Abstract
Purpose
This study aims at profiling the expression of dysregulated genes in circulating monocytes of patients with cancer-related lower limb lymphedema before and after treatment with supermicrosurgical lymphaticovenous anastomosis (LVA).
Materials and Methods
This prospective longitudinal cohort study enrolled 51 women with post-treatment gynecological cancer, including those with unilateral lymphedema (study group, n = 25) and those without (control group, n = 26). Venous blood samples obtained from the study group before and after LVA and those from the controls were sent for next-generation sequencing, which was validated by real-time PCR. Dysregulated gene expression in the study group, relative to expression in the controls, was recorded before LVA. After one month, postoperative changes in the expression of the identified genes were evaluated. Protein-protein interaction (PPI) was used to investigate dysregulated genes whose expression returned to baseline levels after LVA.
Results
Of the 148 preoperative dysregulated genes, which comprised 108 up- and 40 down-regulated genes, 78 genes, consisting of 69 up- and 9 down-regulated genes, showed post-LVA recovery to baseline levels. Through PPI analysis, five functional modules involving immunity, lipid metabolism, oxidative stress, transcriptional regulators, and tumor suppression, as well as six hub genes (CCL2, LPL, PDK4, FOXO3, EGR1, and DUSP5), were identified. Cross-linking and co-regulated genes between modules were also identified.
Conclusion
Localized lymphedema leads to dysregulated gene expression in circulating monocytes. The current study is the first to identify the hub genes related to lymphedema and demonstrate the recovery of some dysregulated genes after LVA.
Introduction
Lymphedema is a chronic, debilitating disease that affects as many as 200 million patients worldwide. In developed countries, cancer is the most frequent secondary cause for lymphedema. The most common risks of cancer-related lymphedema include regional lymphadenectomy, radiotherapy, and chemotherapy.Citation1 Lower limb lymphedema can occur in up to 70% of selected patients diagnosed with gynecological cancers.Citation2 This condition has been linked to edema, inflammation, tissue fibrosis, abnormal adipose deposition,Citation3 and oxidative stress.Citation4,Citation5 Conservative treatments, such as complex decongestive therapy (CDT), have been shown to decrease inflammatory gene expression after lymphedematous volume reduction.Citation6 However, due to limitations of CDT, alternative treatment modalities have been considered. Supermicrosurgical lymphaticovenous anastomosis (LVA) is effective in reducing excess lymphedematous volume by channeling the stagnant lymph into the recipient vein.Citation7,Citation8 Moreover, post-LVA lymphedematous limb volume reduction is correlated with a decrease in oxidative stress, as we recently demonstrated.Citation9
Currently, the systemic impact of cancer-related lower limb lymphedema on gene expression has not been assessed. Some studies have investigated the local dysregulation of gene expression, but these reports specifically focused on adipose-derived mesenchymal stem cells from breast cancer-related lymphedema (BCRL).Citation10,Citation11 Koc et al demonstrated the effects of lymphedema on miRNA expression in patients with BCRL.Citation12 Additionally, whether the expression of dysregulated genes is reversible after lymphatic surgical intervention requires further investigation. In the present study, we aimed to profile the expression of dysregulated genes in circulating monocytes of patients with cancer-related lower limb lymphedema, before and after LVA treatment.
Materials and Methods
Study Participants
The institutional review board of Kaohsiung Chang Gung Memorial Hospital (KCGMH) approved this prospective longitudinal cohort study (IRB No.: 201702190B0/201702190B0C501) and this study has complied with the Declaration of Helsinki. Written informed consent was obtained from all patients. The patient cohort was enrolled between November 2019 and September 2021, from KCGMH. The four recruitment criteria were as follows: female patients, 18 years and older, with gynecological cancer; unilateral lower limb lymphedema; lymphedema duration greater than 2 years; and lymphedema confirmed with indocyanine green (ICG) lymphography before surgery. Patients were excluded from the study group based on the following four exclusion criteria: a diagnosis of primary lymphedema, upper limb lymphedema, or bilateral lower limb lymphedema; a history of previous LVA, lymph node transfer, liposuction, or excisional therapy such as the Charles procedure; patients lost to follow-up; or patients with incomplete data. Details of the operative techniques have been previously described.Citation7 Serum samples from venous blood were collected from the arm before and one month after LVA from the study group. Patients without lymphedema who received treatments for gynecological cancers at the same institution were enrolled as controls with venous serum samples collected for comparison.
Definitions
The severity of the lymphedema was classified based on the International Society of Lymphology (ISL) staging system, namely mild (Stages 0-I) and moderate-to-severe (Stages II–III). Supermicrosurgical LVA was performed by a single senior surgeon with 11–0 nylon sutures (Ethilon, Ethicon, New Jersey, USA) using a high-power surgical microscope (Pentero 900, Carl Zeiss AG, Oberkochen, Germany). The positivity of ICG and flow of a lymphatic vessel (LV) were defined based on microscopic observations. ICG-positive LVs were defined as those that were positive for fluorescence observed at an excitation peak at 789 nm and an emission peak at 814 nm with a microscope-integrated–near-infrared imaging system (Pentero 900, Carl Zeiss AG, Oberkochen, Germany). Microscopically discernible lymphatic flow from the distal opening of a transected LV were defined as flow positive LVs. The diameters of the LVs were determined to a precision of 0.01 mm. The classification of lymphosclerosis was based on intraoperative findings of the four criteria reported in a previous study: wall thickness, appearance, wall expandability, and lumen.Citation13 All LVs were classified into four categories: s0 (very thin, translucent, expandable with identifiable lumen), s1 (thin, white, expandable with identifiable lumen), s2 (thick, white, not expandable with identifiable lumen), and s3 (very thick, white, not expandable with unidentifiable lumen). For LVs, categories s0 and s1 were considered ideal, s2 suboptimal, and s3 not suitable.
