinases, RSK1, and NF-κB activating kinase/TNFR-associated factor NF-κB activator (TANK)-binding kinase 1 (TBK1).Citation36–Citation38 While phosphorylation of Ser536 in the cytosolic p65 by IKK or RSK1 promotes its nuclear translocation,Citation37 cyclin-dependent kinase-6 (CDK-6) phosphorylates Ser-536 in the nucleus and facilitates p65 binding to specific promoter sequences. Inhibition of CDK-6 catalytic activity suppressed NF-κB p65-mediated inflammatory gene expressions.Citation39,Citation40 Interestingly Ser-536 has also been shown to be phosphorylated by TBK1 (TANK-binding kinase 1), a Ser-Thr kinase, belonging to the noncanonical IκB kinases, suggesting that the two NF-κB pathways are interrelated at multiple levels.Citation41 Taken together, modulation of the Ser-536 phosphorylation status is another mechanism for self-limited activation of NF-κB.
Acetylation
Lysine acetylation is a reversible event regulated by his-tone acetyltransferases (HATs) and histone deacetylases (HDACs).Citation42 Seven acetylation sites are identified in p65: Lys122, Lys123, Lys218, Lys221, Lys310, Lys314, and Lys315. The HATs p300 and p300/CBP-associated factor (PCAF) acetylate lysines 122 and 123 of p65. The p300 has also been described to acetylate lysines 310, 314, and 315.Citation28,Citation43 Acetylation of specific lysine residue has been shown to regulate NF-κB p65 activation or deactivation. While acetylation on Lys122 and Lys123 decreases DNA binding, Lys221 and Lys218 acetylation enhances the DNA binding of NF-κB p65 by impairing its association with IκBα.Citation44,Citation45 Although acetylation at Lys310 is critical for full transcriptional activity of p65, it does not affect the DNA binding or assembly with IκBα.Citation43,Citation46 Lys314 and Lys315 acetylation increases promoter selectivity without affecting the NF-κB p65 shuttling or the DNA binding.Citation43,Citation47 The acetylated Lys310 is specifically recognized by the two bromodomains of Brd4, which recruits activated CDK9 to phosphorylate RNA polymerase II for the transcription of a subset of NF-κB target genes.Citation28,Citation48
Tip60 (HIV Tat-interacting protein, 60 kDa), originally identified as a binding partner for the HIV-1 Tat protein, has been shown to function as a coactivator of NF-κB p65, enhance acetylation on Lys310, and upregulate p65 transcriptional activity through a protein–protein interaction.Citation49 The Tip60 has been shown to bind DNA prior to p65 and potentially modulate other cofactor:p65 interactions. This suggests that the site-specific early association of Tip60 could represent another mechanism that regulates p65-mediated gene expression.
Methylation
The protein methyltransferases include two enzymes, the protein lysine methyltransferases and the protein arginine methyltransferases (PRMTs). Both enzymes use small-molecule cofactor, S-adenosyl-l-methionine, as the universal methyl donor for the enzymatic methylation of lysine and arginine side chains.Citation50,Citation51 Methylations of both lysine and arginine on NF-κB p65 has been shown to regulate its transactivation potential.Citation51–Citation53
It has been suggested that the methylation of p65 in the nucleus potentially facilitates binding to specific promoters when the local chromatin remodeling machinery is active.Citation50 Set9, a histone methyltransferase, has been shown to methylate p65 on Lys314 and Lys315 and negatively regulate the NF-κB function by inducing the ubiquitination and degradation of DNA-bound p65.Citation54 In contrast, methylation of Lys37 by Set9 or that of Lys218 and Lys221 by nuclear receptor-binding SET domain-containing protein 1, a histone H3K36 methyltransferase, enhances the transcriptional activity by stabilizing binding of p65 to its enhancers.Citation52,Citation55 Apart from lysine residues, demethylation of Arg 30 by protein arginine methyltransferase 5 (PRMT5) has been shown to activate p65 suggesting that the methylation of both lysine and arginine residues regulate NF-κB-mediated transcriptional activity.Citation50,Citation53 Molecular modeling suggests that methylated R30 can mediate Van der Waals contacts and increase the affinity of p65 for DNA and consequent transactivation of target gene expressions.Citation53 Hence, differential methylation could represent another regulatory mechanism for differential activation of individual genes based on the cell type, stimulus, and other factors.Citation50,Citation51
In summary, the sequential post-translational modifications modulate the strength and duration of NF-κB activity, contribute to the target gene specificity, and thereby fine-tune the biologic responses.Citation28,Citation38,Citation56 Indeed, it has been observed that the phosphorylation of Ser276 followed by acetylation of Lys310 of p65 facilitated recruitment of specific elongation factors Brd4 and P-TEFb to proinflammatory target genes promoting efficient mRNA processing.Citation57 Furthermore, cross-talk mechanisms link post-translational modification-mediated regulations. For example, phosphorylation at Ser-276 and Ser-536 enhances the p300- and PCAF-mediated acetylation of Lys122, Lys123, and Lys310 and upregulates transcriptional activation by p65.Citation58 Thus, by virtue of modifying the interaction of p65 with select cofactors, the post-translational modifications at specific sites contribute to specific biologic responses.Citation54,Citation57
The varied and differential outcomes following individual site phosphorylation or acetylation have been attributed to the induced conformational changes in p65 that direct subsequent cofactor interactions required for transcriptional activation.Citation4,Citation59 In this context it is of interest to note that the p65-TAD critical for its transcriptional ability hosts multiple phosphorylation sites and is largely unstructured.Citation4 The intrinsic disorder of the unstructured p65-TAD confers intramolecular flexibility that allows it to adopt an “induced fit” secondary structure stabilized by phosphorylation of specific residue in the context of an interacting cofactor.Citation28,Citation59 Such intrinsically unstructured protein regions could serve as potential targets for rational drug design based on the transition from disordered to ordered conformation through intermolecular interaction.Citation60 Interestingly synthetic peptides derived from the p65-TAD have been shown to inhibit NF-kB p65-mediated cellular responses.Citation61
Termination of activated p65
To avoid excessive transactivation and deleterious cellular responses, the activated NF-κB p65 orchestrates a highly coordinated termination program that is rapid and self-limiting.
