Abstract
Objective
The roles of long non-coding RNA (lncRNAs) in ischemic stroke (IS) have been widely illustrated. Here, we focused on the function and mechanism of lncRNA SNHG7 in IS.
Methods
Middle cerebral artery occlusion (MCAO) was used for inducing mice to establish IS models in vivo. Oxygen and glucose deprivation/reoxygenation (OGD/R) was used for treating PC12 cells to establish IS models in vitro. Relative expression of SNHG7 and miR-9 was determined by qRT-PCR. The neuronal injury was assessed by measuring relative activity of ROS, malondialdehyde (MDA) level and cell viability. Cell viability was determined by MTT assay. Dual-luciferase reporter (DLR) assay was employed to test the target of SNHG7 or miR-9. Western blot was used to determine the protein expression of SIRT1. Apoptosis rate was measured by flow cytometry.
Results
SNHG7 was down-regulated and miR-9 was up-regulated by MCAO treatment in brain tissues of mice and by OGD/R treatment in PC12 cells. Overexpression of SNHG7 or suppression of miR-9 decreased the relative activity of ROS and the MDA level as well as enhancing cell viability, and SNHG7 reduced apoptosis rate in OGD/R-induced PC12 cells (IS cells). MiR-9 was targeted by SNHG7 and SIRT1 was targeted by miR-9. The protein expression of SIRT1 was reduced by OGD/R treatment in PC12 cells. The suppressive effects of SNHG7 on the relative activity of ROS, the MDA level and apoptosis rate as well as the promotion effect of SNHG7 on cell viability were reversed by miR-9 mimics or sh-SIRT1 in IS cells.
Conclusion
LncRNA SNHG7 alleviated OGD/R-induced neuronal injury by mediating miR-9/SIRT1 axis in vitro.
Introduction
Ischemic stroke (IS) is considered as a cerebrovascular disease with high disability and mortality, and it has high prevalence in the elderly.Citation1 The major symptoms of IS include facial numbness, impair of memory, trouble speaking and partial paralysis.Citation2,Citation3 In the past few years, there is an increasing incidence rate of IS in people, which may result from multitudinous factors, such as hypercholesterolemia, hypertension, obesity and diabetes.Citation4 Although traditional clinical treatments for ISCitation5 including thrombolytic therapy, percutaneous intravascular interventions and medication have achieved effectiveness, the unsatisfactory result of IS therapy urges us seeking for new strategies to relieve IS.
Long non-coding RNAs (lncRNAs), an abundant component of the mammalian transcriptome, are a subtype of RNA with a length of more than 200 nucleotides and lack protein-coding ability.Citation6 Previous studies have reported the potential roles of lncRNAs in IS.Citation7 Chen et al have reported that silencing of lncRNA ROR can alleviate injury of PC12 cells in IS cell models induced by hypoxia/reoxygenation (H/R).Citation8 Lv et al have revealed that lncRNA SNHG1 alleviates apoptosis and inflammation in IS cell model.Citation9 Xiang et al have indicated that inhibition of lncRNA MEG3 markedly enhances cell viability and reduces cell apoptosis in IS cell model induced by oxygen-glucose deprivation and reoxygenation (OGD/R).Citation10 Specifically, lncRNA small nucleolar RNA host gene 7 (SNHG7), located on chromosome 9q34.3, has been reported to make imperative impacts on diverse diseases.Citation11–Citation13 SNHG7 suppresses cell viability of cardiac fibroblasts in cardiac fibrosis.Citation11 SNHG7 can inhibit the high glucose-induced proliferation and migration of human retinal endothelial cells in diabetic retinopathy (DR).Citation12 SNHG7 can promote viability of cartilage cells and suppress apoptosis of cartilage cells in osteoarthritis.Citation13 However, the role and mechanism of SNHG7 in IS have not been elucidated till now.
MicroRNAs (miRNAs), a class of small intracellular molecules with 18–23 nucleotides in length, can regulate gene expression at the post-transcriptional level.Citation14 Many miRNAs are emerging as important molecular mediators of IS, such as miR-143,Citation15 miR-27bCitation16 and miR-191.Citation17 As one of the most widely explored miRNAs, miR-9 has also been revealed to exert vital roles in IS.Citation18–Citation20 The suppression of miR-9 can reduce inflammation and neuronal apoptosis to alleviate neuronal injury in IS cell models induced by OGD/R.Citation18 Overexpression of miR-9 promotes post-ischemic cell viability of the neuronal cells in IS cell models induced by OGD.Citation19 Overexpression of miR-9 inhibits the apoptosis of neurons in IS cell models induced by OGD.Citation20 Nonetheless, the regulatory effect of SNHG7 on miR-9 in IS has not been clarified.
Sirtuins (SIRTs) are a family of nicotinamide adenine dinucleotide (NAD+)-dependent histone deacetylases.Citation21 According to the report, seven closely-related SIRT family members have been identified, including silent information regulator factor 2-related enzyme 1 (SIRT1), SIRT2, SIRT3, SIRT4, SIRT5, SIRT6 and SIRT7.Citation22,Citation23 Abundant studies have revealed the roles of lncRNAs and miRNAs in these sirtuin genes, such as the miR-140-5p/SIRT2 axis in myocardial oxidative stress,Citation24 the miR-421/SIRT3 axis in H/R-induced oxidative stress,Citation25 the lncRNA HMMR-AS1/miR-138/SIRT6 axis in lung adenocarcinomaCitation26 and the miR-20b/SIRT7 axis in high glucose-induced podocyte apoptosis.Citation27 SIRT1 is a NAD+-dependent deacetylase.Citation28 Given that the expression of SIRT1 reduces with aging and SIRT1 slows down cellular senescence, researchers have focused on the involvement of SIRT1 in IS.Citation29,Citation30 Teertam et al have reported that miR-149-5p plays a regulatory role in neuronal cell death through SIRT1/p53 axis during IS.Citation31 Rao et al have revealed that repression of miR-217 protects neurons against OGD/R-induced injury by targeting SIRT1.Citation32 However, the regulatory mechanism between SIRT1 and miR-9 has not been explored in IS. In the current study, the expression and role of SNHG7 and miR-9 were probed in IS cell models. Furthermore, to further understand the molecular mechanism of SNHG7 in IS, the regulatory interrelation among SNHG7, miR-9 and SIRT1 was explored in IS cell models. Our finding might provide a novel target for the prevention and treatment of IS.