Operative Technique
The first and third interdigital toe webbing and the medial and lateral malleoli were intradermally injected with 0.1 mL of ICG. The linear dermal backflow (DB) pattern of the LV was detected with a handheld near-infrared imaging device (Fluobeam, FluoOptic, Grenoble, France) to guide incision placement. For patients with a diffuse DB pattern where linear patterns could not be observed, incisions were placed along the anatomical location of the great saphenous vein. The incision was usually 3 cm in length and extended when necessary. LVA was performed using only antegrade LV. The operative techniques were as previously described.Citation7
Experimental Design and Sample Preparation
Isolation of Peripheral Blood Mononuclear Cells
Venous blood samples were obtained from the controls and the study group. Peripheral blood mononuclear cells (PBMCs) were separated from whole blood by density gradient centrifugation using Ficoll–Paque Premium (17-5442-02, Merck, Kenilworth, New Jersey, U.S.A.). Specifically, 4 mL of Ficoll solution was added to the bottom of a 15 mL tube before the addition of 8 mL of whole blood. After centrifugation at 400 g for 40 min at 25 °C, PBMCs were collected from the plasma/Ficoll interface.
After PBMC separation, CD14+ cells were isolated using anti-human CD14 magnetic particles (557769, BD Bioscience, Franklin Lakes, New Jersey, U.S.A.). PBMCs were diluted with equal amounts of phosphate-buffered saline and centrifuged at 240 g for 5 min. The supernatant was discarded, and the cell pellets were resuspended in 1X BD IMag™ buffer. Cell numbers were determined, and cell suspensions were centrifuged at 200 g for 10 min. For every 107 cells, 50 μL of BD IMag™ anti-human CD14 magnetic particles was added to the supernatant. The cell-magnetic particle mixture was incubated at room temperature for 30 min. After washing with 1X BD IMag™ buffer, pellets of CD14+ cells were collected and resuspended in 500 μL of QIAzol lysis reagent (RLT buffer, RNeasy Mini Kit, Qiagen, Venlo, Netherlands) for further use.
RNA Extraction, Quantification, and Quality Determination
Total RNA was extracted from CD14+ cells using the RNeasy Mini Kit (74104, Qiagen, Venlo, Netherlands) following the manufacturer’s instructions. RNA samples were quantified with the NanoDrop 2000 spectrophotometer (Thermo Scientific, Waltham, Massachusetts, USA) and Qubit RNA Assay Kit (Q10210, Thermo Scientific, Waltham, Massachusetts, U.S.A.). RNA quality was assessed using the RNA integrity number (RIN) using a Caliper LabChip Analyzer (PerkinElmer, Waltham, Massachusetts, U.S.A.).
Library Preparation and Sequencing
Libraries were prepared from 1 μg of total RNA using TruSeq Stranded mRNA (15031047, Illumina, San Diego, California, USA) for Illumina® Platforms in accordance with the manufacturer’s instructions. Library concentrations were measured using the Qubit dsDNA HS Assay Kit (Q32851, Thermo Fisher, Waltham, Massachusetts, U.S.A.). The quality and concentration of the libraries were determined using an Agilent high sensitivity DNA assay kit (5067-4626, Agilent, Santa Clara, California, U.S.A.). Libraries were sequenced using a NextSeq 500 system (Illumina, San Diego, California, U.S.A.).
Data Analysis
The quality of the FASTQ sequence files was evaluated using FastQC (v0.11.8). STAR (v2.7.3a) was used for RNA-seq read alignment and quantification. The reads were aligned to the human reference genome assembly (hg38). Differential gene expression analysis was performed using DESeq2 (v1.24.0). For genes whose changes in differential expression were over two-fold with an adjusted p-value ≤ 0.05, gene ontology and KEGG pathways were analyzed using goseq (v1.36.0). Genes with restored expression after LVA were imported into the STRING database of known and predicted protein-protein interactions (PPIs) (https://string-db.org).
Real-Time Quantitative Polymerase Chain Reaction (PCR)
From the NGS data, genes exhibiting greater than 2-fold changes in expression were selected for real-time PCR validation. For validation, reverse transcription of CD14 RNA to cDNA was performed using a high-capacity cDNA reverse transcription kit (4368814, Applied Biosystems, Waltham, Massachusetts, USA) according to the manufacturer’s instructions. Quantification was conducted using Fast SYBR Green Master Mix (Applied Biosystems, 4385612) and 7500 Real-Time PCR System (Thermo Fisher). Six genes were chosen and the sequences of the primers used were as follows: GAPDH, forward 5’-ACAGTCAGCCGCATCTTCTT-3’ and reverse 5’-GCCCAATACGACCAAATCC-3’; CCL2, forward 5’-GATCTCAGTGCAGAGGC-3’ and reverse 5’-GGTTTGCTTGTCCAGGT-3’; SLC1A3, forward 5’-AGCAATGGAGAAGAGCC-3’ and reverse 5’-ACAATGACAGCGGTGAC-3’; SPRY1, forward 5’-ACCTGCATGTGCTTAGT-3’ and reverse 5’-CTTTAGCAGGAGGATAACAGAGTA-3’; ALOX15B, forward 5’-TGGAGGAGTCTGAATGAGATG-3’ and reverse 5’-GGAAGTTCTTTGGGAGGTAG-3’; ZBTB16, forward 5’-CATGGACTTCAGCACCTAT-3’ and reverse 5’-ACAGCCTCGTTATCAGGA-3’; GPI, forward 5’-CTGCTGGGTATCTGGTACA-3’ and reverse 5’-CACACGGGTTCCAGATT-3’. The real-time PCR master mix was prepared as follows: 10 μL 2X Fast SYBR Green Master Mix, 1 μL forward primer (10 μM), 1 μL reverse primer (10 μM), 1 μL cDNA, and 7 μL nuclease-free water. The default PCR thermal cycling conditions were as follows: 20s at 95°C and 40 cycles of 3s at 95°C and 30s at 60°C.
