Abstract
Lung cancer is a heterogeneous disease, and there is a lack of adequate biomarkers for diagnosis. Long noncoding RNAs (lncRNAs) are emerging as an important set of molecules because of their roles in various key pathophysiological pathways, including cell growth, apoptosis, and metastasis. We review the current knowledge of the lncRNAs in lung cancer. In-depth analyses of lncRNAs in lung cancer have increased the number of potential effective biomarkers, thus providing options to increase the therapeutic benefit. In this review, we summarize the functions, mechanisms, and regulatory networks of lncRNAs in lung cancer, providing a basis for further research in this field.
Introduction
Besides small-cell lung cancer, non-small-cell lung cancer (NSCLC) is any type of epithelial lung cancer and accounts for 85% of all lung cancers. The 5-year survival rate of this heterogeneous disease is 16.6%, and it has only improved slightly in the past few years.Citation1 NSCLC can be classified into discrete subclasses according to histological phenotypes, including squamous cell carcinoma (SCC), adenocarcinoma (ADC), and large-cell carcinoma. The most common type of NSCLC is ADC, which is less associated with smoking and chronic inflammation than SCC.Citation2 The clinical integration of ADC in terms of predictive biomarker signatures is characterized by positive immunostaining for cytokeratin 7 and TTF1; however, SCC is characterized by positivity of cytokeratin 5, cytokeratin 6 and/or SOX2, and p63.Citation3,Citation4 In addition to tissue biopsy for clinical diagnosis, specific gene mutations in tumors have highlighted their usefulness as diagnostic markers and molecular therapy targets. For instance, EGFR, ALK, and MET mutations are always found in ADC patients.Citation2 However, DDR2, FGFR1, and FGFR2 mutations, as well as mutations in genes in the PI3K signaling pathway, are generally found more frequently in SCC.Citation5
Characteristics of lncRNAs
Noncoding RNAs (ncRNAs) are a superclass of endogenous, non-protein-coding RNA transcripts, many of which have essential functions in various cellular processes. Based on their function, ncRNAs can be classified into two subclasses, namely housekeeper ncRNAs (rRNA, tRNA, snRNA, snoRNA) and the regulated ncRNAs. The regulated ncRNAs can be categorized by length as follows: any ncRNA <200 nucleotides (nt) in length is a short ncRNA (siRNA, miRNA, piRNA) and those >200 nt in length are long ncRNAs (lncRNAs). Recently, Iyer et alCitation6 demonstrated that there are ~60,000 ncRNAs in the human genome, and >68% of these are lncRNAs, of which ~80% are not annotated. Approximately 1% of lncRNAs harbor ultraconserved elements, and 7% of lncRNAs harbor disease-associated single-nucleotide polymorphisms.Citation6
An increasing number of lncRNAs have been identified as key regulators of a wide range of cellular processes, including dosage compensation, imprinting, transcription, mRNA splicing, translation, nuclear and cytoplasmic trafficking, and cellular localization. Notably, ectopic expression of lncRNAs is associated with a great variety of diseases.
However, compared with mRNAs, lncRNAs have their own peculiar characteristics. First, the mean length of lncRNAs is shorter than that of mRNAs, with an average of 592 nt compared to 2,453 nt for mRNAs. Second, lncRNAs have fewer exons, although they harbor standard canonical splice sites.Citation7 Third, the methylation level of the transcription start site of lncRNAs is higher than that of mRNAs; therefore, the expression level of lncRNAs is significantly lower. In addition, lncRNAs show strict tissue specificity.Citation8 Furthermore, lncRNAs tend to correlate with transposable elements, especially with endogenous retroviruses, compared with protein-coding genes.Citation9 Moreover, lncRNA conservation includes more than introns or random intergenic regions, but they are less conserved than mRNAs.Citation10 Despite the fact that the conservation of some lncRNAs at the sequence level is not high, they may have the same functions.Citation11,Citation12 DiederichsCitation11 indicated that the conservation of lncRNAs should include four dimensions: sequence, structure, function, and expression from synthetic loci. Despite the differences between lncRNAs and mRNAs, they share certain common characteristics. Large-scale studies revealed that many identified lncRNAs are transcribed by RNA polymerase II, which is the same as that for mRNA. They also share the same posttranscriptional mRNA processing, including 5′-capping, splicing, and poly-adenylation at the 3′ end. Certain transcription factors can bind to the promoter region of lncRNAs, such as c-myc, p53, and Sox2.