Materials and Methods
The Establishment of the Middle Cerebral Artery Occlusion (MCAO) Model
This study was performed after obtaining local ethical committee approval of Zibo First Hospital (No. 2,018,013), and all experiments were performed in accordance with the Guide for the Care and Use of Laboratory Animals. Male C57BL/6 mice (6–8 weeks) were bought from Shanghai Animal Laboratory Center (Shanghai, China). All mice were housed under conditions of constant temperature and humidity. Food and water were available ad libitum. Mice were divided into two groups (the sham group and the MCAO group), with five mice in each group. Mice in the MCAO group were anesthetized with 1.5–3% isoflurane. A 6–0 surgical nylon filament, coated with 1% poly-L-lysine and blunted at the tip, was inserted 10 mm into the internal carotid to occlude the origin of the middle cerebral artery (MCA) in mice. Cerebral blood flow (CBF) was determined in the area of MCA by laser Doppler flowmetry (Moor Instruments, Devon, UK). Only mice with >80% decrease of CBF in the MCA territory were included in the present study. After MCA obstruction for 1 h, the sutures were removed to recover blood flow and mice were allowed to recover for 24 h. In the sham group, all procedures were identical except for inserting an intraluminal filament. Finally, mice were anesthetized with pentobarbital sodium (50 mg/kg) and sacrificed by cervical dislocation. Brain tissues of mice were collected for following experiments.
Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
Total RNAs were extracted from PC12 cells or brain tissues of mice by using TRIzol® reagent (Takara, Dalian, China) and converted to complementary DNAs (cDNAs) with a PrimeScript™ RT reagent kit with gDNA Eraser (Takara). The qRT-PCR was performed by using SYBR Premix Ex Taq II (Takara) and the reaction procedure was shown as follows: initial denaturation at 95°C for 30 s, 40 cycles of 95°C for 10 s, 72°C for 45 s and 60°C for 15 s. The primers used in the current study were shown in . Finally, the relative expression of SNHG7 and miR-9 was calculated by the 2−ΔΔCt method, and GAPDH was chosen as an internal reference.
Table 1 Primers for Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR) in Current Study
Cell Culture
A rat adrenal pheochromocytoma cell line PC12 with specific characteristic of neural cells was used in accordance with previous studies.Citation33–Citation36 PC12 cells were bought from American Type Culture Collection (ATCC, USA). PC12 cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and 1% penicillin-streptomycin and incubated at 37°C under a humidified atmosphere with 5% CO2.
Establishment of OGD/R Model
The OGD/R model in PC12 cells was established as previously reported.Citation37 Briefly, PC12 cells cultured in glucose-free DMEM medium were incubated at 37°C in a hypoxia condition with 5% CO2 and 95% N2. After hypoxia treatment for 4 h, PC12 cells were transferred back to normal DMEM medium containing 10% FBS and 4.5 g/L glucose, and cells were incubated at 37°C under an atmosphere of 95% air and 5% CO2 to recover. Then PC12 cells were harvested after recovery for 4 h, 8 h, 12 h and 24 h for following experiments. PC12 cells in the sham group did not receive the treatment of OGD/R.
Cell Transfection
The pcDNA3.1 (His tag in-frame with the 3ʹ-end of the cDNA; CMV promoter; neomycin resistance), pcDNA-SNHG7 (containing full-length rat SNHG7), short hairpin (sh)-negative control (NC), sh-SNHG7, miR-NC, miR-9 mimics, mimics NC, miR-9 inhibitor, inhibitor NC and sh-SIRT1 were bought from RiboBio Company (Beijing, China). Additionally, we have listed the sequences of shRNAs or miRs as follows: sh-SNHG7 (5ʹ-GCCTGGGTGTTGCTGTGTATT-3ʹ), sh-SIRT1 (5ʹ-CCATTCTTCAAGTTTGCAA-3ʹ), miR-9 mimics (5ʹ-UCAUACAGCUAGAUAACCAAAGA-3ʹ), mimics NC (5ʹ-UCACAGUGAACCGGUCUCUUU-3ʹ), miR-9 inhibitor (5ʹ-GCTAGATAACCAAAG-3ʹ) and inhibitor NC (5ʹ-ACGTCTATACGCCA-3ʹ). When cells grew 80–90% confluence, the above transcripts were transfected into PC12 cells with Lipofectamine3000 (Invitrogen, Carlsbad, CA, USA) for 48 h.
Detection of Reactive Oxygen Species (ROS) and Malondialdehyde (MDA)
A lipid peroxidation product MDA assay kit (ab118970, Abcam, MA, USA) was used for measuring the MDA level under the guidance of the manufacturer’s instructions. A cellular ROS assay kit (ab113851, Abcam) was employed to determine relative activity of ROS.
3-(4, 5-Dimethyl-2-Thiazolyl)-2, 5-Diphenyl-2-H-Tetrazolium Bromide (MTT) Assay
PC12 cells were plated in 96-well plates at a density of 5 × 103/well and then incubated with 20 µL MTT solution (Beyotime, Shanghai, China). After incubation for 4 h, dimethyl sulfoxide (DMSO; 150 µL) was added to dissolve the remaining formazan crystals. Cell viability was evaluated by measuring the optical density (OD) at 450 nm with a microplate reader (BMG LABTECH, Durham, NC, USA).