Statistical Analysis
All statistical analyses were performed using GraphPad Prism 6 (GraphPad Software Inc., San Diego, CA, USA). Quantitative interoperative findings are presented as the median with interquartile range (IQR). Real-time PCR data are presented as mean ± standard error. Data analysis of the differences between groups means was performed using one-way analysis of variance, followed by Tukey’s honestly significant difference test. Statistical significance was set at p < 0.05.
Results
Demographic Data
A total of 51 women with post-treatment gynecological cancer included those with unilateral lymphedema (study group, n=25) and their lymphedema-free counterparts (control group, n=26). When comparing patient characteristics (), the patients in the study group were found to have a higher incidence of cervical cancer (36.0% vs 0.0%) and a significantly (p = 0.002) lower occurrence of ovarian cancer (16.0% vs 46.2%) than the patients in the control group. The study group also had a higher prevalence of radiotherapy than the control group (44.0% vs 7.7%, p = 0.008). No significant intergroup differences were noted in sex (p = 1.000), age (p = 0.166), body mass index (p = 0.840), diabetes (p = 0.941), hypertension (p > 0.999), or chemotherapy (p = 0.651), and the duration of post-cancer treatment follow-up (p = 0.423).
Table 1 Patient Demographics (n=51)
Intraoperative Findings
Supermicrosurgical LVA was performed on the patients of the study group and the intraoperative findings are presented in . In total, 144 LVs were identified with a median of 6.0 LVAs per patient (IQR 4.0–7.0). The median LV diameter was 0.68 mm (IQR 0.51–0.80 mm). The total number of ICG-positive LVs was 109 (75.7%) with a median diameter of 0.75 mm (IQR 0.57–0.81). Based on the lymphosclerosis classification, 10 s0 (6.9%), 98 s1 (68.1%), 29 s2 (20.1%), and 7 s3 (4.9%) LVs were identified. A total of 83 recipient veins were identified with a median of 3.0 (IQR 2.0–4.0) recipient veins per patient and a median diameter of 0.97 mm (IQR 0.8–1.2).
Table 2 Intraoperative Findings
Gene Expression Analysis
Next-generation sequencing (NGS) was used to evaluate gene expression before and one month after LVA. Expression analysis was conducted to compare the study and control groups, with 10 patients from each group, to identify genes that were dysregulated prior to LVA and those that were restored to baseline levels following LVA. Before LVA, a total of 148 dysregulated genes were identified by NGS, which included 108 up- and 40 downregulated genes, compared to the controls. After LVA, 78 out of the 148 previously dysregulated genes restored to baseline expression, including 69 upregulated genes (63 protein-coding, six non-coding) and nine genes with downregulated expression (). Six of these genes exhibited greatest fold changes in expression were selected, namely GPI, SPYR1, SLC1A3, ZBTB16, ALOX15B, and CCL2 for real-time PCR validation of NGS data. These genes exhibited fold changes in expression ranging from 2.3 to 8.8. Venous blood samples from the study group (15 patients) and the controls (16 patients) were used for the validation. Of the six selected genes, four with significantly upregulated expression prior to LVA (SPYR1, SLC1A3, ZBTB16, and ALOX15B) were found to return to baseline levels following LVA, however GPI remained upregulated after LVA. CCL2 was downregulated before LVA, but did not change significantly following LVA, although there was a slight increase. These findings are compatible with the expression patterns observed using NGS (). In compliance with Reviewer’s opinion regarding the correlations between the changes in gene expression and limb volume before and after LVA, the percentage reduction of post-LVA limb volume were calculated from nine out of 15 patients in the study group who have completed their pre- and 6 months post-LVA MR volumetry. These data were compared with six genes selected for real-time PCR validation of NGS data (). These results were shown in Supplemental Figures 1 and 2, demonstrating correlations between the changes in gene expressions and limb volume reduction. The results of Gene Ontology were shown in Supplemental Figures 3 and 4, and KEGG analysis were shown as Supplemental Table 1 and Supplemental Figures 5–8.
Figure 1 Summary. Fifty-one women with post-treatment gynecological cancer, including those with unilateral lymphedema (study group, n=25) and those without (control group, n=26). Venous blood samples obtained from the study group before and after LVA and those from the controls were sent for next-generation sequencing, which was validated by real-time PCR. Before LVA, 148 dysregulated genes were identified by NGS. Compared to the controls, 108 and 40 up- and downregulated genes were detected, respectively. After LVA, expression of 78 of the genes was restored to near baseline levels. Of these, 69 had been upregulated (63 protein coding genes, six non-coding genes) and nine had been downregulated.
