Abstract
Purpose
The goal of this study was to identify the crucial autophagy-related genes (ARGs) in periodontitis and construct mRNA-miRNA-lncRNA networks to further understand the pathogenesis of periodontitis.
Methods
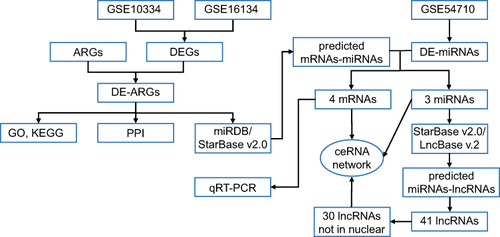
We used the Gene Expression Omnibus (GEO) database and Human Autophagy Database (HADb) to identify differentially expressed mRNAs, miRNAs, and ARGs. These ARGs were subjected to Gene Ontology (GO), KEGG (Kyoto Encyclopedia of Genes and Genomes) pathway, and PPI (protein–protein interaction) network analysis. Two databases (miRDB and StarBase v2.0) were used to reverse-predict miRNAs while the miRNA-lncRNA interaction was predicted using the StarBase v2.0 and LncBase Predicted v.2 databases. After excluding the lncRNAs only present in the nucleus, a competing endogenous RNA (ceRNA) network was built. Finally, we used quantitative real-time PCR (qRT-PCR) to confirm the levels of mRNA expression in the ceRNA network.
Results
The differential expression analysis revealed 10 upregulated and 10 downregulated differentially expressed ARGs. After intersecting the reverse-predicted miRNAs with the differentially expressed miRNAs, a ceRNA network consisting of 4 mRNAs (LAMP2, NFE2L2, NCKAP1, and EGFR), 3 miRNAs (hsa-miR-140-3p, hsa-miR-142-5p, and hsa-miR-671-5p), and 30 lncRNAs was constructed. In addition, qRT-PCR results revealed that EGFR expression was downregulated in diseased gingival tissue of periodontitis patients.
Conclusion
Four autophagy-related genes, especially EGFR, may play a key role in periodontitis progression. The novel ceRNA network may aid in elucidating the role and the mechanism of autophagy in periodontitis, which could be important in developing new therapeutic options.
Introduction
Periodontitis, a chronic infectious disease initiated by dental plaque biofilm and characterized by gingival bleeding and destruction of periodontal supporting tissue, is the main cause of tooth loss in adults.Citation1 It is widely recognized that periodontitis has two-way relationships with a variety of systemic disorders, including cardiovascular diseases and diabetes.Citation2 Microbial infection and host immune-inflammatory responses are the key factors affecting the pathogenesis of periodontitis. However, there are still many problems about the molecular mechanism of periodontitis pathogenic processes, which need further researches.
Autophagy is an evolutionarily highly conserved physiological metabolic process, which can keep the intracellular environment in a state of equilibrium.Citation3 The autophagy process necessitates the regulation of a group of autophagy-related genes (ARG) as well as the recognition of corresponding proteins. These proteins form bilayer membrane-structured autophagosomes, which then fuse with lysosomes to remove intracellular invading pathogens and degrade proteins or damaged organelles.Citation4 Autophagy dysfunction contributes to the pathological process of many diseases such as neurodegenerative disorders and tumors.Citation5,Citation6 Furthermore, the role of autophagy in immune defense and inflammatory regulation has received a lot of attention.Citation7 Periodontitis is also an inflammatory disease. Autophagy may influence the onset and progression of periodontitis by regulating periodontal pathogen infection, host immune-inflammatory responses, and alveolar bone metabolism.Citation8,Citation9 However, the details of the interaction between periodontitis and autophagy remain unclear.
In recent decades, sequencing techniques and bioinformatics have been highly developed and widely used to identify biomarkers of diseases. Recent investigations have revealed critical functions of mRNAs, miRNAs, and lncRNAs in the pathogenesis of periodontitis.Citation10,Citation11 The competing endogenous RNA (ceRNA) hypothesis, suggested by Salmena et al, revealed a new mechanism of interaction between RNAs.Citation12 For instance, miRNA can lead to gene silencing by binding to the microRNA response elements (MREs) of mRNA, while lncRNA, pseudogenes, and circular RNA can regulate gene expression by competitively sponging miRNA. The ceRNA network has been linked to the onset and progression of a range of diseases, including cancer, cardiovascular disease, and neurodegenerative disease, according to numerous research.Citation13 Therefore, the ceRNA network could be a novel way to investigate the pathophysiology of periodontitis.
This research identified differentially expressed ARGs and used bioinformatics tools to construct a ceRNA network. Furthermore, mRNAs in the ceRNA network were validated using quantitative real-time PCR (qRT-PCR). The findings may provide fresh light on the role of autophagy in periodontitis and lay a theoretical foundation for subsequent studies on periodontitis.