Dual-Luciferase Reporter (DLR) Assay
The 3ʹ-UTR fragment of wild type (WT) SNHG7 or WT SIRT1 containing the complementary sequence of miR-9 was synthesized and introduced into a pGL3 Basic Vector (Promega, Madison, WI) to generate SNHG7 WT vector or SIRT1 WT vector. In a similar way, the 3ʹ-UTR fragment of mutant type (mut) SNHG7 or mut SIRT1 including the mutant sequence of miR-9 was synthesized and inserted into a pGL3 Basic Vector (Promega) to form SNHG7 mut vector or SIRT1 mut vector. Then PC12 cells were co-transfected with a WT vector/mut vector and miR-NC/miR-9 mimics for 48 h. Then relative luciferase activity was measured by Dual-Luciferase Reporter Assay System (Promega).
Western Blot
Total proteins were extracted from PC12 cells and brain tissues of mice by RIPA buffer (Beyotime). Equal proteins of each sample were separated by 10% sodium dodecyl sulphate-polyacrylamide gels (SDS-PAGE). Next, separated proteins were transferred onto polyvinylidene fluoride (PVDF) membranes (Merck Millipore, Billerica, MA, USA) and blocked with 5% non-fat milk for 1 h. Then protein samples were incubated with the following primary antibodies overnight at 4°C: anti-SIRT1 (1:1000, ab189494, Abcam) and anti-β-actin (1:6000, ab115777, Abcam). After the membranes were washed with tris buffered saline Tween (TBST), the horseradish-conjugated secondary antibody (1:10,000, ab205718, Abcam) was added to incubate at 37°C for 2 h. The immune-reactivity was visualized with a Chemiluminescence Detection Kit (Thermo Fisher Scientific, Shanghai, China) and the relative protein expression of SIRT1 over the β-actin was quantified by Gel-pro Analyzer software (Media Cybernetics, Maryland, USA).
Flow Cytometry
Apoptosis rate was analyzed by an Annexin V-FITC/PI kit (Beyotime) on flow cytometry. Apoptotic cells (PC12 cells) were labeled with FITC-AnnexinV and PI for 30 min in the dark. The fluorescence was determined on cytoFLEX LX flow cytometer (Beckman-Counter Electronics, Jiangsu, China) using CytExpert software. Quadrants were positioned on Annexin V/PI plots to distinguish apoptotic cells
Bioinformatic Analysis
The targets of SNHG7 were predicted by starbase (http://starbase.sysu.edu.cn/agoClipRNA.php?), and a total of 82 targets for SNHG7 were predicted. Among these targets, miR-9 plays an important role in IS, and the regulatory effect of SNHG7 on miR-9 in IS has not been investigated. Thus, we chose miR-9 as a target in this study.
The targets of miR-9 were predicted by starbase (http://starbase.sysu.edu.cn/agoClipRNA.php?), and a total of 5, 413 targets for miR-9 were predicted. Among these targets, SIRT1 is important in IS, and the regulatory mechanism between miR-9 and SIRT1 has not been explored in IS. Additionally, we have verified the targeting relationship between SIRT1 and miR-9 by miRDB. Therefore, we chose SIRT1 as a target in this study.
Statistical Analysis
Experiments of the present study were implemented in triplicate. The SPSS Statistics 22.0 was utilized for the statistical analysis of data. All data were shown as the mean ± standard deviation. Student’s t-test was employed to assess the differences between two groups. The One-way ANOVA method was used for multiple comparisons, followed by a Tukey’s multiple comparisons test for pairwise comparisons. The P value less than 0.05 was regarded as statistically significant.
Results
SNHG7 Was Down-Regulated in Brain Tissues of MCAO-Induced Mice and OGD/R-Induced PC12 Cells, and It Mitigated OGD/R-Induced Neuronal Injury in PC12 Cells
In order to explore the function of SNHG7 in IS, we firstly determined the relative expression of SNHG7 in brain tissues of MCAO mice and OGD/R-induced PC12 cells by qRT-PCR. The result showed that SNHG7 was markedly decreased in brain tissues of MCAO mice compared with brain tissues of mice without MCAO treatment (P < 0.001, ). SNHG7 was also remarkably decreased by OGD/R treatment at 4 h, 8 h and 12 h in PC12 cells (P < 0.001, ). Then we silenced and overexpressed SNHG7. As shown in , SNHG7 was up-regulated by transfection of pcDNA-SNHG7 and down-regulated by transfection of sh-SNHG7 in PC12 cells (all P < 0.001). The results of functional assays demonstrated that the relative activity of ROS and the MDA level of PC12 cells in the OGD/R group were increased compared with the NC group, while cell viability of PC12 cells in the OGD/R group was notably decreased in comparison with the NC group (all P < 0.001, –). Of note, the relative activity of ROS and the MDA level were reduced by overexpression of SNHG7 in OGD/R-induced PC12 cells, and cell viability was enhanced by overexpression of SNHG7 in OGD/R-induced PC12 cells (all P < 0.001, –). In comparison to overexpression of SNHG7, knockdown of SNHG7 had opposite effects on cell viability, relative activity of ROS and the MDA level in OGD/R-induced PC12 cells (all P < 0.001, –).
Figure 1 SNHG7 is down-regulated in brain tissues of middle cerebral artery occlusion (MCAO)-induced mice and oxygen and glucose deprivation/reoxygenation (OGD/R)-induced PC12 cells, and it mitigates OGD/R-induced neuronal injury in PC12 cells. (A) Relative expression of SNHG7 was detected by quantitative real-time polymerase chain reaction (qRT-PCR) in brains tissues of MCAO mice. ***P < 0.001, vs Sham. n = 5. (B) Relative expression of SNHG7 was determined by qRT-PCR in OGD/R-induced PC12 cells. ***P < 0.001, vs Sham. There are three replicates in each experimental group. (C) Relative expression of SNHG7 was determined by qRT-PCR in PC12 cells after transfection of pcDNA-SNHG7 or sh-SNHG7. ***P < 0.001, vs pcDNA3.1. ###P < 0.001, vs sh-NC. There are three replicates in each experimental group. (D) Relative activity of ROS was determined in OGD/R-induced PC12 cells. ***P < 0.001, vs NC. ###P < 0.001, vs OGD/R + pcDNA3.1. &&&P < 0.001, vs OGD/R + sh-NC. (E) The MDA level was determined in OGD/R-induced PC12 cells. ***P < 0.001, vs NC. ###P < 0.001, vs OGD/R + pcDNA3.1. &&&P < 0.001, vs OGD/R + sh-NC. (F) The cell viability was determined by MTT assay in OGD/R-induced PC12 cells. ***P < 0.001, vs NC. ###P < 0.001, vs OGD/R+ pcDNA3.1. &&&P < 0.001, vs OGD/R + sh-NC. The experiments in were implemented at 12 h after OGD/R treatment, and there are three replicates in each experimental group.