Role of IκBα
As stated above, NF-κB activity is predominantly controlled by association with IκBα. Following translocation, the nuclear p65 binds on the κB sequence in IκBα promoter and increases its transcription. The increased nuclear IκBα in turn removes the NF-κB p65 complex from the DNA terminating its transcriptional activity.Citation19,Citation22 As nuclear p65:DNA interaction exhibits fast dissociation kinetics, the newly synthesized IκBα may not only reverse the binding of p65 to DNA, but also capture p65 molecules released from DNA and transport them back to the cytoplasm.Citation20,Citation38 The iterative cycles of IκBα degradation and replenishment by p65-mediated transactivation provide sustained supply of p65:p50 dimers to the nucleus at a basal level.Citation3,Citation19 In addition, strong oscillations of the negative feedback regulation between the activated p65 and IκBα are monitored by the IκBβ and IκBγ, the inhibitory proteins that undergo slow degradation, exhibit limited activation potential, and prevent abundance of activated p65 in the nucleus.Citation2,Citation62
Dephosphorylation of p65: It is another mechanism that facilitates termination of NF-κB signaling after stimulant removal or target gene expression and reestablish normal responsiveness. Dephosphorylation of Ser-536 by protein phosphatase 2A or by wild-type p53-induced phosphatase 1 has been shown to reduce the interaction of p65 with p300 and thereby inhibit gene expression.Citation4,Citation63,Citation64 However, much like phosphorylation, dephosphorylation of p65 by phosphatases regulates the functions of NF-κB p65 in a context- and signal-dependent manner. For example, following cisplatin stimulation, phosphatase 4-mediated dephosphorylation of Thr-435 upregulated NF-κB p65 activation rather than suppression.Citation65
Deacetylation of p65: Multiple HDACs deacetylate p65 and regulate NF-κB functions depending on the specific acetylation site. While HDAC3-mediated deacetylation of p65 at Lys122, Lys123, Lys314, and Lys315 upregulated inflammatory gene expression, deacetylation at Lys218 and Lys221 promotes the binding of p65 with IκBα leading to the nuclear export and downregulation of NF-κB.Citation43
Demethylation of p65: Similar to phosphorylation and acetylation, methylation is also a reversible event mediated by demethylases. Demethylation of methylated Lys-218 and Lys-221 by F-box and leucine-rich repeat protein 11, an H3K36 demethylase, has been shown to negatively regulate the transcriptional activity of NF-κB p65 and decrease cell proliferation.Citation55,Citation66
Ubiquitination and degradation: They have been suggested as mechanisms that control the strength and duration of NF-κB activation.Citation28,Citation36 Nuclear p65 is degraded by the proteasome in a DNA-binding-dependent manner. Copper metabolism domain protein, a physiologic inhibitor of NF-κB, binds to an E3 ligase SOCS1 (suppressor of cytokine signaling 1) and p65, stabilizing the p65:SOCS1 interaction and thereby promote ubiquitination and degradation of p65.Citation4,Citation67 PDLIM2, a nuclear LIM domain containing ligase, mediates polyubiquitination and proteosomal degradation of nuclear p65 and contributes to the termination of p65-mediated transactivation.Citation10 Interestingly, point mutations in p65 that inhibit DNA binding impaired degradation, suggesting that the sequence-specific interactions with DNA may be critical for transcriptional termination.
In conclusion, while cytoplasmic NF-κB p65 in complex with the IκBs are stable and do not undergo much turnover, the stability of nuclear p65 proteins is short-lived. Several interrelated not mutually exclusive mechanisms regulate the stability and termination of nuclear p65. In general, the postinduction repression of NF-κB p65 by newly synthesized IκBα can be considered upstream events that assist in maintaining the homeostasis between the cytoplasmic and nuclear p65. The nucleocytoplasmic shuttling mediated by the differential binding kinetics of p65 with different importin/exportin proteins or modulations by dephosphorylation, demethylation, or deacetylation are intermediate mechanisms that contribute to effective p65 activation. For example, in conditions of continuous stimulation, oscillations in Ser-536 phosphorylation have been shown to reflect oscillations in NF-κB cytosolic–nuclear translocation, suggesting potential dephosphorylation of Ser-536 in the nucleus.Citation37,Citation68 Ubiquitination and proteosomal degradation of DNA-bound p65 constitute downstream events that play major roles in limiting the intensity and duration of NF-κB p65 activity. In addition, the nature and extent of regulation by post-translational modification can vary with different stimulators and even the same modifications can facilitate different effects.Citation50
Therapeutic targeting of NF-κB p65 in inflammation
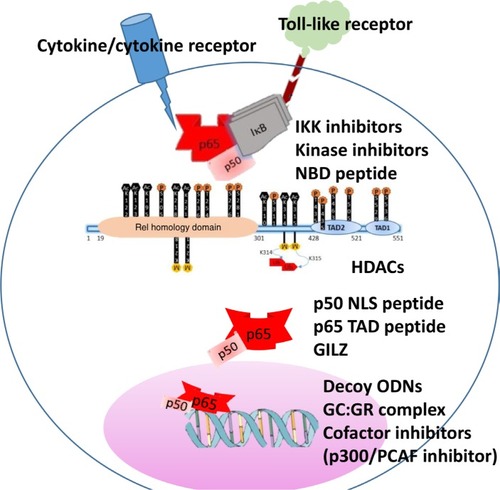
The literature is replete with a vast number of candidate therapeutics designed to target the NF-κB pathway. These compounds belong to different chemical classes such as naturetics, peptidomimetics, small molecules, small interfering RNAs, and microbial products ().Citation69–Citation72 This review is restricted to therapeutic approaches that specifically target the interactions of the p65 subunit of NF-κB with a focus on inflammation, discussing representative compounds and illustrating emerging strategies to overcome the limitations of conventional NF-κB therapeutics ().
Table 1 Strategies and agents developed and evaluated for selective targeting of NF-kB p65
Figure 2 Schematic representation of NF-κB p65 activation and potential inhibitors along the signaling pathway.
Notes: The inset shows phosphorylation and acetylation sites of p65 as sites of kinase inhibitors and HDACs. Representative inhibitory strategies along the pathway are shown.
Abbreviations: NBD, NeMO binding peptide; GC, glucocorticoid; GILZ, glucocorticoid-induced leucine zipper; GR, glucocorticoid receptor; HDAC, histone deacytelases; NLS, nuclear localization signal; ODN, oligodeoxynucleotide; PCAF, p300/CBP-associated factor; TAD, transactivation domain.