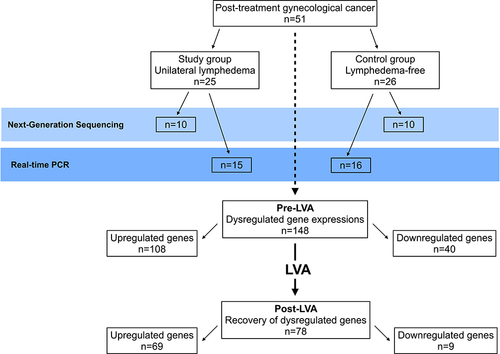
Figure 2 Next-Generation Sequencing (NGS) validation with real-time PCR. From the 72 genes that demonstrated recovery to baseline levels after LVA, gene expressions with fold changes greater than two were selected for NGS validation with real-time PCR. GPI, SPYR1, SLC1A3, ZBTB16, ALOX15B, and CCL2 with gene expression ranging from 2.3- to 8.8-fold changes were selected and analyzed by real-time PCR. The results were (A) GPI was an upregulated gene before LVA and remained upregulated after LVA; (B–E) SPYR1, SLC1A3, ZBTB16, and ALOX15B showed significantly upregulated expressions before LVA and were restored to baseline expression after LVA; (F) CCL2 was down-regulated before LVA but demonstrated no significant post-LVA change. These findings validated the NGS data. *In the figure represented statistical significant difference.
Protein–Protein Interaction and Hub Genes
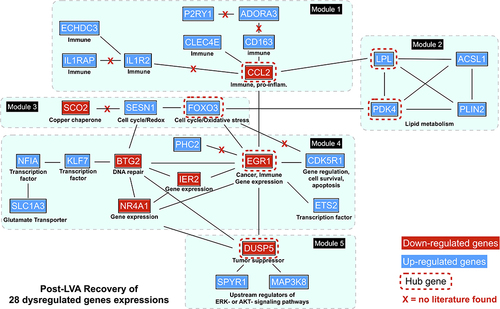
A schematic representation of the PPI network consisting of 28 interconnecting genes is presented in . The approach revealed five functional modules involving immunity, lipid metabolism, oxidative stress, transcriptional regulators, and tumor suppression (). Six hub genes, namely genes carrying the highest degree of connectivity, were also identified. Module 1 (immunity), contained eight genes: C-C motif chemokine ligand 2 (CCL2, hub gene); scavenger receptor cysteine-rich type 1 protein M130 (CD163); adenosine receptor A3 (ADORA3); P2Y puriceptor 1(P2RY1); C-type lectin domain family 4 member E (CLEC4E); interleukin 1 receptor type 2 (IL1R2); interleukin 1 receptor accessory protein (IL1RAP); and enoyl-CoA hydratase domain containing 3 (ECHDC3). Module 2 (lipid metabolism) comprised four genes: lipoprotein lipase (LPL, hub gene); pyruvate dehydrogenase kinase 4 (PDK4, hub gene); long-chain-fatty-acid-CoA ligase 1 (ACSL1); and perilipin-2 (PLIN2). Module 3 (oxidative stress), contained three genes: forkhead box O3 (FOXO3, hub gene); sestrin-1 (SESN1); and protein SCO2 homolog, mitochondrial (SCO2). Module 4 (transcriptional regulators), comprised seven genes: early growth response protein 1 (EGR1, hub gene); immediate early response gene 2 protein (IER2); protein BTG2 (BTG2); nuclear receptor subfamily 4 group A member 1 (NR4A1); cyclin-dependent kinase 5 activator 1 (CDK5R1); protein C-ets-2 (ETS2); Kruppel-like factor 7 (KLF7); nuclear factor I A-type (NFIA); solute carrier family 1 member 3 gene (SLC1A3); and polyhomeotic-like protein 2 (PHC2). Module 5 (tumor suppression), contained three genes: dual specificity phosphatase 5 (DUSP5, hub gene); mitogen-activated protein kinase kinase kinase 8 (MAP3K8); and protein sprouty homolog 1 (SPYR1). Downregulated hub genes comprised CCL2, EGR1, and DUSP5, while upregulated hub genes consisted of LPL, PDK4, and FOXO3.
Table 3 Functional Modules and Hub Genes Identified from 28 Interconnected Genes
Figure 3 The protein-protein interaction (PPI), functional modules, and hub genes. Five functional modules and six hub genes were identified from the PPI networks that comprised 28 interconnecting genes. Module 1, which is involved with immunity, contained the hub gene, CCL2; Module 2, which is associated with lipid metabolism, included the hub genes LPL and PDK4; Module 3, which is linked to oxidative stress, contained the hub gene FOXO3; Module 4, which is associated with transcriptional regulators, contained the hub gene EGR1; Module 5, which is related to tumor suppression, contained the hub gene DUSP5. Downregulated genes are colored in red; upregulated genes are colored in blue.
Discussion
The current study is the first to identify the hub genes related to lymphedema and demonstrate the recovery of some dysregulated genes after LVA. In contrast to most previous studies that investigated the changes in gene expression in lymphedematous adipose tissue, we focused on the systemic effects of lower limb lymphedema on circulating monocytes. Of the 148 genes with dysregulated expression in circulating monocytes, 78 (69 upregulated and nine downregulated) showed restoration of expression levels after LVA. For 28 of these genes, we were able to classify them using PPI interconnections into five functional modules with six hub genes. A particularly interesting observation is that the six identified hub genes are reportedly associated with oxidative stress.Citation14–26 This finding is in accordance with our recent study regarding the alleviation of oxidative stress in lower limb lymphedema following LVA.Citation9
The discovery of functional modules and hub genes may shed light on the mechanisms underlying lymphedema. In Module 1, which consisted of immune-related genes, the hub gene CCL2 is a pro-inflammatory chemokine that recruits monocytesCitation14 and is associated with the development of breast cancerCitation15–17 and colorectal cancer.Citation18 Oxidative stress is known to increase CCL2 mRNA level.Citation19,Citation20 The ablation or blockade of the CCL2 gene has been reported to reduce oxidative stress and increase the expression of anti-inflammatory macrophage markers.Citation21,Citation22 Although upregulation of CCL2 was expected during lymphedema in the presence of elevated oxidative stress, it was instead found to be downregulated in the local lower limb adipose tissues of patients with lymphedema.Citation11 Our observation of downregulated CLL2 expression in circulating monocytes may suggest a different oxidative stress regulatory mechanism in the context of secondary lymphedema that deserves further investigation.