Materials and Methods
Data Sources
The sequence data of periodontitis were retrieved from the Gene Expression Omnibus (GEO) in NCBI (http://www.ncbi.nlm.nih.gov/geo/). Three datasets, GSE10334,Citation14 GSE16134,Citation15,Citation16 and GSE54710Citation17 were obtained using “Homo sapiens [porgn:_txid9606] and periodontitis and gingival tissue” and limiting the sample count to more than 100. The characteristics of the selected GEO database data were shown in . The HADb (Human Autophagy Database, http://www.autophagy.lu/) was used to download 222 autophagy-related genes (ARGs) (Supplementary Table 1).
Table 1 Characteristics of the Selected GEO Database Data
Differential Expression Analysis
GEO2R (http://www.ncbi.nlm.nih.gov/geo/geo2r/) was used to find differentially expressed genes (DEGs) in GSE10334 and GSE16134. The DEGs were given a threshold of adjusted P-value < 0.05 and |log2 FC (fold change)| > 0.58. Differentially expressed miRNAs (DE-miRNAs) in GSE54710 were defined by the same criterion. To obtain the differentially expressed ARGs, a Venn diagram of the DEGs and ARGs was constructed using the web application “Calculate and draw custom Venn diagrams”.
Functional Analysis of Differentially Expressed ARGs
To further clarify the potential functional annotation with the differentially expressed ARGs, the Database for Annotation, Visualization, and Integrated Discovery v6.8 (DAVID) (http://david.ncifcrf.gov) was used to perform Gene Ontology (GO) analyses, including biological process (BP), cellular component (CC), and molecular function (MF).Citation18 Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment was performed based on the KOBased Annotation System (KOBAS) web tool (http://kobas.cbi.pku.edu.cn/index.php).Citation19 The ClueGO plug-in in Cytoscape v3.8.2 was used to perform interrelation analysis between pathways.Citation20 P<0.05 was regarded as statistically significant.
Construction of the Protein–Protein Interaction (PPI) Network
The PPI network was constructed using the Search Tool for the Retrieval of Interacting Genes v11.0 (STRING) database (http://string-db.org/),Citation21 with a confidence score of ≥0.4 as the cutoff. Protein nodes that did not interact with other proteins were deleted from the network. The PPI networks were drawn using Cytoscape v3.8.2.
Construction of the ceRNA Network
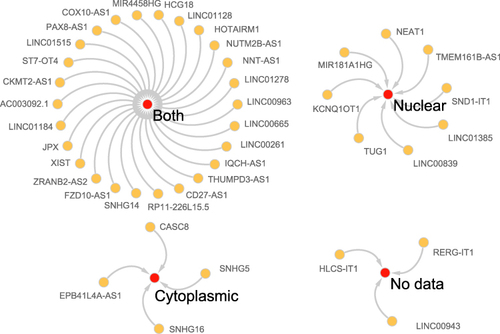
Differentially expressed ARGs were performed with miRDB (http://mirdb.org/)Citation22 and StarBase v2.0 (http://starbase.sysu.edu.cn/)Citation23 to reverse-predict miRNAs. Two databases and DE-miRNAs were used to do intersection. The miRNA-lncRNA interaction was predicted in the StarBase v2.0 and LncBase Predicted v.2Citation24 databases using the miRNAs mentioned above. Only those miRNA-lncRNA relationship pairs found in both databases were chosen to ensure the accuracy of the results. Considering that lncRNAs can serve as nodes of the ceRNA network only in the cytoplasm, we used the lncATLAS database (https://lncatlas.crg.eu/)Citation25 to investigate the intracellular localization of the lncRNAs, excluding those that were only found in the nucleus. The obtained mRNAs, miRNAs, lncRNAs were processed using Cytoscape to construct the ceRNA network.
Validation by qRT-PCR
5 periodontitis patients (average age, 29.8 years old; range 26–33 years old) were recruited from The Second Affiliated Hospital, College of Medicine, Zhejiang University, and were excluded if they had cardiovascular diseases, diabetes, cancer, HIV, or HBV infection. The details of the participants were shown in . A total of 10 samples were obtained from healthy and diseased gingival tissues in patients. Diseased tissue was collected from periodontal pathology sites (bleeding on probing, probing pocket depth ≥5 mm, and clinical attachment loss ≥3 mm), while healthy tissue was obtained from a neighboring site (no bleeding on probing, probing pocket depth ≤ 3 mm, and no clinical attachment loss). This study was conducted according to the ethical guidelines of the Declaration of Helsinki and was approved by the Research Medical Ethics Committee of The Second Affiliated Hospital, College of Medicine, Zhejiang University. All the participants were made aware of the study and gave their written informed consent.
Table 2 The Details of the Participants
Gingival tissue samples were collected from each participant during surgery. Subsequently, total RNA was extracted using RNAiso Reagent 9180 (TaKaRa, Kyoto, Japan). 4 mRNAs in the ceRNA network were validated by qRT-PCR using TB Green Premix Ex Taq™ RR420A (TaKaRa, Kyoto, Japan) and LightCycler 480 System (Roche, Basel, Switzerland). The primer sequences were designed using Primer 5.0 (Sunya, Hangzhou, China) and reported in . β-actin was applied as an internal control, and the relative gene expression was determined using the 2−ΔΔCt method.