Inhibitors of the NF-κB p65:IκB complex
As stated above, canonical activation of NF-κB p65 involves intracellular signaling networks that utilize adapter protein interactions, and phosphorylation and ubiquitination processes that hinge on signal-induced proximity and appropriate orientation of the multimeric subunits. Several chronic inflammatory pathologies are associated with dysregulated phosphorylation/activation of the NF-kB:IkB complex. Hence, preventing phosphorylation by using kinase inhibitors constitutes attractive therapeutic strategy. A vast number of kinase inhibitors target either the IKK complex or the adapter proteins in the NF-kB signal transduction pathway (please refer to excellent reviewsCitation3,Citation71,Citation73,Citation74). The therapeutic potential of these kinase inhibitors has been predominantly evaluated in multiple cancers.Citation3,Citation74 While none of these have been shown to specifically target p65, a few affect the post-translational modifications in p65 by inhibiting the associated cofactors. For example, p38 mitogen-activated protein kinase (MAPK) inhibitor has been shown to inhibit phosphorylation of the coactivator p300 and preclude acetylation on Lys310 of p65, thereby preventing DNA binding and transcriptional activity.Citation75 Second-generation p38 MAPK inhibitors are being evaluated in clinical trials for chronic inflammatory diseases such as COPD, Crohn’s disease, and Alzheimer’s disease.Citation76,Citation77
Inhibitors that prevent p65 nuclear translocation
NLS inhibitors: As stated above, nucleocytoplasmic shuttling of p65 is critical for transcriptional regulation of target genes. Binding of the p65 NLSs with the adapter proteins importins enables its translocation through the NPC.Citation11,Citation25 Within the nucleus, importin is dissociated by GTPases and the activated p65 is released to initiate transcription.Citation62 This suggests that inhibitors of the p65:importin adapter interactions may be selective NF-κB inhibitors. It was proposed that rationally designed peptides derived from the NLS of the NF-κB subunits would suppress nuclear translocation of p50:p65 dimers by competitively inhibiting their interaction with importins. A linear peptide of the p50 NLS sequence (SN50) covalently synthesized with a cell-permeable compound inhibited the nuclear import of NF-κB p65 in human monocytic cells and murine endothelial cells stimulated with the proinflammatory agonists such as lipopolysaccharide (LPS) or the cytokine TNF-α.Citation78,Citation79 Interestingly a short peptide mimic (RLRWR) of the DNA binding motif of p50 designated as anti-inflammatory peptide-6 exhibited cell-penetrating ability, interacted directly with the p65 thereby inhibiting DNA binding and transactivation of inflammatory mediators.Citation80 A cell-permeable peptide made up of NLS of c-myc has also been shown to inhibit nuclear translocation of p65 and suppress T-cell responses in models of inflammation.Citation81,Citation82
Many inhibitors that directly target select importin-α, improtin-β, or exportin proteins have been developed to inhibit nuclear translocation of p65 and subsequent cellular responses. Examples of this class of inhibitors include small molecules such as inhibitor of nuclear import-43 and selinexor, a selective inhibitor of nuclear export compound that inhibits the exportin XPO1.Citation83,Citation84 The efficacy of these agents has been largely investigated in tumor models and is yet to be evaluated in inflammatory pathologies.
Blocking p65 TAD: This includes strategies that inhibit p65 independent of other rel proteins.
Competing p65 peptide: The p65 nuclear translocation and consequent transactivation of target genes is facilitated by interactions of p65-TAD with other transcription factors and cofactors. A conceptually simple strategy for development of inhibitors of interprotein interactions is to take advantage of the evolutionary selection of residues for individual protein:protein interaction and design interface peptides derived from the primary sequence of one of the binding partners.Citation85,Citation86 As stated above, phosphorylation on Ser276, Ser529, Ser536, and Ser471 induces conformational changes in p65 and facilitates its binding with other transcriptional cofactors such as p300 and PCAF.Citation59 Synthetic peptides encompassing these phosphorylation sites in p65 fused with a protein transduction domain have been shown to suppress NF-κB activation induced by a variety of stimuli such as LPS, interleukin-1, okadaic acid, phorbol 12-myristate 13-acetate, and cigarette smoke condensate inflammatory stimuli.Citation61
Glucocorticoids-mediated cytoplasmic sequestration of p65: The classical model for the therapeutic potential of glucocorticoids suggests that the direct binding of the glucocorticoid receptor on specific glucocorticoid receptor elements (GREs) in the promoter region upregulates transactivation of anti-inflammatory genes.Citation87,Citation88 With six GRE in its promoter, glucocorticoid-induced leucine zipper is strongly upregulated by glucocorticoids.Citation89 GILZ has been shown to physically bind the p65 TAD via its proline-rich region and inhibit NF-κB signaling.Citation90,Citation91 In protein interfaces, proline-rich regions that adopt extended polyproline type II helical conformation constitute excellent drug templates with the specificity of the interaction determined by the residues adjacent to the interacting moieties in each partner.Citation60,Citation92 Synthetic peptide mimics of the p65 binding motif of GILZ suppressed nuclear translocation of p65, inflammation, and apoptosis.Citation93,Citation94
The expanding network of NF-κB interactors has increased the potential for identifying newer targets for specific inhibition. Among the proteins that bind p65, a subset directly binds the TAD such as the p300,Citation95,Citation96 silencing mediator of retinoic acid and thyroid hormone receptorsCitation97 and the Smad proteins. Smads are a family of structurally similar proteins belonging to the TGF-β superfamily involved in regulating cell development and growth. In humans, eight Smads are known and are classified into three subtypes: five receptor-regulated Smads (R-Smads), one common partner Smad (co-Smad), and two inhibitory Smads (I-Smad). While the R-Smads mediate signaling from the TGF-β receptor and I-Smads suppress R-Smads, the co-Smad recruits coregula-tors to the transcriptional factors and modules gene expressions. Structurally, the Smad proteins have two conserved globular domains called the mad homology (MH) 1 and MH2 domains.Citation98 Recently, using co-immunoprecipitation assays, it has been reported that the TA2 domain of p65 physically interacted with the MH1 domain of Smad4.Citation99 A peptide derived from the amino-terminal region of TA2 that associates with the MH1 domain of Smad4 covalently synthesized with a cell-permeable peptide prevented NF-κB signaling-mediated inhibition of osteoblast differentiation by blocking the association of p65 with Smad4 and allowing increased bone morphogenetic protein-2 signaling.Citation100
Inhibitors of nuclear p65
Decoy oligodeoxynucleotides (decoy ODNs): They constitute a type of gene therapy and are made up of double-stranded DNA fragments of the same sequence as the binding site of the transcription factor on DNA.Citation101 Decoy ODNs possessing κB consensus sequence bind NF-κB p65, prevent it from interacting with the cis-element of specific target gene, and thereby inhibit initiation of the transcription process.Citation102 Transfection of NF-κB decoy ODNs to the arteries by hemagglutinating virus of Japan liposomes prevented ischemia-induced myocardial infarction by suppressing expression of cytokines and adhesion molecules.Citation103 Similar observation of protection against inflammation following decoy NF-κB ODNs has been reported in models of arthritis, colitis, cystic fibrosis, and atopic dermatitis.Citation104–Citation106 The relative ease of design and construction is a significant advantage of decoy ODNs as therapeutic agents. However, limited cellular uptake due to the negative charge and large size as well as severe toxicities due to lack of cellular restrictions is a potential drawback. Recently, several strategies have been developed for successful and targeted delivery of decoy ODNs such as the use of nanoparticles and ultrasound-targeted microbubbles.Citation101,Citation107 NF-κB decoy ODNs in such formulation are currently in clinical trials for atopic dermatitis and discogenic low back pain, and they appear to be well tolerated, nontoxic, and potentially efficaciousCitation108 (www.anges.co.jp).