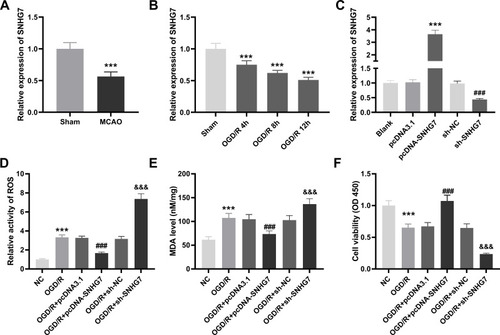
SNHG7 Could Serve as a Competing Endogenous RNA (ceRNA) for miR-9
To further explore the mechanism of SNHG7 in IS, we predicted the target genes of SNHG7 by starbase2.0. We observed that SNHG7 had potential binding sites for miR-9 (). Then the interaction between SNHG7 and miR-9 was tested by DLR assay. The result showed that relative luciferase activity of PC12 cells was significantly reduced by co-transfection of SNHG7 WT vector and miR-9 mimics (P < 0.001, ), whereas relative luciferase activity of PC12 cells showed no statistical change after co-transfection of SNHG7 mut vector and miR-9 mimics (). Relative expression of miR-9 was increased in brain tissues of MCAO mice in relative to that in brain tissues of mice without MCAO treatment (P < 0.001, ). The result of Pearson’s correlation analysis showed a negative correlation between SNHG7 and miR-9 in brain tissues of MCAO mice (P < 0.0001, ). In addition, relative expression of miR-9 was elevated by OGD/R treatment at 4 h, 8 h, 12 h and 24 h in PC12 cells (P < 0.001, ).
Figure 2 SNHG7 can serve as a competing endogenous RNAs (ceRNA) for miR-9. (A) The binding sequences between SNHG7 and miR-9 were predicted by starbase2.0. (B) The interaction between SNHG7 and miR-9 in PC12 cells was validated by dual-luciferase reporter (DLR) assay. ***P < 0.001, vs miR-NC. There are three replicates in each experimental group. (C) Relative expression of miR-9 was detected by quantitative real-time polymerase chain reaction (qRT-PCR) in brain tissues of middle cerebral artery occlusion (MCAO) mice. ***P < 0.001, vs Sham. n = 5. (D) The relationship between SNHG7 and miR-9 in brain tissues of MCAO mice was analyzed by Pearson’s correlation analysis. P < 0.0001. n = 5. (E) Relative expression of miR-9 in oxygen and glucose deprivation/reoxygenation (OGD/R)-induced PC12 cells was detected by qRT-PCR. ***P < 0.001, vs Sham. There are three replicates in each experimental group.
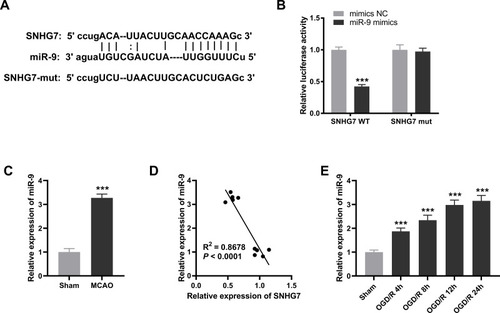
Inhibition of miR-9 Alleviated OGD/R-Induced Neuronal Injury in PC12 Cells
Afterwards, we explored the expression and function of miR-9 in PC12 cells. We observed that the expression of miR-9 was obviously elevated by OGD/R treatment in PC12 cells (P < 0.001, ). MiR-9 was evidently increased by transfection of miR-9 mimics and decreased by transfection of miR-9 inhibitor in OGD/R-induced PC12 cells (all P < 0.001, ). The relative activity of ROS and the MDA level of PC12 cells in the OGD/R group were increased in comparison with the NC group, whereas the viability of PC12 cells in the OGD/R group was significantly decreased in comparison with the NC group (all P < 0.001, –). Importantly, the relative activity of ROS and the MDA level were reduced by inhibition of miR-9 in OGD/R-induced PC12 cells, while cell viability was enhanced by inhibition of miR-9 in OGD/R-induced PC12 cells (all P < 0.001, –). In contrast to inhibition of miR-9, up-regulation of miR-9 had opposite effects on cell viability, the relative activity of ROS and the MDA level in OGD/R-induced PC12 cells (all P < 0.001, –).
Figure 3 Inhibition of miR-9 alleviates oxygen and glucose deprivation/reoxygenation (OGD/R)-induced neuronal injury in PC12 cells. (A) Relative expression of miR-9 was determined by real-time polymerase chain reaction (qRT-PCR) in OGD/R-induced PC12 cells. ***P < 0.001, vs.NC. ###P < 0.001, vs OGD/R + mimics NC. &&&P < 0.001, vs OGD/R + inhibitor NC. (B) The cell viability was determined by MTT assay in OGD/R-induced PC12 cells. ***P < 0.001, vs NC. ###P < 0.001, vs OGD/R + mimics NC. &&&P < 0.001, vs OGD/R+ inhibitor NC. (C) Relative activity of ROS was determined in OGD/R-induced PC12 cells. ***P < 0.001, vs NC. ###P < 0.001, vs OGD/R + mimics NC. &&&P < 0.001, vs OGD/R + inhibitor NC. (D) The MDA level was determined in OGD/R-induced PC12 cells. ***P< 0.001, vs NC. ###P < 0.001, vs OGD/R + mimics NC. &&&P < 0.001, vs OGD/R + inhibitor NC. The experiments in were implemented at 12 h after OGD/R treatment, and there are three replicates in each experimental group.