Table 3 The Primer Sequences for PCR
Statistical Analysis
qRT-PCR results were presented as the mean ± standard deviation. Kolmogorov–Smirnov test was employed to analyze the data’s normality. For data that conformed to the normal distribution, the Student’s t-test was used, and for data that did not adhere to the normal distribution, the Mann–Whitney U-test was used. P<0.05 was considered statistically significant when using GraphPad Prism 9.1.1 (GraphPad Software, San Diego, California USA, www.graphpad.com).
Results
Identification of DEGs, DE-miRNAs, and Differentially Expressed ARGs in Periodontitis
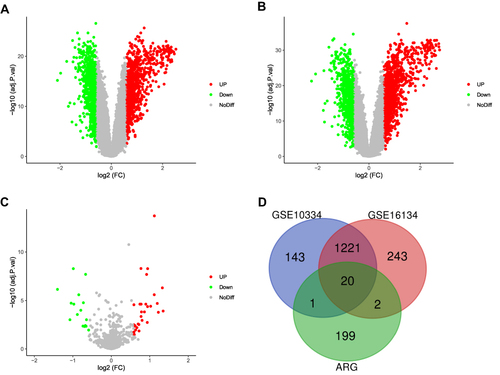
A schematic of the flowchart of this study is shown in . The online analysis tool GEO2R was used to evaluate three datasets acquired from the GEO database. Adjusted P-value < 0.05 and |log2 FC (fold change)| > 0.58 were considered as criteria to screen the DEGs and DE-miRNAs. The volcano plots of DEGs in GSE10334 and GSE16134, as well as DE-miRNAs in GSE54710, were made in –. 222 ARGs were downloaded from the HADb. The intersections of the two groups of DEGs and ARGs obtained 20 differentially expressed ARGs (), with 10 genes upregulated and 10 genes downregulated ().
Table 4 Differentially Expressed ARGs of Periodontitis
Figure 2 Screening of differentially expressed mRNAs, miRNAs, and ARGs. (A) Volcano plot of differentially expressed mRNAs in GSE10334. Upregulated mRNAs are represented by the red dot, whereas downregulated mRNAs are represented by the green dot. (B) Volcano plot of differentially expressed mRNAs in GSE16134. (C) Volcano plot of differentially expressed miRNAs in GSE54710. (D) A Venn diagram between GSE10334, GSE16134, and ARGs. The coincident section indicates the 20 differentially expressed genes that are shared across the three series.
Enrichment Analysis and PPI Network Construction of the Differentially Expressed ARGs
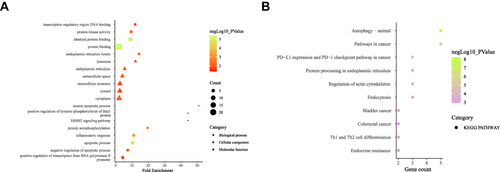
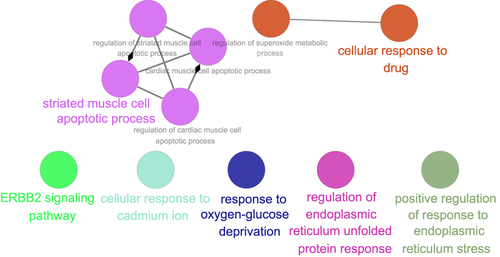
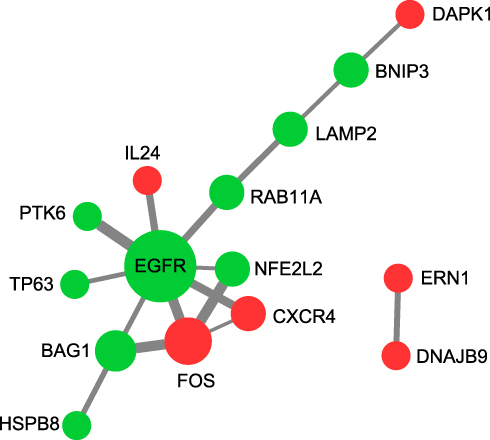
The DAVID v6.8 online tool was used to conduct GO analyses of differentially expressed ARGs. The enriched GO terms with P < 0.05 were presented in , including apoptotic process, inflammatory response, protein autophosphorylation, negative regulation of the apoptotic process, positive regulation of transcription from RNA polymerase II promoter, neuron apoptotic process, positive regulation of tyrosine phosphorylation of Stat3 protein and ERBB2 signaling pathway in the BP category. For CC, differentially expressed ARGs were mainly clustered in the cytoplasm, endoplasmic reticulum, extracellular space, cytosol, endoplasmic reticulum lumen, extracellular exosome, and lysosome. About MF, differentially expressed ARGs are primarily enriched in processes including protein binding, identical protein binding, protein kinase activity, and transcription regulatory region DNA binding. As demonstrated in , the top 10 KEGG were screened out according to the P-value < 0.05. The interaction network of BP was visualized using the Cytoscape ClueGo plug-in (). In addition, a PPI network was built based on the STRING online database and Cytoscape software in .