Selective glucocorticoid receptor agonists (SEGRA): Cross talk between the NF-κB and the GR signaling occurs at multiple levels such as the physical interaction between the DNA binding domain of GR and the nuclear p65, direct interference of GR with p65 and associated basal transcription machinery, and competitive binding of co-activators such as PKAc or CBP.Citation87,Citation109,Citation110 Many SEGRA or dissociated agonists of glucocorticoid receptor based on steroidal scaffold or non-steroidal modulators of the glucocorticoid receptor (SEGRM) developed to selectively facilitate that transrepressive actions are enhanced have been shown to inhibit NF-κB-mediated inflammatory responses.Citation111–Citation113 In this context it has been reported that coactivation of GR and p65 results in their association at binding sites that cluster with p65 target genes suggesting competition between GR and p65 for binding specific response elements.Citation87 Interestingly, SEGRM of the family of diabenzoxepane or dibenzosuperane (compound 10, PF-802, Fosdagocorat) that bind GR at sites other than the ligand (glucocorticoid)-binding domain induce conformational changes precluding GR binding the GRE.Citation113,Citation114 Together with the enhanced potential for the nuclear GR to compete for the co-activators, the anti-inflammatory effects of such SEGRM could be attributed to specific inhibition of NF-κB p65.Citation113,Citation115,Citation116 Elucidation of these mechanisms would also assist in adopting the information from the GR:p65 interactome in developing agents that provide beneficial (predominantly transrepressive) effects of glucocorticoids while avoiding the serious adverse responses.
Inhibitors of post-translational modifications
The dynamic nature of the PTMs in regulating gene expression patterns makes this system particularly amenable for epigenetic drug discovery. Considerable evidence suggests that the NF-κB p65 PTMs are deregulated in conditions of chronic inflammation and autoimmune diseases.Citation28,Citation58 Targeting the p65 PTM sites or the protein:protein interactions represents powerful approaches for amelioration of these conditions. Although all PTMs are important components of the epigenome, acetylation network consists of a large number of druggable targets.
HAT inhibitors: As stated above, HATs are epigenetic enzymes that add acetyl groups onto lysine residues of proteins including NF-κB p65. The following include few examples of HAT inhibitors that selectively target lysine residues on p65.
PCAF inhibitors: The PCAF inhibitors specifically acetylate only Lys-122 of p65 and blocking of this acetylation could diminish the nuclear retention and transcriptional activity of p65.Citation42,Citation45 Specific inhibitors of PCAF developed using structure-based design and molecular docking have been shown to inhibit p65-induced transactivation of inflammatory mediators in stimulated macrophages and glial cells.Citation117
Tip60, a member of the MYST family of co-activators, has been shown to activate p65-mediated transcription by maintaining the Lys310 in the acetylated state.Citation49 Analogs of the naturally occurring anacardic acid (6-pentadecylsalicylic acid/MG 149) that inhibit Tip60 have been shown to reduce proinflammatory gene expression in murine precision cut lung slices (PCLS).Citation42,Citation118
HDAC inhibitors: Several studies have reported the potential of small-molecule HDAC inhibitors to regulate p65 acetylation.Citation43,Citation119 The functional efficacy of pan-HDAC inhibitors in multiple cancers has been reviewed extensively.Citation120–Citation122 The following are few examples of HADC inhibitors that have been shown to modulate inflammation.
NF-κB p65 acetylation on Lys 310 by a HDAC1–3 inhibitor, MS-275, has been shown to exert mixed effects depending on the cell type and the nature of the stimuli. In LPS-stimulated macrophages and murine PCLS, treatment with MS-275 upregulated both anti-inflammatory and pro-inflammatory cytokines.Citation123 Similarly, in poly(I-C)-induced dendritic cells, MS-275 reduced the release of both the proinflammatory TNF-α, IL-6, and IL-12 cytokines and the anti-inflammatory IL-10.Citation124 In contrast, in a model of cigarette smoke-exposed lung inflammation that mimics the human COPD, MS-275 treatment robustly attenuated the expression of inflammatory chemokines and decreased neutrophil influx in the lungs.Citation123 Another HDAC 1–2 inhibitor, KBH-A42, reduced LPS-induced endotoxemia.Citation125 With both NLS and nuclear export sequence, HDAC-3 exerts proinflammatory effects.Citation126 Silencing HDAC-3 reduced vascular cell adhesion molecule 1, monocyte recruitment, proinflammatory transcriptional activity, and disease progression in an allergic skin inflammation model.Citation126 In human macrophages, HDAC3 siRNA inhibited cytokine response to LPS.Citation127 In contrast to the multiple HDAC inhibitor MS 275, treatment with RGFP966, a selective and potent inhibitor of HDAC-3, upregulated IL-10 and exhibited anti-inflammatory effects in response to LPS/IFNγ in macrophages and in mouse PCLS.Citation128 Deacetylation of Lys-310 by the HDAC SIRT1 has been shown to inhibit p65-mediated transactivation of cytokines and apoptosis mediators.Citation46 Inhibiting the activity of SIRT1 markedly increased the acetylation of Lys-310 and the transcriptional activity of NF-κB.Citation44,Citation56
The arginine methyltransferase, PRMT5, that demethylates R30 of p65 plays a key role in regulating endothelial cell inflammation, proliferation, and differentiation.Citation66,Citation123 Increased expression of PRMT5 that correlates with the enhanced inflammatory cytokines has been observed in the synovial tissues in rheumatoid arthritis. Inhibition of PRMT5 with a specific inhibitor EP015666, has been shown to decrease phosphorylation of IKK and IKBα, reduce trans-activation of inflammatory cytokines, and suppress fibroblast migration potentially ameliorating disease progression in arthritis.Citation129 Another PRMT5 inhibitor, HLCL65, has been shown to selectively inhibit pathogenic T cells in a mouse model of multiple sclerosis.Citation130 Taken together these observations suggest that the manipulation of specific acetylation sites in p65 can lead to resultant proinflammatory or anti-inflammatory effects.