Module 2 appears to be involved in lipid metabolism and was composed of abnormally upregulated genes controlling the adipocytokine signaling pathways involving the hub genes LPL and PDK4. Both genes were upregulated, but baseline expression was restored after LVA. A previous study demonstrated that tissue swelling in BCRL, which is attributed to both fluid accumulation and fat deposition,Citation23 is associated with a significantly higher expression of LPL,Citation10 which is known to be involved in lipid uptake, lipolysis, and fatty acid utilization.Citation24–26 Several studies have suggested that oxidative stress is positively correlated with LPL production in macrophages.Citation27–30 In addition, exercise-induced oxidative stress has been found to increase PDK4 gene expression.Citation31 Consistent with this, a previous RNA sequencing study showed a strong association between positive LPL and PDK4 expression and oxidative stress.Citation32 Moreover, our recent study demonstrated that elevated oxidative stress in lower limb lymphedema can be reduced by LVA.Citation9 The decline in oxidative stress after LVA may, therefore, have contributed to the observed restoration of LPL and PDK4 expression.
Module 3 contained FOXO3 as a hub gene, which is a hypoxic stress-responsive transcription factor known to protect against oxidative stress in the kidneyCitation33–35 and the heart.Citation36,Citation37 Oxidative stress has been shown to induce FOXO3 and peroxiredoxin 2 (PRDX2) gene expression.Citation38 PRDX2 is a thioredoxin-dependent peroxidase that functions as a defense system against reactive oxygen species.Citation39 In the present study, the abnormally upregulated preoperative FOXO3 expression was restored after LVA. Furthermore, our previous study demonstrated an association between a reduction in PRDX2 expression and a decrease in oxidative stress after LVA.Citation9 Therefore, the co-expression of FOXO3 and PRDX2 may be related to the level of oxidative stress in lower limb lymphedema.
Module 4 contained the key hub gene, EGR1, which not only plays a vital role in regulating cell differentiation, growth, and survival, but is also a mediator of inflammatory gene expression and tumor cell metastasis.Citation40–42 Oxidative stress downregulates EGR1 expression in cardiomyocytes,Citation43 which is consistent with the findings of our study. Module 5 was composed of genes coding for upstream regulators of the ERK or AKT signaling pathway, including DUSP5.Citation44–46 It is involved in T-cell activation,Citation47 anti-inflammation,Citation48 and acts as a negative regulator of the mitogen-activated protein kinase pathway.Citation49,Citation50 DUSP5 also serves as a tumor suppressor in human cancers.Citation51–53 Consistently, its downregulation is associated with increased cardiac oxidative stressCitation45 and observed in cervical, ovarian, and breast cancers.Citation49,Citation54,Citation55
Interestingly, a few sets of co-regulated genes were found within different modules in the present investigation, as well as in other studies. For example, co-regulated LPL, PLIN2, and ACSL1 gene expression in Module 2 is also related to lipolysis and fatty acid utilization.Citation25 FOXO3 and SESN1 in Module 3 also work synergistically to regulate the responses to oxidative stress.Citation56 Moreover, cross-linking among different modules by gene co-regulation was observed in the present study. Furthermore, concomitant CCL2 (Module 1) downregulation and LPL (Module 2) upregulation has been shown to be associated with an increased body mass index.Citation57
The limitations of this study include the lack of normal healthy controls to show the difference between patients with lymphedema and those with good health. The extent of oxidative stress was not measured in the present study due to the limited sample availability. Although six genes were analyzed by real-time PCR, the validation of all dysregulated genes would provide more comprehensive evidence. In summary, although the reduction in post-LVA oxidative stress may be linked to the post-LVA recovery of dysregulated genes, more evidence is required to validate this hypothesis. Whether there is a causal correlation between oxidative stress and lymphedema requires further investigation.
Conclusion
Localized lymphedema leads to dysregulated gene expression in circulating monocytes. The current study identified hub genes related to lymphedema and demonstrated the restoration of some dysregulated genes after LVA. A thorough understanding of the pathophysiological changes that may result in lymphedema requires further investigation. The present study may provide some basic information that can help in the advancement of lymphedema research at the molecular level.