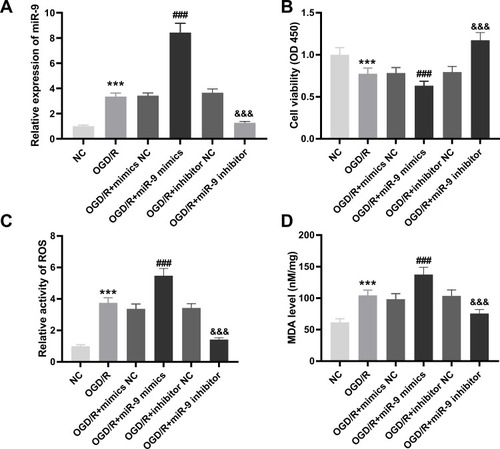
SIRT1 Was Targeted by miR-9
Subsequently, we predicted the target genes of miR-9 with starbase2.0. It was observed that SIRT1 had a conserved binding region of miR-9 (). The result of DLR revealed that relative luciferase activity of PC12 cells was reduced by co-transfection of SIRT1 WT vector and miR-9 mimics (P < 0.001, ), whereas relative luciferase activity of PC12 cells showed no significant change after co-transfection of SIRT1 mut vector and miR-9 mimics (). The result of Western blot showed that SIRT1 was down-regulated in brain tissues of MCAO mice in relative to that in brain tissues of mice without MCAO treatment (P < 0.001, ). There was a negative correlation between SIRT1 and miR-9 in brain tissues of MCAO mice (P < 0.0001, ). Furthermore, SIRT1 was decreased by OGD/R treatment at 4 h, 8 h, 12 h and 24 h in PC12 cells (P < 0.001, ).
Figure 4 SIRT1 is targeted by miR-9. (A) The binding sequence between SIRT1 and miR-9 was predicted by starbase2.0. (B) The interaction between SIRT1 and miR-9 in PC12 cells was validated by dual-luciferase reporter (DLR) assay. ***P < 0.001, vs miR-NC. There are three replicates in each experimental group. (C) Relative expression of SIRT1 was detected by Western blot in brain tissues of middle cerebral artery occlusion (MCAO) mice. ***P < 0.001, vs Sham. n = 5. (D) The relationship between SIRT1 and miR-9 in brain tissues of MCAO mice was analyzed by Pearson’s correlation analysis. P < 0.0001. n = 5. (E) Relative expression of SIRT1 in oxygen and glucose deprivation/reoxygenation (OGD/R)-induced PC12 cells was detected by Western blot. ***P < 0.001, vs Sham. There are three replicates in each experimental group.
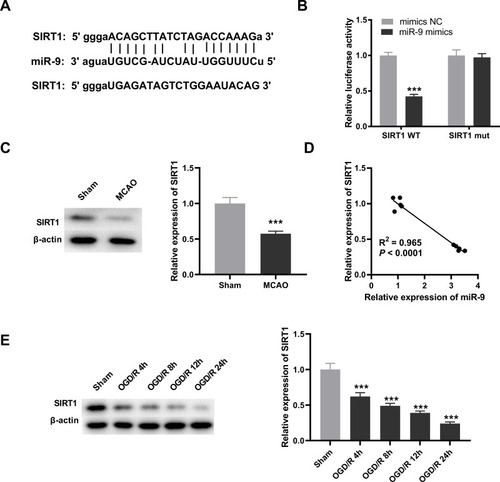
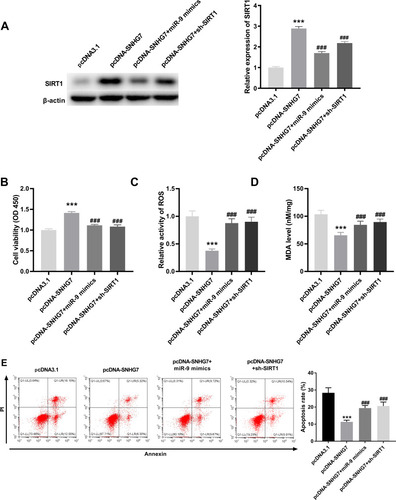
LncRNA SNHG7 Mitigated OGD/R-Induced Neuronal Injury via Regulating miR-9/SIRT1 Axis in PC12 Cells
Based on above results, we hypothesized that SNHG7 might repress the injury of PC12 cells through mediating miR-9/SIRT1 axis. To verify this, we firstly explored the interrelation among SNHG7, miR-9 and SIRT1. The result of Western blot demonstrated that SIRT1 was up-regulated by overexpression of SNHG7, and the promotion effect of SNHG7 on SIRT1 was reversed by miR-9 mimics or sh-SIRT1 in OGD/R-induced PC12 cells (all P < 0.001, ). Then rescue assays were performed. The results indicated that cell viability was promoted by overexpression of SNHG7, and the promotion effect of SNHG7 on cell viability was reversed by overexpression of miR-9 or knockdown of SIRT1 in OGD/R-induced PC12 cells (all P < 0.001, ). The relative activity of ROS and the MDA level were reduced by overexpression of SNHG7, and the suppression effects of SNHG7 on the relative activity of ROS and the MDA level were reversed by overexpression of miR-9 or knockdown of SIRT1 in PC12 cells induced by OGD/R (all P < 0.001, and ). Moreover, apoptosis rate of OGD/R-induced PC12 cells was reduced by up-regulation of SNHG7, while the reduction effect of SNHG7 on apoptosis rate was reversed by up-regulation of miR-9 or knockdown of SIRT1 in OGD/R-induced PC12 cells (all P < 0.001, ).