Conclusion
Overwhelming evidence supports a critical role of increased NF-κB p65-mediated transactivation in the pathogenesis of multiple chronic inflammatory diseases. The beneficial effects of many natural compounds such as the curcumin and that of the synthetic inhibitors of upstream signaling molecules such as the cytokine and cytokine receptor antagonists in chronic inflammatory diseases have been attributed to reduced NF-κB p65 signaling.Citation3,Citation69–Citation71,Citation101 Hence, targeting p65 activation directly for better clinical efficacy has been a long-sought-after goal. As discussed earlier, several strategies to block p65 at each level along the path of signaling have been assessed and novel strategies are being developed. However, a critical hurdle is selective targeting of activated p65 without the off-target effects. In this context, steric blockade of p65-TAD using GILZ mimetics or mimics of the p65:Smad4 interface appears to be a promising strategy to selectively inhibit activated p65.Citation91,Citation100,Citation131 Targeted intracellular delivery in a context- and tissue-specific manner is another drawback that impedes development of effective p65 inhibitors. It is anticipated that advances in nanotechnology-based delivery systems will assist in not only improving the pharmacokinetic profile but also facilitate contextual delivery of drugs and drug-like agents. Recently, a nanocarrier delivery system consisting of a functional cell-penetrating stearoyl-oligopeptide was used for effective delivery of two RNAi agents that silenced p65 mRNA in macrophages and exerted therapeutic effects in a model of atopic dermatitis.Citation132 The properties of biocompatibility and stability in body fluids and lack of immunogenicity make nanocarriers as ideal portals for drug delivery. In future, development of engineered nanocarriers will not only facilitate targeted delivery but also accommodate conformational changes needed for efficacious intracellular p65 inhibitors.Citation1,Citation133
Acknowledgments
The authors sincerely appreciate funding from the National Institutes of Health (1R41 AG053117) and from Provaidya LLC to MS.
Disclosure
The authors report no conflicts of interest in this work.
References
- BasakSBeharMHoffmannALessons from mathematically modeling the NF-κB pathwayImmunol Rev2012246122123822435558
- OeckinghausAGhoshSThe NF-kappaB family of transcription factors and its regulationCold Spring Harb Perspect Biol200914a00003420066092
- KarinMYamamotoYWangQMThe IKK NF-kappa B system: a treasure trove for drug developmentNat Rev Drug Discov200431172614708018
- ChristianFSmithECarmodyRThe regulation of NF-κB subunits by phosphorylationCells20165112
- RazaniBZarnegarBYtterbergAJNegative feedback in noncanonical NF-kappaB signaling modulates NIK stability through IKKalpha-mediated phosphorylationSci Signal20103123ra4120501937
- GrayCMRemouchampsCMccorkellKANoncanonical NF-κB signaling is limited by classical NF-κB activitySci Signal20147311ra1324497610
- OeckinghausAHaydenMSGhoshSCrosstalk in NF-κB signaling pathwaysNat Immunol201112869570821772278
- ShihVF-STsuiRCaldwellAHoffmannAA single NFκB system for both canonical and non-canonical signalingCell Res20112118610221102550
- ChenL-FGreeneWCShaping the nuclear action of NF-κBNat Rev Mol Cell Biol20045539240115122352
- CollinsPMitxitorenaICarmodyRThe ubiquitination of NF-κB subunits in the control of transcriptionCells20165223
- LiangPZhangHWangGKPNB1, XPO7 and IPO8 mediate the translocation of NF-kappaB/p65 into the nucleusTraffic2013141132114323906023
- BaylissRCorbettAHStewartMThe molecular mechanism of transport of macromolecules through nuclear pore complexesTraffic20001644845611208130
- KorwekZTudelskaKNałęcz-JaweckiPImportins promote high-frequency NF-κB oscillations increasing information channel capacityBiol Direct20161116127835978
- PhelpsCBSengchanthalangsyLLMalekSGhoshGMechanism of kappa B DNA binding by Rel/NF-kappa B dimersJ Biol Chem2000275243922439910825175
- SchmitzMLBaeuerlePAThe p65 subunit is responsible for the strong transcription activating potential of NF-kappa BEmbo J19911012380538171935902
- van EssenDEngistBNatoliGSaccaniSTwo modes of transcriptional activation at native promoters by NF-kappaB p65PLoS Biol200973e7319338389
- KoyanagiMHijikataMWatashiKMasuiOShimotohnoKCentrosomal P4.1-associated protein is a new member of transcriptional coactivators for nuclear factor-kappaBJ Biol Chem200528013124301243715687488
- UranishiHTetsukaTYamashitaMInvolvement of the pro-oncoprotein TLS (translocated in liposarcoma) in nuclear factor-kappa B p65-mediated transcription as a coactivatorJ Biol Chem200127616133951340111278855
- HuangTTKudoNYoshidaMMiyamotoSA nuclear export signal in the N-terminal regulatory domain of Ikappa Balpha controls cytoplasmic localization of inactive NF-kappa B/Ikappa Balpha complexesProc Natl Acad Sci20009731014101910655476
- HuxfordTHuangDBMalekSGhoshGThe crystal structure of the IkappaBalpha/NF-kappaB complex reveals mechanisms of NF-kappaB inactivationCell19989567597709865694
- JacobsMDHarrisonSCStructure of an IkappaBalpha/NF-kappaB complexCell1998957497589865693
- Arenzana-SeisdedosFThompsonJRodriguezMSBachelerieFThomasDHayRTInducible nuclear expression of newly synthesized I kappa B alpha negatively regulates DNA-binding and transcriptional activities of NF-kappa BMol Cell Biol1995155268926967739549
- BaldwinASRegulation of cell death and autophagy by IKK and NF-κB: critical mechanisms in immune function and cancerImmunol Rev2012246132734522435564
- YdePMengelBJensenMHKrishnaSTrusinaAModeling the NF-κB mediated inflammatory response predicts cytokine waves in tissueBMC Syst Biol20115111521771307
- FagerlundRKinnunenLKohlerMJulkunenIMelenKNF-{kappa} B is transported into the nucleus by importin {alpha}3 and importin {alpha}4J Biol Chem2005280159421595115677444
- KöhlerMSpeckCChristiansenMEvidence for distinct substrate specificities of importin alpha family members in nuclear protein importMol Cell Biol199919117782779110523667