Abbreviations
ACSL1, Acyl-CoA Synthetase Long Chain Family Member 1; ADORA3, Adenosine A3 Receptor; BCRL, breast cancer-related lymphedema; BTG2, B-cell translocation gene 2; CCL2, C-C Motif Chemokine Ligand 2; CD163, Cluster of Differentiation 163; CDK5R1, Cyclin Dependent Kinase 5 Regulatory Subunit 1; CLEC4E, C-Type Lectin Domain Family 4 Member E; DB, dermal backflow; DUSP5, Dual Specificity Phosphatase 5; ECHDC3, Enoyl-CoA Hydratase Domain Containing 3; EGR1, Early Growth Response 1; ETS2, ETS Proto-Oncogene 2 Transcription Factor; FOXO3, Forkhead box O3; ICG, indocyanine green; IER2, Immediate Early Response 2; IL1R2, Interleukin 1 Receptor Type 2; IL1RAP, Interleukin 1 Receptor Accessory Protein; IQR, interquartile range; ISL, International Society of Lymphology; LPL, Lipoprotein lipase; LV, lymphatic vessel; LVA, lymphaticovenous anastomosis; MAP3K8, Mitogen-Activated Protein Kinase Kinase Kinase 8; NGS, next-generation sequencing; NR4A1, Nuclear Receptor Subfamily 4 Group A Member 1; P2RY1, Purinergic Receptor P2Y1; PBMC, peripheral blood mononuclear cell; PBS, phosphate-buffered saline; PDK4, Pyruvate Dehydrogenase Kinase 4; PHC2, Polyhomeotic Homolog 2; PLIN2, Perilipin 2; PPI, protein-protein interaction; RIN, RNA integrity number; SCO2, Supercritical Carbon Dioxide; SESN1, Sestrin 1; SPYR1, Mycobacterium sp Spyr1.
Data Sharing Statement
Data is available upon request.
Ethics Approval and Informed Consent
This study was approved by the Institutional Review Board of Chang Gung Memorial Hospital (Approval number: 201702190B0/201702190B0C501).
Consent for Publication
The images and tables can be published, and that the person(s) providing consent have been shown the article contents to be published.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Disclosure
None of the authors has a financial interest in any of the products, devices, or drugs mentioned in this manuscript.
Acknowledgments
Special thanks to the International Supermicrosurgical Lymphedema Center at Kaohsiung Chang Gung Memorial Hospital, Ms. Sherry Hsin-Miao Shih, Hsiu-Ling Wu (RN), Lili Su (RN), Shu-Hsia Chang (RN), Yi-Chun Lin (RN), Shu-Hui Peng (RN), and all colleagues for their help.
Additional information
Funding
References
- Grada AA, Phillips TJ. Lymphedema: pathophysiology and clinical manifestations. J Am Acad Dermatol. 2017;77(6):1009–1020. doi:10.1016/j.jaad.2017.03.022
- Dessources K, Aviki E, Leitao MM Jr. Lower extremity lymphedema in patients with gynecologic malignancies. Int J Gynecol Cancer. 2020;30(2):252–260. doi:10.1136/ijgc-2019-001032
- Sung CJ, Wang SX, Hsu JF, Yu RP, Wong AK. Current understanding of pathological mechanisms of lymphedema. Adv Wound Care. 2021. doi:10.1089/wound.2021.0041
- Beier A, Siems W, Brenke R, Grune T. [Increased formation of free radicals in chronic lymphedema]. Z Lymphol. 1994;18(1):8–11. German.
- Siems WG, Brenke R, Beier A, Grune T. Oxidative stress in chronic lymphoedema. QJM. 2002;95(12):803–809. doi:10.1093/qjmed/95.12.803
- Foldi E, Sauerwald A, Hennig B. Effect of complex decongestive physiotherapy on gene expression for the inflammatory response in peripheral lymphedema. Lymphology. 2000;33(1):19–23.
- Yang JC, Wu SC, Chiang MH, Lin WC, Chiang PL, Hsieh CH. Supermicrosurgical lymphaticovenous anastomosis as an alternative treatment option for moderate-to-severe lower limb lymphedema. J Am Coll Surg. 2020;230(2):216–227. doi:10.1016/j.jamcollsurg.2019.10.007
- Yang JC, Yen YH, Wu SC, Lin WC, Chiang MH, Hsieh CH. Supermicrosurgical lymphaticovenous anastomosis as an alternative treatment option for patients with lymphorrhea. Plast Reconstr Surg. 2019;144(5):1214–1224. doi:10.1097/PRS.0000000000006169
- Yang JC, Huang LH, Wu SC, et al. Lymphaticovenous anastomosis supermicrosurgery decreases oxidative stress and increases antioxidant capacity in the serum of lymphedema patients. J Clin Med. 2021;10(7):1540. doi:10.3390/jcm10071540
- Levi B, Glotzbach JP, Sorkin M, et al. Molecular analysis and differentiation capacity of adipose-derived stem cells from lymphedema tissue. Plast Reconstr Surg. 2013;132(3):580–589. doi:10.1097/PRS.0b013e31829ace13
- Xiang Q, Xu F, Li Y, et al. Transcriptome analysis and functional identification of adipose-derived mesenchymal stem cells in secondary lymphedema. Gland Surg. 2020;9(2):558–574. doi:10.21037/gs.2020.02.09
- Koc M, Wald M, Varaliova Z, et al. Lymphedema alters lipolytic, lipogenic, immune and angiogenic properties of adipose tissue: a hypothesis-generating study in breast cancer survivors. Sci Rep. 2021;11(1):8171. doi:10.1038/s41598-021-87494-3
- Yamamoto T, Yamamoto N, Yoshimatsu H, Narushima M, Koshima I. Factors associated with lymphosclerosis: an analysis on 962 lymphatic vessels. Plast Reconstr Surg. 2017;140(4):734–741. doi:10.1097/PRS.0000000000003690
- Zollbrecht C, Grassl M, Fenk S, et al. Expression pattern in human macrophages dependent on 9p21.3 coronary artery disease risk locus. Atherosclerosis. 2013;227(2):244–249. doi:10.1016/j.atherosclerosis.2012.12.030
- Qian BZ, Li J, Zhang H, et al. CCL2 recruits inflammatory monocytes to facilitate breast-tumour metastasis. Nature. 2011;475(7355):222–225. doi:10.1038/nature10138