Classification and functions of lncRNAs
The most recent classification of lncRNAs is based on their location relative to that of target protein-coding genes. According to these criteria, lncRNAs can be classified as exonic, intronic, overlapping, or intergenic. Moreover, based on the transcriptional direction with respect to protein-coding genes, lncRNAs are divided into two groups, namely sense and antisense.Citation7
According to transcriptional modes, lncRNAs can be categorized as cis-acting and trans-acting lncRNAs. Cis-acting lncRNAs mediate gene expression based on their position in the vicinity of the target gene transcriptional site. However, trans-acting lncRNAs can control the expression of genes at any loci based on the recruitment of proteins to the target sites to participate in transcriptional regulation. lncRNAs hybridize with DNA or RNA molecules to form triple-stranded RNA–DNA structures that play essential roles in transcription.Citation12,Citation13
lncRNAs can be classified into various discrete subclasses on the basis of their function as follows ().Citation14
Figure 1 Overview of the five molecular functions of lncRNAs.
Abbreviations: lncRNAs, long noncoding RNAs; TF, transcription factor; Pol, polymerase.
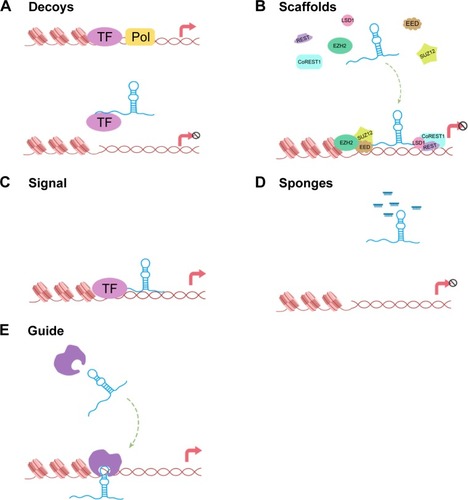
Decoys
lncRNA decoys exert biological functions by binding to proteins indirectly and playing a role in multiple processes of life.
Scaffolds
The second set of lncRNAs function as scaffolds, as lncRNAs act as platforms to bind relevant molecular complexes. Typically, HOTAIR acts as a bridging scaffold for the PRC2 and LSD1/CoREST/REST complex to suppress gene expression. PRC2 binds to an 89 bp fragment in the 5′ end of HOTAIR, and the LSD1/CoREST/REST complex binds to a 646 bp fragment in its 3′ end.Citation15,Citation16 Genome-wide RNA immunoprecipitation analysis has shown that ~20% of the various types of lncRNAs are bound to PRC2.Citation17 Another example of a molecular scaffold is Kcnq1ot1, which combines PRC2 and G9a to generate H3K27me3 and H3K9me3, leading to epigenetic silencing of genes.Citation18 Indeed, several studies show that not only proteins but also lncRNAs play critical roles in bridging molecular components.
Signal
lncRNAs play critical roles in signal regulation and in the responses to various stimuli. lncRNAs are often expressed in a spatial- and temporal-specific pattern. Upon specific expression, they can modulate translation and integrate developmental cues.Citation19 For instance, lincRNA-p21, which binds to hnRNP-K, promotes the proper localization of hnRNP-K and results in the silencing of p53-regulated genes.Citation20 Certain lncRNAs are emerging as signals of functionally significant biological events because of their roles in regulating transcriptional activity or pathways.
Sponges
PTEN1 (pseudogene of PTEN) upregulates the tumor suppressor PTEN by attracting miRNAs to its 3′-untranslated region.Citation21 The lncRNA GAPLINC acts as a “sponge” and modulates CD44 expression by attracting miR211-3p.Citation22 The lncRNA antisense ncRNA in the INK4 locus (ANRIL) can function as a “sponge” to titrate miR-99a/miR-449a, thus activating CDK6 and inactivating p15INK4B/p16INK4A. Consequently, E2F1 is released inappropriately and contributes to gastric cancer cell proliferation.Citation23 lncRNAs therefore function as sponges by interacting with miRNAs and suppressing their effects on target sites. Recent evidence highlights a classification of circRNAs as miRNA sponges that contribute to the downregulation of target genes. Compared to linear RNAs, the half-life of circRNAs is longer.Citation24 It is plausible that the duration and the effect of circRNAs have more advantages.