- SteinBCogswellPCBaldwinASFunctional and physical associations between NF-kappa B and C/EBP family members: a Rel domain-bZIP interactionMol Cell Biol1993137396439748321203
- HuangBYangX-DLambAChenL-FPosttranslational modifications of NF-κB: another layer of regulation for NF-κB signaling pathwayCell Signal20102291282129020363318
- BarnesPJKinases as novel therapeutic targets in asthma and chronic obstructive pulmonary diseasePharmacol Rev201668378881527363440
- ZhongHSuyangHErdjument-BromageHTempstPGhoshSThe transcriptional activity of NF-kappaB is regulated by the IkappaB-associated PKAc subunit through a cyclic AMP-independent mechanismCell19978934134249150141
- ReberLVermeulenLHaegemanGFrossardNSer276 phosphorylation of NF-kB p65 by MSK1 controls SCF expression in inflammationPLoS One200942e439319197368
- RyoASuizuFYoshidaYRegulation of NF-kappaB signaling by Pin1-dependent prolyl isomerization and ubiquitin-mediated proteolysis of p65/RelAMol Cell20031261413142614690596
- BussHDörrieASchmitzMLPhosphorylation of serine 468 by GSK-3beta negatively regulates basal p65 NF-kappaB activityJ Biol Chem200427948495714957415465828
- MattioliIGengHSebaldAInducible phosphorylation of NF-kappa B p65 at serine 468 by T cell costimulation is mediated by IKK epsilonJ Biol Chem2006281106175618316407239
- MsakiASánchezAMKohLFThe role of RelA (p65) threonine 505 phosphorylation in the regulation of cell growth, survival, and migrationMol Biol Cell201122173032304021737676
- KarinMBen-NeriahYPhosphorylation meets ubiquitination: the control of NF-κB activityAnnu Rev Immunol200018162166310837071
- SakuraiHChibaHMiyoshiHSugitaTToriumiWIkappaB kinases phosphorylate NF-kappaB p65 subunit on serine 536 in the transactivation domainJ Biol Chem199927443303533035610521409
- ViatourPMervilleM-PBoursVChariotAPhosphorylation of NF-κB and IκB proteins: implications in cancer and inflammationTrends Biochem Sci2005301435215653325
- BussHHandschickKJurrmannNCyclin-dependent kinase 6 phosphorylates NF-κB P65 at serine 536 and contributes to the regulation of inflammatory gene expressionPLoS One2012712e5184723300567
- HandschickKBeuerleinKJuridaLCyclin-dependent kinase 6 is a chromatin-bound cofactor for NF-κB-dependent gene expressionMol Cell201453219320824389100
- ShanmugasundaramKNayakBShimEHLiviCBBlockKSudarshanSThe oncometabolite fumarate promotes pseudohypoxia through noncanonical activation of NF-κB signalingJ Biol Chem201428935246912469925028521
- DekkerFJvan den BoschTMartinNISmall molecule inhibitors of histone acetyltransferases and deacetylases are potential drugs for inflammatory diseasesDrug Discov Today201419565466024269836
- CalaoMBurnyAQuivyVDekoninckAvan LintCA pervasive role of histone acetyltransferases and deacetylases in an NF-κB-signaling codeTrends Biochem Sci200833733934918585916
- ChenL-FMuYGreeneWCAcetylation of RelA at discrete sites regulates distinct nuclear functions of NF-kappaBEmbo J200221236539654812456660
- KiernanRBrèsVNgRWMPost-activation turn-off of NF-κB-dependent transcription is regulated by acetylation of p65J Biol Chem200327842758276612419806
- YeungFHobergJERamseyCSModulation of NF-κB-dependent transcription and cell survival by the SIRT1 deacetylaseEmbo J200423122369238015152190
- BuerkiCRothgiesserKMValovkaTFunctional relevance of novel p300-mediated lysine 314 and 315 acetylation of RelA/p65Nucleic Acids Res20083651665168018263619
- HuangBYangX-DZhouM-MOzatoKChenL-FBrd4 coactivates transcriptional activation of NF- B via specific binding to acetylated RelAMol Cell Biol20092951375138719103749
- KimJ-WJangS-MKimC-HAnJ-HKangE-JChoiK-HNew molecular bridge between RelA/p65 and NF-κB target genes via histone acetyltransferase TIP60 cofactorJ Biol Chem2012287107780779122249179
- LuTStarkGRNF-κB: regulation by methylationCancer Res201575183692369526337909
- LuTStarkGRUsing sequential immunoprecipitation and mass spectrometry to identify methylation of NF-kappaBMethods Mol Biol2015128038339325736762
- CkEBaltimoreDRegulation of NF-kappaB activity through lysine monomethylation of p65Proc Natl Acad Sci U S A2009106189721897719864627
- WeiHWangBMiyagiMPRMT5 dimethylates R30 of the p65 subunit to activate NF- BProc Natl Acad Sci U S A201311033135161352123904475
- YangX-DHuangBLiMLambAKelleherNLChenL-FNegative regulation of NF-κB action by Set9-mediated lysine methylation of the RelA subunitEmbo J20092881055106619262565
- LuTJacksonMWWangBRegulation of NF- B by NSD1/FBXL11-dependent reversible lysine methylation of p65Proc Natl Acad Sci U S A20101071465120080798
- QuivyVvan LintCRegulation at multiple levels of NF-κB-mediated transactivation by protein acetylationBiochem Pharmacol20046861221122915313420
- BhattDGhoshSRegulation of the NF-κB-mediated transcription of inflammatory genesFront Immunol201457124611065
- ChenL-FWilliamsSAMuYNF- B RelA phosphorylation regulates RelA acetylationMol Cell Biol200525187966797516135789
- GaoJXuDCorrelation between posttranslational modification and intrinsic disorder in proteinPac Symp Biocomput20129410322174266
- SrinivasanMDunkerAKProline rich motifs as drug targets in immune mediated disordersInt J Pept201220127 Article ID 63476914
- TakadaYSinghSAggarwalBBIdentification of a p65 peptide that selectively inhibits NF-kappa B activation induced by various inflammatory stimuli and its role in down-regulation of NF-kappaB-mediated gene expression and up-regulation of apoptosisJ Biol Chem200427915150961510414711835
- MacaraIGTransport into and out of the nucleusMicrobiol Mol Biol Rev200165457059411729264
- ChewJBiswasSShreeramSWIP1 phosphatase is a negative regulator of NF-κB signallingNat Cell Biol200911565966619377466
- YangJFanGHWadzinskiBESakuraiHRichmondAProtein phosphatase 2A interacts with and directly dephosphorylates RelAJ Biol Chem200127651478284783311591705
- YehPYYehKHChuangSESongYCChengALSuppression of MEK/ERK signaling pathway enhances cisplatin-induced NF-kappaB activation by protein phosphatase 4-mediated NF-kappaB p65 Thr dephosphorylationJ Biol Chem200427925261432614815073167