Construction of the ceRNA Network
Reverse Prediction of miRNAs Targeted by mRNAs
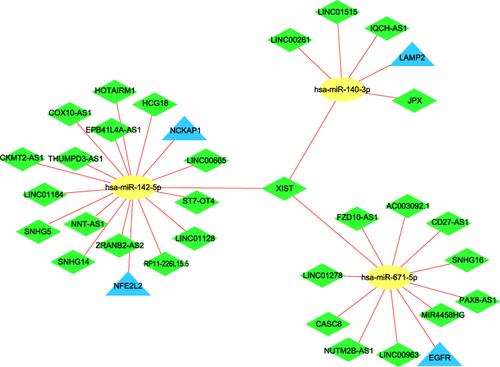
The miRDB and StarBase v2.0 databases were performed to reverse-predict miRNA of 20 differentially expressed ARGs. The intersecting genes between the predicted miRNAs and 46 DE-miRNAs were then discovered. Finally, mRNA-miRNA interaction pairs involving 4 mRNAs (LAMP2, NFE2L2, NCKAP1, and EGFR) and 3 miRNAs (hsa-miR-140-3p, hsa-miR-142-5p, and hsa-miR-671-5p) were identified. In addition, 4 selected mRNAs were downregulated and 3 miRNAs were upregulated, which agreed with the ceRNA hypothesis.
Prediction of lncRNAs Targeted by miRNAs
We reverse predicted their upstream lncRNAs using 3 miRNAs. A total of 41 lncRNAs was identified using the intersection of the StarBase v2.0 and LncBase Predicted v.2 databases (). According to the endogenous competition role of lncRNAs is primarily manifested in the cytoplasm, 11 lncRNAs which were located only in the nucleus were excluded. Cytoscape was used to visualize the distribution information for 30 lncRNAs (). Finally, a ceRNA of 4 mRNAs, 3miRNAs, 30lncRNAs were identified ().
Table 5 The miRNA-lncRNA Network
Validation by qRT-PCR
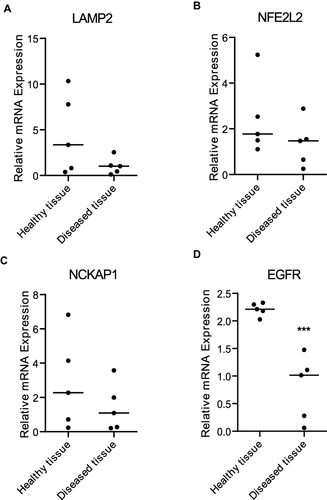
LAMP2, NFE2L2, NCKAP1, and EGFR were validated by qRT-PCR. The result revealed that the expression level of EGFR in diseased gingival tissue of periodontitis patients was significantly lower than that of healthy controls (). There was no significant difference in the remaining three mRNAs between the two groups.
Discussion
Periodontitis, the most common oral infectious disease, not only affects periodontal health and occlusal function by destructing periodontal supporting tissue but also interferes with general health through periodontal pathogens and inflammatory mediators.Citation1 A great number of studies have shown that genetic factors are strongly linked to periodontitis, and may even affect the speed and severity of disease progression.Citation26 Although simple genetic factors do not cause periodontitis, they can increase the susceptibility of hosts to periodontitis. Therefore, it is meaningful to explore the pathogenesis of periodontitis from a genetic perspective.
Sequencing techniques and bioinformatics have been highly developed and frequently employed to identify biomarkers of diseases in recent decades. The absolute value of log2FC greater than 1 is commonly used as the screening criterion for differential genes in the literature, whereas periodontitis-related bioinformatics analysis articles frequently choose a lesser value. For instance, Song et alCitation27 chose 0.58 while Jin et alCitation11 selected 0.45 as the screening index. Considering that periodontitis is not only a polygenic disease, but also affected by many factors such as microorganisms, smoking, stress, and systemic diseases.Citation28 If the screening criterion is too stringent, generous potential key genes may be missed, so we also set 0.58 as the screening criterion.
In published bioinformatics articles related to periodontitis, some researchers have analyzed GSE16134 and identified five genes (CTSS, PLEK, IRF-8, PTGS2, and FOSB) that may be implicated in the formation and progression of periodontitis.Citation27 Suzuki et alCitation29 have analyzed three datasets (GSE10334, GSE16134, and GSE23586) and screened several key genes which may be related to chronic periodontitis pathogenesis such as CSF3, CXCL12, IL1B, TAGLN, CD19, IL8, and CD79A. What’s more, upstream genes of biomarker candidates like TNF and FGF2 were predicted. However, there are few bioinformatics analyses on the relationship between periodontitis and autophagy.