Guide
Multiple studies indicate that nuclear-retained lncRNAs function in guiding chromatin modifiers to specific genomic loci. Typical is PRC2, which contains a histone methyltransferase (enhancer of zeste 2, EZH2) that inhibits gene expression via trimethylation of histone H3Lys27 (H3K27me3).Citation25 In addition, chromatin conformation changes induced by nuclear-enriched lncRNAs can promote gene expression.Citation26
Regulation modes of lncRNAs
lncRNAs involved in epigenetic regulation
An example of the chromatin-modifying capabilities of lncRNAs is dosage compensation in mammals, which requires the preferential silencing of one parental allele. Xist gives rise to stable epigenetic silencing of large-scale genes in the X-chromosome by tethering PRC2 to the transcriptional site, inducing the formation of H3K27me3 to inactivate heterochromatin.Citation27 Another example is lncRNA p21, which can change DNA methylation levels by promoting histone methyltransferase and DNA methyltransferase binding to target sites, thus affecting the expression of reprogramming genes.Citation28
lncRNAs participate in transcriptional regulation
Evidence to date indicates that lncRNAs, which are transcribed from enhancers or promoters, can act in cis pattern to control transcriptional efficiency. lncRNAs transcribed from enhancers can affect their activity or help recruit protein factors. Among lncRNAs in prostate cancer, two overexpressed lncRNAs bind to the androgen receptor, promoting androgen receptor binding to an enhancer.Citation29
lncRNAs affect posttranscriptional processing
Most genes are transcribed via tissue-specific and cell-specific alternative-splicing patterns in humans.Citation30 Noteworthy is that the majority of lncRNAs are often similarly expressed in a spatial- and temporal-specific manner. Accumulating evidence indicates that lncRNAs may regulate alternative splicing by cis-acting mechanisms or by recruiting regulatory splicing factors.Citation31,Citation32 Recently, Gonzalez et al showed that a conserved antisense lncRNA transcribed from the FGFR2 locus in humans regulates alternative splicing by recruiting the histone demethylase KDM2a and polycomb group proteins. These findings suggested that lncRNAs can regulate alternative splicing through the establishment of a splicing-specific chromatin signature.Citation33
Dysregulation and functional roles of lncRNAs in lung cancer
Lung cancer is often associated with aberrant lncRNA transcriptomes, including onco-lncRNAs and tumor suppressor lncRNAs. Here, we discuss recent discoveries that implicate aberrant lncRNAs in lung cancer (). In addition, we provide a framework of systematically functionalized lncRNAs and integrate them with the protein-coding RNA dimension in complex networks ().
Figure 2 Overview of the regulatory network of lncRNAs in lung cancer.
Abbreviations: lncRNAs, long noncoding RNAs; miR, microRNA; MMPs, matrix metalloproteinases; EMT, epithelial-to-mesenchymal transition.
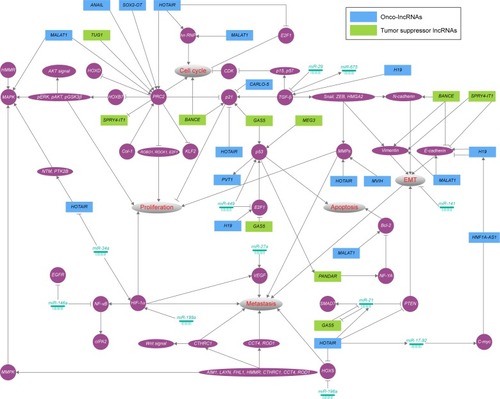
Table 1 Summary of lncRNAs in lung cancer
Onco-lncRNAs
MALAT1
Characteristics of MALAT1
MALAT1, also known as NEAT2, is located on chromosome 6p24.3 in humans with a length of 8.7 kb.Citation11,Citation12 MALAT1 is a well-characterized lncRNA with markedly high expression in most NSCLC types. MALAT1 generates a primary noncoding transcript that is enriched in the nucleus.Citation34 Furthermore, the MALAT1 gene shows strong sequence conservation from humans to zebra fish.Citation35 MALAT1 transcription is initiated from multiple promoters and produces different MALAT1 transcript variants. However, most MALAT1 transcripts, which harbor two distinct nuclear speckle localizational elements, are enriched in nuclear speckles, indicating essential role of MALAT1 in RNA metabolism. MALAT1 is modified at the 3′ end and forms a triple-helical structure, which may be the reason why MALAT1 appears as a very stable lncRNA.Citation36,Citation37 The half-life of MALAT1 ranges from 9 to 16.5 hours.Citation38 After modification, MALAT1 generates a second small mascRNA that is localized to the cytoplasm.Citation39 MALAT1 binds to the unmethylated PRC2 protein, therefore contributing to PRC2 preferential binding to H2AK5ac and H2AK13ac, marking transcriptional activation.Citation40 At the transcriptional level, several studies have shown that MALAT1 regulates gene expression, such as that of growth control genes. MALAT1 affects the phosphorylation levels of serine/arginine (SR) splicing factors, such as B-MYB, leading to changes in gene expression.Citation12 MALAT1 also regulates gene expression at the posttranscriptional level.Citation41 For instance, MALAT1 affects a minor subset of transcripts in the process of alternative splicing, such as RNPS1, PRP6, and SON.Citation42 In summary, MALAT1 can modulate gene expression both at the transcriptional and posttranscriptional level.