- LuTYangMHuangDBRole of lysine methylation of NF-κB in differential gene regulationProc Natl Acad Sci U S A201311033135101351523904479
- GengHWittwerTDittrich-BreiholzOKrachtMSchmitzMLPhosphorylation of NF-κB p65 at Ser468 controls its COMMD1-dependent ubiquitination and target gene-specific proteasomal eliminationEMBO Rep200910438138619270718
- BussHDörrieASchmitzMLHoffmannEReschKKrachtMConstitutive and interleukin-1-inducible phosphorylation of p65 NF-{kappa}B at serine 536 is mediated by multiple protein kinases including I{kappa}B kinase (IKK)-{alpha}, IKK{beta}, IKK{epsilon}, TRAF family member-associated (TANK)-binding kinase 1 (TBK1), and an unknown kinase and couples p65 to TATA-binding protein-associated factor II31-mediated interleukin-8 transcriptionJ Biol Chem200427953556335564315489227
- CalzadoMBacherSSchmitzMLNF-kappaB inhibitors for the treatment of inflammatory diseases and cancerCurr Med Chem200714336737617305539
- GaspariniCFeldmannMNF-kappaB as a target for modulating inflammatory responsesCurr Pharm Des201218355735574522726116
- GuptaSCSundaramCReuterSAggarwalBBInhibiting NF-kappaB activation by small molecules as a therapeutic strategyBiochim Biophys Acta2010179977578720493977
- LiuTZhangLJooDSunSCNF-κB signaling in inflammationSignal Transduct Target Ther201721702329158945
- UweSAnti-inflammatory interventions of NF-kappaB signaling: potential applications and risksBiochem Pharmacol20087581567157918070616
- UehlingDEHarrisPARecent progress on MAP kinase pathway inhibitorsBioorg Med Chem Lett201525194047405626298497
- SahaRNJanaMPahanKMAPK p38 regulates transcriptional activity of NF-kappaB in primary human astrocytes via acetylation of p65J Immunol2007179107101710917982102
- AlamJBlackburnKPatrickDNeflamapimod: clinical phase 2b-ready oral small molecule inhibitor of p38alpha to reverse synaptic dysfunction in early Alzheimer’s diseaseJ Prev Alzheimers Dis2017427327829181493
- BühlerSLauferSAp38 MAPK inhibitors: a patent review (2012 – 2013Expert Opin Ther Pat201424553555424611721
- FujiharaSMCleavelandJSGrosmaireLSA D-amino acid peptide inhibitor of NF-kappa B nuclear localization is efficacious in models of inflammatory diseaseJ Immunol200016521004101210878377
- LinYZYaoSYVeachRATorgersonTRHawigerJInhibition of nuclear translocation of transcription factor NF-kappa B by a synthetic peptide containing a cell membrane-permeable motif and nuclear localization sequenceJ Biol Chem19952702414255142587782278
- WangYFXuXFanXA cell-penetrating peptide suppresses inflammation by inhibiting NF-κB signalingMol Ther201119101849185721556052
- NadlerSGTritschlerDHaffarOKBlakeJBruceAGCleavelandJSDifferential expression and sequence-specific interaction of karyopherin alpha with nuclear localization sequencesJ Biol Chem19972727431043159020149
- van der WattPJChiAStelmaTTargeting the nuclear import receptor Kpn 1 as an anticancer therapeuticMol Cancer Ther201615456057326832790
- KashyapTArguetaCAboukameelASelinexor, a selective inhibitor of nuclear export (SINE) compound, acts through NF-kappaB deactivation and combines with proteasome inhibitors to synergistically induce tumor cell deathOncotarget2016748788837889527713151
- StelmaTLeanerVDKPNB1-mediated nuclear import is required for motility and inflammatory transcription factor activity in cervical cancer cellsOncotarget2017820328333284728427184
- OlivaBFernandez-FuentesNKnowledge-based modeling of peptides at protein interfaces: PiPreDBioinformatics20153191405141025540186
- SrinivasanMInterface peptide mimetics: rationale and applications as therapeutic agentsMed Chem (Los Angeles)201663189194
- RaoNASMccalmanMTMoulosPCoactivation of GR and NFKB alters the repertoire of their binding sites and target genesGenome Res20112191404141621750107
- YamamotoKRSteroid receptor regulated transcription of specific genes and gene networksAnnu Rev Genet19851912092523909942
- RiccardiCGILZ (glucocorticoid-induced leucine zipper), a mediator of the anti-inflammatory and immunosuppressive activity of glucocorticoidsAnn Ig201022535920701225
- AyroldiERiccardiCGlucocorticoid-induced leucine zipper (GILZ): a new important mediator of glucocorticoid actionFaseb J200923113649365819567371
- SrinivasanMBayonBChopraNLahiriDKNovel nuclear factor-kappaB targeting peptide suppresses β-amyloid induced inflammatory and apoptotic responses in neuronal cellsPLoS One20161110e016031427764084
- CubellisMVCaillezFBlundellTLLovellSCProperties of polyproline II, a secondary structure element implicated in protein-protein interactionsProteins200558488089215657931
- SrinivasanMBlackburnCLahiriDKFunctional characterization of a competitive peptide antagonist of p65 in human macrophage-like cells suggests therapeutic potential for chronic inflammationDrug Des Devel Ther2014824092421
- SrinivasanMJanardhanamSNovel p65 binding glucocorticoid-induced leucine zipper peptide suppresses experimental autoimmune encephalomyelitisJ Biol Chem201128652447994481021965677
- O’SheaJMPerkinsNDRegulation of the RelA (p65) transactivation domainBiochem Soc Trans200836460360818631125
- GerritsenMEWilliamsAJNeishASMooreSShiYCollinsTCREB-binding protein/p300 are transcriptional coactivators of p65Proc Natl Acad Sci U S A1997947292729329096323
- LeeSKKimJHLeeYCCheongJLeeJWSilencing mediator of retinoic acid and thyroid hormone receptors, as a novel transcriptional corepressor molecule of activating protein-1, nuclear factor-kappaB, and serum response factorJ Biol Chem200027517124701247410777532
- DennlerSDijkeTSmad proteins in TGF-beta signalingSchwabMEncyclopedia of CancerBerlinSpringer2011
- Hirata-TsuchiyaSFukushimaHKatagiriTInhibition of BMP2-induced bone formation by the p65 subunit of NF-κB via an interaction with Smad4Mol Endocrinol20142891460147025029242
- UrataMKokabuSMatsubaraTA peptide that blocks the interaction of NF-κB p65 subunit with Smad4 enhances BMP2-induced osteogenesisJ Cell Physiol201823397356736629663368