- Li D, Ji H, Niu X, et al. Tumor-associated macrophages secrete CC-chemokine ligand 2 and induce tamoxifen resistance by activating PI3K/Akt/mTOR in breast cancer. Cancer Sci. 2020;111(1):47–58. doi:10.1111/cas.14230
- Szekely B, Bossuyt V, Li X, et al. Immunological differences between primary and metastatic breast cancer. Ann Oncol. 2018;29(11):2232–2239. doi:10.1093/annonc/mdy399
- De la Fuente Lopez M, Landskron G, Parada D, et al. The relationship between chemokines CCL2, CCL3, and CCL4 with the tumor microenvironment and tumor-associated macrophage markers in colorectal cancer. Tumour Biol. 2018;40(11):1010428318810059. doi:10.1177/1010428318810059
- Uchiyama T, Itaya-Hironaka A, Yamauchi A, et al. Intermittent hypoxia up-regulates CCL2, RETN, and TNFalpha mRNAs in adipocytes via down-regulation of miR-452. Int J Mol Sci. 2019;20(8):1960. doi:10.3390/ijms20081960
- Kumar A, Shalmanova L, Hammad A, Christmas SE. Induction of IL-8(CXCL8) and MCP-1(CCL2) with oxidative stress and its inhibition with N-acetyl cysteine (NAC) in cell culture model using HK-2 cell. Transpl Immunol. 2016;35:40–46. doi:10.1016/j.trim.2016.02.003
- Luciano-Mateo F, Cabre N, Fernandez-Arroyo S, et al. Chemokine (C-C motif) ligand 2 gene ablation protects low-density lipoprotein and paraoxonase-1 double deficient mice from liver injury, oxidative stress and inflammation. Biochim Biophys Acta Mol Basis Dis. 2019;1865(6):1555–1566. doi:10.1016/j.bbadis.2019.03.006
- Seok SJ, Lee ES, Kim GT, et al. Blockade of CCL2/CCR2 signalling ameliorates diabetic nephropathy in db/db mice. Nephrol Dial Transplant. 2013;28(7):1700–1710. doi:10.1093/ndt/gfs555
- Brorson H, Svensson H. Complete reduction of lymphoedema of the arm by liposuction after breast cancer. Scand J Plast Reconstr Surg Hand Surg. 1997;31(2):137–143. doi:10.3109/02844319709085480
- Mulumba M, Jossart C, Granata R, et al. GPR103b functions in the peripheral regulation of adipogenesis. Mol Endocrinol. 2010;24(8):1615–1625. doi:10.1210/me.2010-0010
- Bi Y, Yuan X, Chen Y, Chang G, Chen G. Expression analysis of genes related to lipid metabolism in peripheral blood lymphocytes of chickens challenged with reticuloendotheliosis virus. Poult Sci. 2021;100(5):101081. doi:10.1016/j.psj.2021.101081
- Rogers C, Moukdar F, McGee MA, et al. EGF receptor (ERBB1) abundance in adipose tissue is reduced in insulin-resistant and type 2 diabetic women. J Clin Endocrinol Metab. 2012;97(3):E329–E340. doi:10.1210/jc.2011-1033
- Renier G, Desfaits AC, Lambert A, Mikhail R. Role of oxidant injury on macrophage lipoprotein lipase (LPL) production and sensitivity to LPL. J Lipid Res. 1996;37(4):799–809. doi:10.1016/S0022-2275(20)37578-7
- Maingrette F, Renier G. Leptin increases lipoprotein lipase secretion by macrophages: involvement of oxidative stress and protein kinase C. Diabetes. 2003;52(8):2121–2128. doi:10.2337/diabetes.52.8.2121
- Peng J, Lv YC, He PP, et al. Betulinic acid downregulates expression of oxidative stress-induced lipoprotein lipase via the PKC/ERK/c-Fos pathway in RAW264.7 macrophages. Biochimie. 2015;119:192–203. doi:10.1016/j.biochi.2015.10.020
- Oliveira JVB, Lima RPA, Pordeus Luna RC, et al. The direct correlation between oxidative stress and LDL-C levels in adults is maintained by the Friedewald and Martin equations, but the methylation levels in the MTHFR and ADRB3 genes differ. PLoS One. 2020;15(12):e0239989. doi:10.1371/journal.pone.0239989
- Townsend LK, Weber AJ, Barbeau PA, Holloway GP, Wright DC. Reactive oxygen species-dependent regulation of pyruvate dehydrogenase kinase-4 in white adipose tissue. Am J Physiol Cell Physiol. 2020;318(1):C137–C149. doi:10.1152/ajpcell.00313.2019
- Andersen E, Ingerslev LR, Fabre O, et al. Preadipocytes from obese humans with type 2 diabetes are epigenetically reprogrammed at genes controlling adipose tissue function. Int J Obes. 2019;43(2):306–318. doi:10.1038/s41366-018-0031-3
- Li L, Kang H, Zhang Q, D’Agati VD, Al-Awqati Q, Lin F. FoxO3 activation in hypoxic tubules prevents chronic kidney disease. J Clin Invest. 2019;129(6):2374–2389. doi:10.1172/JCI122256
- Deng A, Ma L, Zhou X, Wang X, Wang S, Chen X. FoxO3 transcription factor promotes autophagy after oxidative stress injury in HT22 cells. Can J Physiol Pharmacol. 2021;99(6):627–634. doi:10.1139/cjpp-2020-0448
- Nlandu-Khodo S, Osaki Y, Scarfe L, et al. Tubular beta-catenin and FoxO3 interactions protect in chronic kidney disease. JCI Insight. 2020;5(10). doi:10.1172/jci.insight.135454
- Wang LF, Huang CC, Xiao YF, et al. CD38 deficiency protects heart from high fat diet-induced oxidative stress via activating Sirt3/FOXO3 pathway. Cell Physiol Biochem. 2018;48(6):2350–2363. doi:10.1159/000492651
- Lv W, Jiang J, Li Y, Fu L, Meng F, Li J. MiR-302a-3p aggravates myocardial ischemia-reperfusion injury by suppressing mitophagy via targeting FOXO3. Exp Mol Pathol. 2020;117:104522. doi:10.1016/j.yexmp.2020.104522
- Miyamoto N, Izumi H, Miyamoto R, et al. Nipradilol and timolol induce Foxo3a and peroxiredoxin 2 expression and protect trabecular meshwork cells from oxidative stress. Invest Ophthalmol Vis Sci. 2009;50(6):2777–2784. doi:10.1167/iovs.08-3061
- Lee YJ. Knockout mouse models for peroxiredoxins. Antioxidants. 2020;9(2):182.