MALAT1 in lung cancer
MALAT1, which is found in abundance in various tissues and cell lines, is successfully knocked out by the zinc finger nuclease technique in A549 cells. MALAT1 has been shown to be a critical regulator of the metastasis phenotype in lung cancer cells.Citation43 In A549 cells, inhibition of MALAT1 downregulates the expression of motility-associated genes, including HMMR, AIM1, SLC26A2, LAYN, CCT4, ROD1, CTHRC, and FHL1.Citation44,Citation45 Xenograft models have confirmed this effect. The prevailing view is that MALAT1 participates in the regulation of motility-related genes to enhance the motility of lung ADC cells.Citation45 Recently, Shen et alCitation46 demonstrated that MALAT1 overexpression promotes lung cancer brain metastasis by inducing epithelial-to-mesenchymal transition (EMT). Another study suggested that MALAT1 is involved in cell cycle regulation at the level of G2/M phase progression. MALAT1 interacts with nuclear hnRNP C and promotes hnRNP C translocation to the cytoplasm in the G2/M phase.Citation47 In addition to its effect on cell cycle progression, MALAT1 affects the expression of the proapoptotic factor Bcl-2, which is specifically associated with prognosis in NSCLC.Citation48 Moreover, a meta-analysis and various studies have shown that high MALAT1 expression is related to poor overall survival in NSCLC.Citation49
HOTAIR
Characteristics of HOTAIR
HOTAIR is located on chromosome 12q13.13 in humans, and has a length of 2.1 kb. There are four gene clusters (HOXA, HOXB, HOXC, and HOXD) and 39 HOX genes in the genome.Citation25 These clusters generate numerous lncRNAs that are often expressed in a spatial- and temporal-specific pattern.Citation50 Although HOTAIR is located in the HOXC locus, it has been shown to repress gene expression in the HOXD locus. In addition, the secondary structure of HOTAIR contains four independently folding modules, two of which are evolutionarily conserved protein-binding domains.Citation51 HOTAIR is enriched in the nucleus but is also expressed in the cytoplasm.Citation52 HOTAIR has little sequence conservation in humans and mice, but the molecular mechanism is similar in the two species. Reciprocally, the conserved lncRNA sequence does not always possess the same function in other species as expected.