- HeckerMWagnerAHTranscription factor decoy technology: a therapeutic updateBiochem Pharmacol2017144293428642036
- MorishitaRSugimotoTAokiMIn vivo transfection of cis element “decoy” against nuclear factor-kappaB binding site prevents myocardial infarctionNat Med1997388948999256281
- SawaYMorishitaRSuzukiKA novel strategy for myocardial protection using in vivo transfection of cis element ‘decoy’ against NFkappaB binding site: evidence for a role of NFkappaB in ischemia-reperfusion injuryCirculation1997969 Suppl280284
- DajeeMMuchamuelTSchryverBBlockade of experimental atopic dermatitis via topical NF-kappaB decoy oligonucleotideJ Invest Dermatol200612681792180316628194
- de StefanoDOligonucleotides decoy to NF-kappaB: becoming a reality?Discov Med201112639710521878187
- TaharaKSamuraSTsujiKOral nuclear factor-κB decoy oligonucleotides delivery system with chitosan modified poly(D,L-lactide-co-glycolide) nanospheres for inflammatory bowel diseaseBiomaterials201132387087820934748
- FarahmandLDarvishiBMajidzadeh-AKSuppression of chronic inflammation with engineered nanomaterials delivering nuclear factor κB transcription factor decoy oligodeoxynucleotidesDrug Deliv20172411249126128870118
- TamaiKKanedaYMorishitaRKatayamaIDevelopment of NF-kappa B decoy ointment and clinical trial for atopic dermatitisDermatitis200819293294
- DoucasVShiYMiyamotoSWestAVermaIEvansRMCytoplasmic catalytic subunit of protein kinase A mediates cross-repression by NF-kappa B and the glucocorticoid receptorProc Natl Acad Sci U S A20009722118931189811027313
- RayAPrefontaineKEPhysical association and functional antagonism between the p65 subunit of transcription factor NF-kappa B and the glucocorticoid receptorProc Natl Acad Sci U S A19949127527568290595
- LesovayaEYemelyanovASwartACSwartPHaegemanGBudunovaIDiscovery of compound A--a selective activator of the glucocorticoid receptor with anti-inflammatory and anti-cancer activityOncotarget2015631307303074426436695
- SchäckeHBergerMRehwinkelHAsadullahKSelective glucocorticoid receptor agonists (SEGRAs): novel ligands with an improved therapeutic indexMol Cell Endocrinol20072751–210911717630119
- SundahlNBridelanceJLibertCde BosscherKBeckIMSelective glucocorticoid receptor modulation: new directions with non-steroidal scaffoldsPharmacol Ther2015152284125958032
- CarsonMWLuzJGSuenCGlucocorticoid receptor modulators informed by crystallography lead to a new rationale for receptor selectivity, function, and implications for structure-based designJ Med Chem201457384986024446728
- StockTFleishakerDWangXMukherjeeAMebusCImproved disease activity with fosdagrocorat (PF-04171327), a partial agonist of the glucocorticoid receptor, in patients with rheumatoid arthritis: a Phase 2 randomized studyInt J Rheum Dis201720896097028328159
- CatleyMDissociated steroidsScientificWorldJournal2007742143017450306
- ParkSYKimMJKimYJSelective PCAF inhibitor ameliorates cognitive and behavioral deficits by suppressing NF-κB-mediated neuroinflammation induced by Aβ in a model of Alzheimer’s diseaseInt J Mol Med20153541109111825672970
- van den BoschTLeusNGJWapenaarHA 6-alkylsalicylate histone acetyltransferase inhibitor inhibits histone acetylation and pro-inflammatory gene expression in murine precision-cut lung slicesPulm Pharmacol Ther201744889528323055
- LegartováSStixováLStrnadHBasic nuclear processes affected by histone acetyltransferases and histone deacetylase inhibitorsEpigenomics20135437939623895652
- XuJSunJWangPMaXLiSPendant HDAC inhibitor SAHA derivatised polymer as a novel prodrug micellar carrier for anticancer drugsJ Drug Target2018265–644845729251528
- LiXZhangJXieYJiangYYingjieZXuWProgress of HDAC inhibitor panobinostat in the treatment of cancerCurr Drug Targets201415662263424597570
- GaoSLiXZangJXuWZhangYPreclinical and clinical studies of chidamide (CS055/HBI-8000), an orally available subtype-selective HDAC inhibitor for cancer therapyAnticancer Agents Med Chem201717680281227592546
- LeusNGvan den BoschTvan der WoudenPEHDAC1-3 inhibitor MS-275 enhances IL10 expression in RAW264.7 macrophages and reduces cigarette smoke-induced airway inflammation in miceSci Rep201774504728344354
- NencioniABeckJWerthDHistone deacetylase inhibitors affect dendritic cell differentiation and immunogenicityClin Cancer Res200713133933394117606727
- ChoiYParkSKKimHMHistone deacetylase inhibitor KBH-A42 inhibits cytokine production in RAW 264.7 macrophage cells and in vivo endotoxemia modelExp Mol Med200840557458118985016
- LeusNGZwindermanMRDekkerFJHistone deacetylase 3 (HDAC 3) as emerging drug target in NF-κB-mediated inflammationCurr Opin Chem Biol20163316016827371876
- WinklerARNockaKNWilliamsCMSmoke exposure of human macrophages reduces HDAC3 activity, resulting in enhanced inflammatory cytokine productionPulm Pharmacol Ther201225428629222613758
- LeusNGvan der WoudenPEvan den BoschTHDAC 3-selective inhibitor RGFP966 demonstrates anti-inflammatory properties in RAW 264.7 macrophages and mouse precision-cut lung slices by attenuating NF-κB p65 transcriptional activityBiochem Pharmacol2016108587426993378
- ChenDZengSHuangMXuHLiangLYangXRole of protein arginine methyltransferase 5 in inflammation and migration of fibroblast-like synoviocytes in rheumatoid arthritisJ Cell Mol Med201721478179027860244
- WebbLMAmiciSAJablonskiKAPRMT5-selective inhibitors suppress inflammatory T cell responses and experimental autoimmune encephalomyelitisJ Immunol201719841439145128087667
- SrinivasanMLahiriDKSignificance of NF-κB as a pivotal therapeutic target in the neurodegenerative pathologies of Alzheimer’s disease and multiple sclerosisExpert Opin Ther Targets201519447148725652642
- KanazawaTHamasakiTEndoTFunctional peptide nanocarriers for delivery of novel anti-RelA RNA interference agents as a topical treatment of atopic dermatitisInt J Pharm20154891–226126725956048
- DeciMBLiuMDinhQTNguyenJPrecision engineering of targeted nanocarriersWiley Interdiscip Rev Nanomed Nanobiotechnol Epub2018213