Figure 5 LncRNA SNHG7 lightens oxygen and glucose deprivation/reoxygenation (OGD/R)-induced neuronal injury via regulating miR-9/SIRT1 axis in PC12 cell. (A) Relative expression of SIRT1 was determined by Western blot in OGD/R-induced PC12 cells. ***P < 0.001, vs pcDNA3.1. ###P < 0.001, vs pcDNA-SNHG7. (B) The cell viability was determined by MTT assay in OGD/R-induced PC12 cells. ***P < 0.001, vs pcDNA3.1. ###P < 0.001, vs pcDNA-SNHG7. (C) Relative activity of ROS was determined in OGD/R-induced PC12 cells. ***P < 0.001, vs.pcDNA3.1. ###P < 0.001, vs pcDNA-SNHG7. (D) The MDA level was determined in OGD/R-induced PC12 cells. ***P < 0.001, vs pcDNA3.1. ###P < 0.001, vs.pcDNA-SNHG7. (E) Apoptosis rate was determined by flow cytometry in OGD/R-induced PC12 cells. ***P < 0.001, vs pcDNA3.1. ###P < 0.001, vs pcDNA-SNHG7. The experiments in were implemented at 12 h after OGD/R treatment, and there are three replicates in each experimental group.
Discussion
IS is a severe neurological disease with serious impairment of the neurological function.Citation38 Growing literature has shown that down-regulation of lncRNAs is implicated in the pathological process of IS, such as lncRNA Oprm1,Citation39 lncRNA ZFAS1Citation40 and lncRNA ANRIL.Citation34 Coincident with expression trend of previous studies, we also observed that SNHG7 was down-regulated by MCAO treatment in brain tissues of mice and by OGD/R treatment in PC12 cells, implying that SNHG7 might involve in the pathologies of IS. Besides, numerous studies have revealed the key role of SNHG7 in many diseases.Citation11–Citation13 SNHG7 can inhibit the high glucose-induced proliferation and migration of human retinal endothelial cells in DR.Citation12 SNHG7 promotes cell viability and represses cell apoptosis in osteoarthritis.Citation13 SNHG7 suppresses cell viability of cardiac fibroblasts in cardiac fibrosis.Citation11 In the present study, we found that the relative activity of ROS, apoptosis rate and the MDA level were reduced by SNHG7, while the viability was stimulated by SNHG7 in PC12 cells induced by OGD/R, suggesting that SNHG7 mitigated OGD/R-induced neuronal injury in PC12 cells. Based on these outcomes, we inferred that SNHG7 might be a promising target for IS therapy.
In recent years, the expression and function of miR-9 in IS have drawn wide attention.Citation18,Citation19,Citation41 Xue et al have revealed that miR-9 is up-regulated by OGD treatment in neurons, and the suppression of miR-9 remarkably alleviates neuronal injury in IS cell models by decreasing OGD-induced neuronal apoptosis.Citation18 Ji et al have revealed that the level of serum miR-9 is markedly higher in acute IS patients than that in non-stroke volunteers.Citation41 Nampoothiri et al have reported that inhibition of miR-9 enhances viability of neuronal cells in IS cell models.Citation19 Consistently, we also observed that miR-9 was increased by MCAO treatment in brain tissues of mice and by OGD/R induction in PC12 cells. Moreover, we observed that down-regulation of miR-9 reduced the relative activity of ROS and the MDA level in PC12 cells induced by OGD/R, while promoted viability of PC12 cells induced by OGD/R. These results of functional assays revealed that inhibition of miR-9 alleviated neuronal injury in IS cell models induced by OGD/R, and the role of miR-9 in our study was similar to previous studies. Besides, miR-9 has been revealed to interact with SNHG7 in skeletal fractureCitation42 and in malignant melanoma.Citation43 Similarly, we also found that miR-9 was targeted by SNHG7 and negatively correlated with SNHG7. Meanwhile, we discovered that overexpression of miR-9 reversed the suppression effects of SNHG7 on the relative activity of ROS and the MDA level as well as the promotion effect of SNHG7 on cell viability in OGD/R-induced PC12 cells. Collectively, SNHG7 might alleviate neuronal injury by sponging miR-9 in IS cell models.
Previously, many studies have reported the expression and role of SIRT1 in IS.Citation29,Citation30 Yan et al have revealed that SIRT1 is down-regulated in the stroke group compared with the normal group, and rosuvastatin ameliorates cerebral infarction condition of rats with cerebral IS by SIRT1/NF-κB pathway.Citation44 Teertam et al have indicated that the expression of SIRT1 is down-regulated in the MCAO group in comparison with the sham group, and overexpression of SIRT1 decreases the loss of pyramidal neuron in brains of IS mice.Citation31 In line with previous literature, we found that SIRT1 was down-regulated by MCAO treatment in brain tissues of mice and by OGD/R treatment in PC12 cells, suggesting that SIRT1 might alleviate IS. Furthermore, it has been reported that SIRT1 can act as the downstream target of miR-199a in cerebral ischemiaCitation45 and miR-149-5p in acute IS.Citation31 Here, we observed that SIRT1 was targeted by miR-9 and negatively correlated with miR-9. Based on above results, we speculated that inhibition of miR-9 alleviated neuronal injury by targeting SIRT1 in IS cell models. Meantime, we discovered that the knockdown of SIRT1 reversed the inhibition effects of SNHG7 on the relative activity of ROS, the MDA level and apoptosis rate as well as the promotion effect of SNHG7 on cell viability in OGD/R-induced PC12 cells. At length, we deduced that SNHG7 might attenuate OGD/R-induced neuronal injury by mediating miR-9/SIRT1 axis in vitro.
Conclusions
To sum up, SNHG7 was strikingly down-regulated by MCAO treatment in brain tissues of mice and by OGD/R treatment in PC12 cells. DLR assay verified that SNHG7 could act as the sponge of miR-9 and SIRT1 was the downstream target of miR-9 in brain tissues of mice. Overall, our results indicated that SNHG7 mitigated neuronal injury through competition with miR-9 to regulate the expression of SIRT1 in IS cell models induced by OGD/R. SNHG7 was a promising therapeutic target for attenuating IS. However, the functional experiments of SNHG7 in vivo were not performed in our study, and further study is needed to verify our findings.