HOTAIR-mediated epigenetic gene silencing is dependent on its function as a bridge scaffold for PRC2 and LSD1/CoREST/REST. PRC2 binds to the 5′ end of HOTAIR and regulates chromosome occupancy by EZH2 (a subunit of PRC2), which leads to histone H3 lysine 27 trimethylation of the HOXD locus. Meanwhile, the LSD1/CoREST/REST complex binds to the 3′ end of HOTAIR.Citation53 HOTAIR represses gene expression by increasing the occupation of H3K27me3 and decreasing the occupation of H3K4me3 on the promoter in a trans-acting manner.Citation54 In addition to its role in the PRC2 and LSD1/CoREST/REST complexes, HOTAIR acts as a scaffold for E3 ubiquitin ligases and their corresponding substrates, namely E3 ubiquitin ligase Dzip3 and its substrate Ataxin-1, resulting in proteolysis.Citation55 Furthermore, HOTAIR harbors a target site for miR-34aCitation56 and miR-141 in its exon 6, both of which can attenuate the expression of HOTAIR. Exon 6 of HOTAIR contains a target site for let-7i that mediates the formation of a hetero-tetramer containing let-7i, Ago2, HOTAIR, and HuR.Citation57
HOTAIR in lung cancer
Ectopic expression of HOTAIR has been reported in a plethora of cancerous tissues. High HOTAIR levels are associated with invasion and metastases and linked to an advanced stage of disease and poor survival in patients with lung cancer.Citation58–Citation60 EMT mediates the invasive phenotype of lung cancer cells. HOTAIR may promote the EMT process of lung epithelial cells by distinct mechanisms. One of these mechanisms is transcriptional repression of HOXA5 gene. HOXA5 is related to postnatal lung development. It is likely that HOTAIR and miR-196a act in common to repress the expression of HOXA5, therefore contributing to dedifferentiation during lung tumorigenesis.Citation61 Moreover, HOTAIR facilitates the process of EMT by inhibiting the expression of cell adhesion-related genes in small-cell lung cancer epithelial cells.Citation59 Meanwhile, tumor-promoting type 1 collagen, which is a potent inducer of EMT, can modulate the translational control of HOTAIR in NSCLC cells.Citation62 In addition to the repression of EMT inhibitors and the promotion of EMT, HOTAIR also affects the expression of gelatinase, a matrix metalloproteinase (MMP) that plays a role in triggering invasion in lung cancer cells.Citation58 HOTAIR can promote resistance to cisplatin via downregulation of p21 (WAF1/CIP1) protein levels in lung ADC cells.Citation63 Furthermore, HOTAIR as a direct target of HIF-1α, promotes cell proliferation, migration, and invasion in hypoxic NSCLC. Evidence indicates that HIF-1α binds to HOTAIR via interaction with the upstream region of HOTAIR in NSCLC cells.Citation64 The upregulation of HOTAIR has been shown to have a negative impact on lung cancer by regulating genes involved in invasion, metastasis, and poor survival.Citation59
SOX2-OT
SOX2-OT is located on chromosome 3p26.33 in humans and has a length of 4.2 kb. SOX2-OT is an intronic lncRNA that overlaps with the SOX2 gene, which is a major regulator of pluripotency.Citation65 Recent studies have shown that SOX2 and SOX2-OT have similar expression patterns in lung SCC and ADC tissues.Citation66 There are multiple transcription initiation sites in the human and mouse SOX2-OT locus, leading to many spliced variants.Citation67 In addition, the human genomic region of SOX2-OT is characterized by multiple conserved transcription factor-binding sites. These sites have essential roles in the tumorigenesis process.Citation65 SOX2-OT is expressed at higher levels in human primary lung cancer tissues than in adjacent non-tumor tissues. Typically, SOX2-OT exhibits significantly higher expression in SCC of the lung than in ADC.Citation68 With respect to the mechanism of SOX2-OT, Hou et al showed that knockdown of SOX2-OT expression leads to cell cycle arrest at G2/M phase through the modulation of the expression of EZH2. Meanwhile, its expression level is significantly correlated with cell proliferation and colony formation ability in lung cancer cell lines.Citation66 This study further indicates that high SOX2-OT expression predicts poor survival in lung cancer patients.
HNF1A-AS1
HNF1A-AS1 is located on chromosome 12q24 and has a length of 2.46 kb. Overexpression of HNF1A-AS1 has been reported in lung ADC tissues compared with the corresponding non-tumor tissues. In addition, elevated expression of HNF1A-AS1 is linked to tumor–node–metastasis (TNM) stage, tumor size, and lymph node metastasis. HNF1A-AS1 can regulate EMT-related protein expression via binding to DNMT1, therefore regulating cell growth and metastasis both in vitro and in vivo.Citation1
ANRIL
ANRIL is derived from the p15/CDKN2B-p16/CDKN2A-p14/ARF gene cluster, which maps to human chromosome 9p21.3 with a length of 126 kb. This gene family is associated with cutaneous malignant melanoma and neural system tumors. ANRIL includes several isoforms with tissue-specific expression because it consists of 19 exons.Citation69 Recently, elevated levels of ANRIL have been reported in NSCLC tissues, and its expression level is significantly correlated with poor prognosis. siRNA-mediated knockdown of ANRIL results in the inhibition of cell proliferation and the promotion of apoptosis both in vitro and in vivo.Citation70 An ongoing study indicates that the subcellular localization of ANRIL is mostly in the cell nucleus. ANRIL is indicated as a “decoy”, and it represses KLF2 and p21 transcription by binding to PRC2 in NSCLC PC9 cells, which sheds light on the effect of ANRIL on NSCLC cell proliferation and apoptosis partly in trans.Citation70 Generally, KLF2 as a tumor suppressor is significantly downregulated in various cancers, leading to inhibition of cell proliferation via KRAS.Citation71 ANRIL can regulate the transcription of miR-99a and miR-449a by recruiting the PRC2 complex in gastric cancer.Citation72 Recently, Ren et alCitation73 demonstrated that high expression of miR-449a attenuates lung cancer cell proliferation, and the downregulation of miR-449a is correlated with a shorter disease-free survival of patients. It is plausible that ANRIL modulates the expression of miR-449a, thereby inhibiting the proliferation of lung cancer cells during lung tumorigenesis.