It is widely recognized that autophagy has been a research hotspot in recent years. Bullon et alCitation30 found the level of mitochondrial reactive oxygen species and the expression of autophagy gene LC3 increased consistently in peripheral blood mononuclear cells of patients with periodontitis. An et alCitation31 also found that the expression of ARGs such as LC3, Beclin1, Atg7, and Atg12 in the periodontal ligament of patients with periodontitis was significantly increased. Indeed, autophagy may influence the onset and development of periodontitis in a variety of ways.Citation9 First of all, autophagy can enhance the body’s ability to resist pathogen infection. Secondly, autophagy can regulate inflammation. It is worth noting that moderate inflammation is beneficial to the body, while excessive inflammation can cause damage to the host.Citation32 Last but not least, autophagy can regulate the process of alveolar bone resorption by regulating the differentiation of osteoclasts and osteoblasts.Citation33,Citation34 Nevertheless, the specific mechanism of autophagy on periodontitis remains to be further studied.
In this work, we found 20 differentially expressed ARGs based on two mRNA microarray datasets and the HADb database, including 10 upregulated (ARSA, CXCR4, DAPK1, DNAJB9, ERN1, FOS, IL24, NAMPT, PRKCQ, and SERPINA1) and 10 downregulated (BAG1, BNIP3, EGFR, HSPB8, LAMP2, NCKAP1, NFE2L2, PTK6, RAB11A, and TP63) genes. To investigate interactions among the differentially expressed ARGs, we used GO analysis, KEGG pathway analysis, and PPI network. GO term analysis revealed these ARGs to be mostly enriched in apoptotic process and inflammatory response (BP), cytoplasm and endoplasmic reticulum (CC), protein binding and identical protein binding (MF). Changes in KEGG were mainly enriched in autophagy – animal and pathways in cancer. We also identified three miRNAs, including hsa-miR-140-3p, has-miR-142-5p, and hsa-miR-671-5p, which were upregulated in the gingival tissue of periodontitis compared to controls. Finally, a ceRNA network consisting of 4 mRNAs (LAMP2, NFE2L2, NCKAP1, and EGFR), 3 miRNAs, and 30 lncRNAs was created after removing those lncRNAs located only in the nucleus.
LAMP2 is critical for the process of fusion between phagosome and lysosome.Citation35 LAMP2-deficient mice showed increased susceptibility to periodontitis due to a reduced maturation process of phagosomes in polymorphonuclear leukocytes.Citation36 NFE2L2 plays a vital role in resisting oxidative stress and maintaining oxidative stress homeostasis. It can not only transcriptionally regulate macroautophagy but also modulate chaperone-mediated autophagy through the regulation of LAMP2A.Citation37 Zhang et alCitation38 showed that NCKAP1 is negatively related to monocyte in periodontitis. EGFR is a transmembrane protein that affects bone homeostasis, proliferation, and differentiation of osteoblasts.Citation39 The inhibitory effects of periodontitis and alveolar bone loss were reversed when silenced EGFR with EGFR siRNA.Citation40 In this study, we identified LAMP2, NFE2L2, NCKAP1, and EGFR as key genes between autophagy and periodontitis. These four ARGs were downregulated in periodontitis, which may not be conducive to osteoblast differentiation and increase the host’s susceptibility to periodontitis, thus aggravating periodontitis. Further qRT-PCR results confirmed that the expression of EGFR in the diseased gingival tissue of periodontitis patients was indeed downregulated compared to the healthy controls, which indicated that EGFR may play a significant role in the progression of periodontitis. As a matter of fact, the expression of the other three genes (including LAMP2, NFE2L2, and NCKAP1) in the diseased gingival tissue was also downregulated, but there was no significant difference. The potential reasons need to be taken into account when interpreting these results. On the one hand, the sample size is too small. On the other hand, the diseased tissue and healthy tissue come from the same individual. If tissues are taken from patients with periodontitis and patients without periodontitis, there may be more obvious differences.
Recently, miR-140-3p was manifested as a biomarker for osteoarthritis and osteoporosis.Citation41,Citation42 Another study found that miR-140-3p mimics significantly reduced protein expression of CXCR4, p-JAK2, and p-STAT3 in IL-1β-stimulated chondrocytes.Citation43 They also suggested that overexpression of miR-140-3p could inactivate JAK2/STAT3 signaling via suppressing CXCR4 in osteoarthritis. Moreover, Mao et alCitation44 revealed that overexpression of miR-140-3p inhibited cell survival and differentiation and hastened cell apoptosis in pre-osteoblast. In our research, miR-140-3p was upregulated in periodontitis, which may affect the progress of periodontitis through the above mechanisms. As for mir-142-5p, it has been found that it can promote the osteoclastogenesis of bone marrow-derived macrophages in vitro.Citation45 Besides, Sun et alCitation46 raised XIST/miR-142-5p/SGTB axis as a novel molecular mechanism in osteoarthritis. We speculated that miR-142-5p may affect bone resorption by promoting osteoclast differentiation, thereby aggravating periodontitis. In addition, a 17-miRNA-based diagnostic signature was identified for periodontitis, which included miR-671-5p.Citation11 A previous study has shown that miR-671-5p played a significant role in inhibiting the characteristics of osteosarcoma cells.Citation47 In brief, the above three miRNAs may be linked to the onset of periodontitis.