Abbreviations
Long non-coding RNA, lncRNAs; ischemic stroke, IS; Middle cerebral artery occlusion, MCAO; Oxygen and glucose deprivation/reoxygenation, OGD/R; Malondialdehyde, MDA; Dual-luciferase reporter, DLR.
Ethics Approval
This study was conducted after obtaining local ethical committee approval of Zibo First Hospital (No. 2018013) and We confirm that all experiments were executed in accordance with the Guide for the Care and Use of Laboratory Animals.
Disclosure
The authors report no conflicts of interest for this work.
Additional information
Funding
References
- Rodrigo R, Fernandez-Gajardo R, Gutierrez R, et al. Oxidative stress and pathophysiology of ischemic stroke: novel therapeutic opportunities. CNS Neurol Disord Drug Targets. 2013;12:698–714. doi:10.2174/187152731131205001523469845
- Gupta A, Sattur MG, Aoun RJN, et al. Hemicraniectomy for ischemic and hemorrhagic stroke: facts and controversies. Neurosurg Clin N Am. 2017;28:349–360. doi:10.1016/j.nec.2017.02.01028600010
- Randolph SA. Ischemic Stroke. Workplace Health Saf. 2016;64:444. doi:10.1177/216507991666540027621261
- Grysiewicz RA, Thomas K, Pandey DK. Epidemiology of ischemic and hemorrhagic stroke: incidence, prevalence, mortality, and risk factors. Neurol Clin. 2008;26:871–895, vii. doi:10.1016/j.ncl.2008.07.00319026895
- Hao L, Zou Z, Tian H, et al. Stem cell-based therapies for ischemic stroke. Biomed Res Int. 2014;2014:468748. doi:10.1155/2014/46874824719869
- Dharap A, Pokrzywa C. Vemuganti R Increased binding of stroke-induced long non-coding RNAs to the transcriptional corepressors Sin3A and coREST. ASN Neuro. 2013;5:283–289. doi:10.1042/AN2013002924063527
- Bao MH, Szeto V, Yang BB, et al. Long non-coding RNAs in ischemic stroke. Cell Death Dis. 2018;9:281. doi:10.1038/s41419-018-0282-x29449542
- Chen H, Li X, LncRNA ROR is involved in cerebral hypoxia/reoxygenation-induced injury in PC12 cells via regulating miR-135a-5p/ROCK1/2. Am J Transl Res. 2019;11:6145–6158.31632583
- Lv L, Xi HP, Huang JC. et al. LncRNA SNHG1 alleviated apoptosis and inflammation during ischemic stroke by targeting miR-376a and modulating CBS/H2S pathway. Int J Neurosci;2020 1–11. doi:10.1080/00207454.2020.1782904
- Xiang Y, Zhang Y, Xia Y, et al. LncRNA MEG3 targeting miR-424-5p via MAPK signaling pathway mediates neuronal apoptosis in ischemic stroke. Aging. 2020;12:3156–3174. doi:10.18632/aging.10279032065781
- Wang J, Zhang S, Li X, et al. LncRNA SNHG7 promotes cardiac remodeling by upregulating ROCK1 via sponging miR-34-5p. Aging. 2020;12:10441–10456. doi:10.18632/aging.10326932507765
- Ke N, Pi LH, Liu Q, et al. Long noncoding RNA SNHG7 inhibits high glucose-induced human retinal endothelial cells angiogenesis by regulating miR-543/SIRT1 axis. Biochem Biophys Res Commun. 2019;514:503–509. doi:10.1016/j.bbrc.2019.04.14131056258
- Tian F, Wang J, Zhang Z, et al. LncRNA SNHG7/miR-34a-5p/SYVN1 axis plays a vital role in proliferation, apoptosis and autophagy in osteoarthritis. Biol Res. 2020;53:9. doi:10.1186/s40659-020-00275-632066502
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi:10.1016/j.cell.2009.01.00219167326
- Bai Y, Zhang Y, Han B, et al. Circular RNA DLGAP4 ameliorates ischemic stroke outcomes by targeting mir-143 to regulate endothelial-mesenchymal transition associated with blood-brain barrier integrity. J Neurosci. 2018;38:32–50. doi:10.1523/JNEUROSCI.1348-17.201729114076
- Yuan Y, Zhang Z, Wang Z, et al. MiRNA-27b regulates angiogenesis by targeting AMPK in mouse ischemic stroke model. Neuroscience. 2019;398:12–22. doi:10.1016/j.neuroscience.2018.11.04130513374
- Du K, Zhao C, Wang L, et al. MiR-191 inhibit angiogenesis after acute ischemic stroke targeting VEZF1. Aging. 2019;11:2762–2786. doi:10.18632/aging.10194831064890
- Xue Y, Li M, Liu D, et al. Expression of miR-9 in the serum of patients with acute ischemic stroke and its effect on neuronal damage. Int J Clin Exp Pathol. 2018;11:5885–5892.31949675
- Nampoothiri SS, Rajanikant GK. miR-9 upregulation integrates post-ischemic neuronal survival and regeneration in vitro. Cell Mol Neurobiol. 2019;39:223–240. doi:10.1007/s10571-018-0642-130539420
- Wei N, Xiao L, Xue R, et al. MicroRNA-9 mediates the cell apoptosis by targeting Bcl2l11 in ischemic stroke. Mol Neurobiol. 2016;53:6809–6817. doi:10.1007/s12035-015-9605-426660116
- Sosnowska B, Mazidi M, Penson P, et al. The sirtuin family members SIRT1, SIRT3 and SIRT6: their role in vascular biology and atherogenesis. Atherosclerosis. 2017;265:275–282. doi:10.1016/j.atherosclerosis.2017.08.02728870631
- Giblin W, Skinner ME, Lombard DB. Sirtuins: guardians of mammalian healthspan. Trends Genet. 2014;30:271–286. doi:10.1016/j.tig.2014.04.00724877878