- Banerji R, Saroj SD. Early growth response 1 (EGR1) activation in initial stages of host-pathogen interactions. Mol Biol Rep. 2021;48(3):2935–2943. doi:10.1007/s11033-021-06305-0
- Wang B, Guo H, Yu H, Chen Y, Xu H, Zhao G. The role of the transcription factor EGR1 in cancer. Front Oncol. 2021;11:642547. doi:10.3389/fonc.2021.642547
- Li TT, Liu MR, Pei DS. Friend or foe, the role of EGR-1 in cancer. Med Oncol. 2019;37(1):7. doi:10.1007/s12032-019-1333-6
- Li J, Gong L, Zhang R, et al. Fibroblast growth factor 21 inhibited inflammation and fibrosis after myocardial infarction via EGR1. Eur J Pharmacol. 2021;910:174470. doi:10.1016/j.ejphar.2021.174470
- Gruosso T, Garnier C, Abelanet S, et al. MAP3K8/TPL-2/COT is a potential predictive marker for MEK inhibitor treatment in high-grade serous ovarian carcinomas. Nat Commun. 2015;6:8583. doi:10.1038/ncomms9583
- Xu Z, Tong Q, Zhang Z, et al. Inhibition of HDAC3 prevents diabetic cardiomyopathy in OVE26 mice via epigenetic regulation of DUSP5-ERK1/2 pathway. Clin Sci. 2017;131(15):1841–1857. doi:10.1042/CS20170064
- Ng KY, Chan LH, Chai S, et al. TP53INP1 downregulation activates a p73-dependent dusp10/erk signaling pathway to promote metastasis of hepatocellular carcinoma. Cancer Res. 2017;77(17):4602–4612. doi:10.1158/0008-5472.CAN-16-3456
- Chuang HC, Tan TH. MAP4K family kinases and DUSP family phosphatases in T-cell signaling and systemic lupus erythematosus. Cells. 2019;8(11):1433. doi:10.3390/cells8111433
- Seo H, Cho YC, Ju A, et al. Dual-specificity phosphatase 5 acts as an anti-inflammatory regulator by inhibiting the ERK and NF-kappaB signaling pathways. Sci Rep. 2017;7(1):17348. doi:10.1038/s41598-017-17591-9
- Liu T, Sun H, Liu S, et al. The suppression of DUSP5 expression correlates with paclitaxel resistance and poor prognosis in basal-like breast cancer. Int J Med Sci. 2018;15(7):738–747. doi:10.7150/ijms.24981
- Chen HF, Chuang HC, Tan TH. Regulation of dual-specificity phosphatase (DUSP) ubiquitination and protein stability. Int J Mol Sci. 2019;20(11):2668.
- Png CW, Weerasooriya M, Guo J, et al. DUSP10 regulates intestinal epithelial cell growth and colorectal tumorigenesis. Oncogene. 2016;35(2):206–217. doi:10.1038/onc.2015.74
- Ding J, Li J, Wang H, et al. Long noncoding RNA CRNDE promotes colorectal cancer cell proliferation via epigenetically silencing DUSP5/CDKN1A expression. Cell Death Dis. 2017;8(8):e2997. doi:10.1038/cddis.2017.328
- Kidger AM, Rushworth LK, Stellzig J, et al. Dual-specificity phosphatase 5 controls the localized inhibition, propagation, and transforming potential of ERK signaling. Proc Natl Acad Sci U S A. 2017;114(3):E317–E326. doi:10.1073/pnas.1614684114
- Min H, He W. Long non-coding RNA ARAP1-AS1 promotes the proliferation and migration in cervical cancer through epigenetic regulation of DUSP5. Cancer Biol Ther. 2020;21(10):907–914. doi:10.1080/15384047.2020.1806641
- Wang L, Hu J, Qiu D, et al. Dual-specificity phosphatase 5 suppresses ovarian cancer progression by inhibiting IL-33 signaling. Am J Transl Res. 2019;11(2):844–854.
- Donlon TA, Morris BJ, Chen R, et al. FOXO3 longevity interactome on chromosome 6. Aging Cell. 2017;16(5):1016–1025. doi:10.1111/acel.12625
- Canzoneri R, Naipauer J, Stedile M, et al. Identification of an AP1-ZFP36 regulatory network associated with breast cancer prognosis. J Mammary Gland Biol Neoplasia. 2020;25(2):163–172. doi:10.1007/s10911-020-09448-1