According to our prediction, lncRNA XIST may be associated with hsa-miR-140-3p, has-miR-142-5p, and hsa-miR-671-5p. LncRNA Xist is the master regulator of X-chromosome inactivation, an epigenetic process that may be important for female development and disease susceptibility.Citation48 Furthermore, lncRNA XIST may participate in the development of various cancers and non-cancer diseases by functioning as a ceRNA.Citation49 Periodontitis has a gender disparity in incidence rate, with males being at higher risk.Citation50 Gender and its related factors, such as genetics, smoking and hormone levels, may affect the incidence rate of periodontitis. It is noteworthy that Chen et alCitation51 reported that lncRNA XIST inhibited osteogenic differentiation of bone marrow mesenchymal stem cells. FengCitation52 et al also demonstrated that lncRNA XIST participated in osteogenic differentiation of periodontal ligament stem cells. Therefore, lncRNA XIST may be the most influential lncRNA in the ceRNA network we have established.
To be honest, certain limitations still existed in this study. First of all, due to the lack of datasets on the expression of lncRNA in periodontitis, we cannot determine whether the predicted lncRNA was upregulated or downregulated in periodontitis, therefore the established ceRNA network needed further filtering and validation. Secondly, the sample size of our experiment was small, and the expression levels of miRNAs and lncRNAs were not verified. Lastly, non-tumor diseases are often unable to conduct clinical correlation studies and prognosis analysis on account of lacking clinical and prognosis data. Despite these limitations, our study provided more directions to understand the role of autophagy in periodontitis and lay a theoretical foundation for subsequent studies.
Conclusions
In conclusion, we constructed a ceRNA network consisting of 4 mRNAs, 3 miRNAs, and 30 lncRNAs. Combined with qRT-PCR and prediction results, lncRNA XIST - miR-671-5p - EGFR may play a critical role in the ceRNA network. Our study may shed light on the pathogenesis of periodontitis. However, more researches and trials are still needed to clarify the function of these molecules.
Data Sharing Statement
The datasets of this article were generated from the GEO database (http://www.ncbi.nlm.nih.gov/geo/) and HADb (http://www.autophagy.lu/).
Ethics Approval Statement
This study was conducted according to the ethical guidelines of the Declaration of Helsinki. The experimental design was approved by the Research Medical Ethics Committee of The Second Affiliated Hospital, College of Medicine, Zhejiang University. All the participants were informed of the study and provided written informed consent.
Disclosure
The authors declare that there is no conflict of interest.
Acknowledgments
We would like to thank Dr. Tianchi Tang from the Department of Neurological Surgery, The Second Affiliated Hospital, Zhejiang University School of Medicine (Hangzhou, China) for his help in modifying the manuscript.
Additional information
Funding
References
- Slots J. Periodontitis: facts, fallacies and the future. Periodontol 2000. 2017;75(1):7–23. doi:10.1111/prd.12221
- Genco RJ, Sanz M. Clinical and public health implications of periodontal and systemic diseases: an overview. Periodontol 2000. 2020;83(1):7–13. doi:10.1111/prd.12344
- Levine B, Mizushima N, Virgin HW. Autophagy in immunity and inflammation. Nature. 2011;469(7330):323–335. doi:10.1038/nature09782
- Deretic V. Autophagy in immunity and cell-autonomous defense against intracellular microbes. Immunol Rev. 2011;240(1):92–104. doi:10.1111/j.1600-065X.2010.00995.x
- Nixon RA. The role of autophagy in neurodegenerative disease. Nat Med. 2013;19(8):983–997. doi:10.1038/nm.3232
- Amaravadi RK, Kimmelman AC, Debnath J. Targeting autophagy in cancer: recent advances and future directions. Cancer Discov. 2019;9(9):1167–1181. doi:10.1158/2159-8290.CD-19-0292
- Kocak M, Ezazi Erdi S, Jorba G, et al. Targeting autophagy in disease: established and new strategies. Autophagy. 2021:1–23. doi:10.1080/15548627.2021.1936359.