- Frye RA, Frye RA. Phylogenetic classification of prokaryotic and eukaryotic Sir2-like proteins. Biochem Biophys Res Commun. 2000;273:793–798. doi:10.1006/bbrc.2000.300010873683
- Zhao L, Qi Y, Xu L, et al. MicroRNA-140-5p aggravates doxorubicin-induced cardiotoxicity by promoting myocardial oxidative stress via targeting Nrf2 and Sirt2. Redox Biol. 2018;15:284–296. doi:10.1016/j.redox.2017.12.01329304479
- Liu Y, Qian XM, He QC, et al. MiR-421 inhibition protects H9c2 cells against hypoxia/reoxygenation-induced oxidative stress and apoptosis by targeting Sirt3. Perfusion. 2020;35:255–262. doi:10.1177/026765911987072531469043
- Cai Y, Sheng Z, Chen Y, et al. LncRNA HMMR-AS1 promotes proliferation and metastasis of lung adenocarcinoma by regulating MiR-138/sirt6 axis. Aging. 2019;11:3041–3054. doi:10.18632/aging.10195831128573
- Wang X, Lin B, Nie L, et al. microRNA-20b contributes to high glucose-induced podocyte apoptosis by targeting SIRT7. Mol Med Rep. 2017;16:5667–5674. doi:10.3892/mmr.2017.722428849008
- Meng X, Tan J, Li M, et al. Sirt1: role under the condition of ischemia/hypoxia. Cell Mol Neurobiol. 2017;37:17–28.26971525
- Imai S, Guarente L. Ten years of NAD-dependent SIR2 family deacetylases: implications for metabolic diseases. Trends Pharmacol Sci. 2010;31:212–220. doi:10.1016/j.tips.2010.02.00320226541
- Petegnief V. Planas AM SIRT1 regulation modulates stroke outcome. Transl Stroke Res. 2013;4:663–671. doi:10.1007/s12975-013-0277-y24323420
- Teertam SK, Jha S. Up-regulation of Sirt1/miR-149-5p signaling may play a role in resveratrol induced protection against ischemia via p53 in rat brain. J Clin Neurosci. 2020;72:402–411. doi:10.1016/j.jocn.2019.11.04331866350
- Rao G, Zhang W. MicroRNA217 inhibition relieves cerebral ischemia/reperfusion injury by targeting SIRT1. Mol Med Rep. 2019;20:1221–1229.31173187
- Li G, Zou L, Xie W, et al. The effects of NONRATT021972 lncRNA siRNA on PC12 neuronal injury mediated by P2X7 receptor after exposure to oxygen-glucose deprivation. Purinergic Signal. 2016;12:479–487. doi:10.1007/s11302-016-9513-827100355
- Liu B, Cao W, Xue J. LncRNA ANRIL protects against oxygen and glucose deprivation (OGD)-induced injury in PC-12 cells: potential role in ischaemic stroke. Artif Cells Nanomed Biotechnol. 2019;47:1384–1395. doi:10.1080/21691401.2019.159694431174432
- Yuan Y, Zheng Z. Geniposide protects PC-12 cells against oxygen and glucose deprivation-induced injury by up-regulation of long-noncoding RNA H19. Life Sci. 2019;216:176–182. doi:10.1016/j.lfs.2018.11.04730472296
- Zhao FY, Tang J, Zhang L, et al. [Role of long non-coding RNA BC088414 in hypoxic-ischemic injury of neural cells]. Zhongguo Dang Dai Er Ke Za Zhi. 2015;17:1348–1353. Chinese.26695678
- Wen Y, Yu Y, Fu X. LncRNA Gm4419 contributes to OGD/R injury of cerebral microglial cells via IkappaB phosphorylation and NF-kappaB activation. Biochem Biophys Res Commun. 2017;487:923–929. doi:10.1016/j.bbrc.2017.05.00528476620
- Vijayan M, Reddy PH. Non-coding RNAs based molecular links in type 2 diabetes, ischemic stroke, and vascular dementia. J Alzheimers Dis. 2020;75:353–383.32310177
- Jing H, Liu L, Jia Y, et al. Overexpression of the long non-coding RNA Oprm1 alleviates apoptosis from cerebral ischemia-reperfusion injury through the Oprm1/miR-155/GATA3 axis. Artif Cells Nanomed Biotechnol. 2019;47:2431–2439. doi:10.1080/21691401.2019.162640831187646
- Zhang Y. lncRNA zfas1 improves neuronal injury and inhibits inflammation, oxidative stress, and apoptosis by sponging mir-582 and upregulating NOS3 expression in cerebral ischemia/reperfusion injury. Inflammation. 2020.
- Ji Q, Ji Y, Peng J, et al. Increased brain-specific MiR-9 and MiR-124 in the serum exosomes of acute ischemic stroke patients. PLoS One. 2016;11:e0163645. doi:10.1371/journal.pone.016364527661079
- Chen Z, Liu Z, Shen L, et al. Long non-coding RNA SNHG7 promotes the fracture repair through negative modulation of miR-9. Am J Transl Res. 2019;11:974–982.30899396
- Wang W, Liu G, Liu M, et al. Long non-coding RNA SNHG7 promotes malignant melanoma progression through negative modulation of miR-9. Histol Histopathol. 2020;18225.
- Yan L, Zhu T. Effects of rosuvastatin on neuronal apoptosis in cerebral ischemic stroke rats via Sirt1/NF-kappa B signaling pathway. Eur Rev Med Pharmacol Sci. 2019;23:5449–5455.31298398
- Yin WL, Yin WG, Huang BS, et al. LncRNA SNHG12 inhibits miR-199a to upregulate SIRT1 to attenuate cerebral ischemia/reperfusion injury through activating AMPK signaling pathway. Neurosci Lett. 2019;690:188–195. doi:10.1016/j.neulet.2018.08.02630144542