- Jiang M, Li Z, Zhu G. The role of autophagy in the pathogenesis of periodontal disease. Oral Dis. 2020;26(2):259–269. doi:10.1111/odi.13045
- Yang Y, Huang Y, Li W. Autophagy and its significance in periodontal disease. J Periodontal Res. 2021;56(1):18–26. doi:10.1111/jre.12810
- Li S, Liu X, Li H, et al. Integrated analysis of long noncoding RNA-associated competing endogenous RNA network in periodontitis. J Periodontal Res. 2018;53(4):495–505. doi:10.1111/jre.12539
- Jin SH, Zhou JG, Guan XY, et al. Development of an miRNA-array-based diagnostic signature for periodontitis. Front Genet. 2020;11:577585. doi:10.3389/fgene.2020.577585
- Salmena L, Poliseno L, Tay Y, et al. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146(3):353–358. doi:10.1016/j.cell.2011.07.014
- Ala U. Competing endogenous RNAs, non-coding RNAs and diseases: an intertwined story. Cells. 2020;9(7):1574. doi:10.3390/cells9071574
- Demmer RT, Behle JH, Wolf DL, et al. Transcriptomes in healthy and diseased gingival tissues. J Periodontol. 2008;79(11):2112–2124. doi:10.1902/jop.2008.080139
- Kebschull M, Demmer RT, Grun B, et al. Gingival tissue transcriptomes identify distinct periodontitis phenotypes. J Dent Res. 2014;93(5):459–468. doi:10.1177/0022034514527288
- Papapanou PN, Behle JH, Kebschull M, et al. Subgingival bacterial colonization profiles correlate with gingival tissue gene expression. BMC Microbiol. 2009;9(1):221. doi:10.1186/1471-2180-9-221
- Stoecklin-Wasmer C, Guarnieri P, Celenti R, et al. MicroRNAs and their target genes in gingival tissues. J Dent Res. 2012;91(10):934–940. doi:10.1177/0022034512456551
- Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi:10.1038/nprot.2008.211
- Xie C, Mao X, Huang J, et al. KOBAS 2.0: a web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 2011;39(suppl_2):W316–W322. doi:10.1093/nar/gkr483
- Bindea G, Mlecnik B, Hackl H, et al. ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics. 2009;25(8):1091–1093. doi:10.1093/bioinformatics/btp101
- Szklarczyk D, Morris JH, Cook H, et al. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017;45(D1):D362–D368. doi:10.1093/nar/gkw937
- Chen Y, Wang X. miRDB: an online database for prediction of functional microRNA targets. Nucleic Acids Res. 2020;48(D1):D127–D131. doi:10.1093/nar/gkz757
- Li JH, Liu S, Zhou H, et al. starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014;42(D1):D92–D97. doi:10.1093/nar/gkt1248
- Paraskevopoulou MD, Vlachos IS, Karagkouni D, et al. DIANA-LncBase v2: indexing microRNA targets on non-coding transcripts. Nucleic Acids Res. 2016;44(D1):D231–D238. doi:10.1093/nar/gkv1270
- Mas-Ponte D, Carlevaro-Fita J, Palumbo E, et al. LncATLAS database for subcellular localization of long noncoding RNAs. RNA. 2017;23(7):1080–1087. doi:10.1261/rna.060814
- Loos BG, Papantonopoulos G, Jepsen S, et al. What is the contribution of genetics to periodontal risk? Dent Clin North Am. 2015;59(4):761–780. doi:10.1016/j.cden.2015.06.005
- Song L, Yao J, He Z, et al. Genes related to inflammation and bone loss process in periodontitis suggested by bioinformatics methods. BMC Oral Health. 2015;15(1):105. doi:10.1186/s12903-015-0086-7
- Loos BG, Van Dyke TE. The role of inflammation and genetics in periodontal disease. Periodontol 2000. 2020;83(1):26–39. doi:10.1111/prd.12297
- Suzuki A, Horie T, Numabe Y. Investigation of molecular biomarker candidates for diagnosis and prognosis of chronic periodontitis by bioinformatics analysis of pooled microarray gene expression datasets in Gene Expression Omnibus (GEO). BMC Oral Health. 2019;19(1):52. doi:10.1186/s12903-019-0738-0
- Bullon P, Cordero MD, Quiles JL, et al. Autophagy in periodontitis patients and gingival fibroblasts: unraveling the link between chronic diseases and inflammation. BMC Med. 2012;10(1):122. doi:10.1186/1741-7015-10-122
- An Y, Liu W, Xue P, et al. Increased autophagy is required to protect periodontal ligament stem cells from apoptosis in inflammatory microenvironment. J Clin Periodontol. 2016;43(7):618–625. doi:10.1111/jcpe.12549
- Hasturk H, Kantarci A. Activation and resolution of periodontal inflammation and its systemic impact. Periodontol 2000. 2015;69(1):255–273. doi:10.1111/prd.12105
- Li RF, Chen G, Ren JG, et al. The adaptor protein p62 is involved in RANKL-induced autophagy and osteoclastogenesis. J Histochem Cytochem. 2014;62(12):879–888. doi:10.1369/0022155414551367
- Whitehouse CA, Waters S, Marchbank K, et al. Neighbor of Brca1 gene (Nbr1) functions as a negative regulator of postnatal osteoblastic bone formation and p38 MAPK activity. Proc Natl Acad Sci U S A. 2010;107(29):12913–12918. doi:10.1073/pnas.0913058107
- Tanaka Y, Guhde G, Suter A, et al. Accumulation of autophagic vacuoles and cardiomyopathy in LAMP-2-deficient mice. Nature. 2000;406(6798):902–906. doi:10.1038/35022595
- Beertsen W, Willenborg M, Everts V, et al. Impaired phagosomal maturation in neutrophils leads to periodontitis in lysosomal-associated membrane protein-2 knockout mice. J Immunol. 2008;180(1):475–482. doi:10.4049/jimmunol.180.1.475
- Pajares M, Rojo AI, Arias E, et al. Transcription factor NFE2L2/NRF2 modulates chaperone-mediated autophagy through the regulation of LAMP2A. Autophagy. 2018;14(8):1310–1322. doi:10.1080/15548627.2018.1474992
- Zhang X, Jin Y, Wang Q, et al. Autophagy-mediated regulation patterns contribute to the alterations of the immune microenvironment in periodontitis. Aging. 2021;13(1):555–577. doi:10.18632/aging.202165
- Chunhui Y, Wenjun C, Hui W, et al. Pilose antler peptide protects osteoblasts from inflammatory and oxidative injury through EGF/EGFR signaling. Int J Biol Macromol. 2017;99::15–20. doi:10.1016/j.ijbiomac.2017.02.056
- Kim EN, Kaygusuz O, Lee HS, et al. Simultaneous quantitative analysis of ginsenosides isolated from the fruit of panax ginseng C.A. Meyer and regulation of HO-1 expression through EGFR signaling has anti-inflammatory and osteogenic induction effects in HPDL cells. Molecules. 2021;26(7):2092. doi:10.3390/molecules26072092
- Ntoumou E, Tzetis M, Braoudaki M, et al. Serum microRNA array analysis identifies miR-140-3p, miR-33b-3p and miR-671-3p as potential osteoarthritis biomarkers involved in metabolic processes. Clin Epigenetics. 2017;9(1):127. doi:10.1186/s13148-017-0428-1
- Ramirez-Salazar EG, Carrillo-Patino S, Hidalgo-Bravo A, et al. Serum miRNAs miR-140-3p and miR-23b-3p as potential biomarkers for osteoporosis and osteoporotic fracture in postmenopausal Mexican-Mestizo women. Gene. 2018;679::19–27. doi:10.1016/j.gene.2018.08.074
- Ren T, Wei P, Song Q, et al. MiR-140-3p ameliorates the progression of osteoarthritis via targeting CXCR4. Biol Pharm Bull. 2020;43(5):810–816. doi:10.1248/bpb.b19-00959
- Mao JH, Sui YX, Ao S, et al. miR-140-3p exhibits repressive functions on preosteoblast viability and differentiation by downregulating MCF2L in osteoporosis. In Vitro Cell Dev Biol Anim. 2020;56(1):49–58. doi:10.1007/s11626-019-00405-9
- Lou Z, Peng Z, Wang B, et al. miR-142-5p promotes the osteoclast differentiation of bone marrow-derived macrophages via PTEN/PI3K/AKT/FoxO1 pathway. J Bone Miner Metab. 2019;37(5):815–824. doi:10.1007/s00774-019-00997-y
- Sun P, Wu Y, Li X, et al. miR-142-5p protects against osteoarthritis through competing with lncRNA XIST. J Gene Med. 2020;22(4):e3158. doi:10.1002/jgm.3158
- Ma C, Nie ZK, Guo HM, et al. MiR-671-5p plays a promising role in restraining osteosarcoma cell characteristics through targeting TUFT1. J Biochem Mol Toxicol. 2020;34(7):e22490. doi:10.1002/jbt.22490
- Loda A, Heard E. Xist RNA in action: past, present, and future. PLoS Genet. 2019;15(9):e1008333. doi:10.1371/journal.pgen.1008333
- Wang W, Min L, Qiu X, et al. Biological function of long non-coding RNA (LncRNA) Xist. Front Cell Dev Biol. 2021;9:645647. doi:10.3389/fcell.2021.645647
- Lipsky MS, Su S, Crespo CJ, et al. Men and oral health: a review of sex and gender differences. Am J Mens Health. 2021;15(3):15579883211016361. doi:10.1177/15579883211016361
- Chen X, Yang L, Ge D, et al. Long non-coding RNA XIST promotes osteoporosis through inhibiting bone marrow mesenchymal stem cell differentiation. Exp Ther Med. 2019;17(1):803–811. doi:10.3892/etm.2018.7033
- Feng Y, Wan P, Yin L. Long noncoding RNA X-inactive specific transcript (XIST) promotes osteogenic differentiation of periodontal ligament stem cells by sponging microRNA-214-3p. Med Sci Monit. 2020;26:e918932. doi:10.12659/MSM.918932