H19
Characteristics of H19
H19 is located on chromosome 11q15.5 in humans and has a length of 2.3 kb. H19 is a paternally imprinted gene that is spliced into five exons. The H19 gene locus is complex, harboring conserved miR-675 and antisense protein-encoding transcript (HOTS), which is a tumor suppressor.Citation74 Another pro-tumorigenic antisense transcript, 91H, overlaps with the H19 gene locus.Citation75 H19 and its nearby gene IGF2 show uniparental mono-allelic expression. There is an imprinting control region (ICR) between them. The ICR is unmethylated on maternal chromosomes, where it binds to the transcription factor CTCF and inhibits the enhancer from binding to the ICR. As a result, the enhancer binds to H19 and induces its expression. Conversely, on paternal chromosomes, the ICR is methylated and binds to the enhancer, resulting in H19 downregulation.Citation76 H19 can act in both cis and trans patterns: for instance, H19 regulates the imprinting of IGF2 by silencing the expression of neighboring genes. An example of H19 acting in trans pattern is its role as a molecular sponge for miRNA let-7, which is involved in inducing EMT.Citation77
H19 in lung cancer
Evidence to date indicates that H19 is associated with various tumorigenesis signaling pathways, including the p53 and HIF-1α, TGF-β, Bcr-Abl, Wnt/β-catenin, and HGF pathways.Citation78 In lung cancer cells, H19 is induced by hypoxic stress via a p53-dependent manner. Knockdown of H19 expression in hypoxia has a suppressing effect on cancer cell proliferation, anchorage-independent growth, and colony formation.Citation79,Citation80 Furthermore, knockdown of H19 can reverse the tumorigenic and scattering effect of HGF/SF on A549 cells.Citation79 The overexpression of H19 has a negative impact on lung cancer. Notably, the upregulated H19 is loss of imprinting independent in the airway epithelia of smokers in comparison with nonsmokers.Citation81
CARLO-5
CARLO-5 is located on chromosome 8q24.21 in humans and has a length of 1.6 kb. CARLO-5 is significantly upregulated in NSCLC tissues. Overexpression of CARLO-5 in NSCLC tissues is significantly correlated with advanced TNM stage. The expression of p16, p21, and p27, which are G0/G1 arrest markers, decreases with the downregulation of CARLO-5.Citation82 High level of CARLO-5 expression is a prognostic indicator of poor patient survival. Moreover, elevated expression of CARLO-5 is associated with increased proliferation and invasion ability, partially through the modulation of EMT.Citation83
MVIH
MVIH is located on chromosome 10q22 in humans. Overexpression of MVIH has been reported in NSCLC tissues, and its expression level is significantly correlated with TNM stage and tumor size. High levels of MVIH expression are prognostic indicators of poor survival.Citation84 siRNA-mediated knockdown of MVIH inhibits cell proliferation and invasion, partly via modulating the expression of MMP2 and MMP9. MMPs are involved in multiple biological processes, including remodeling of extracellular matrix, cell proliferation, differentiation, and metastasis.
PVT1
PVT1 is located on chromosome 8q24.21 in humans and has a length of 210 kb. The similar expression patterns of PVT1 and MYC gene might be explained by the shared genomic locus between them. A study identified a p53 transcription factor-binding site in the PVT1 promote region.Citation85 Overexpression of PVT1 in NSCLC tissues is significantly correlated with TNM stage. In addition, patients with high levels of PVT1 expression show poor survival.Citation86 Knockdown of PVT1 expression inhibits lung cancer cell proliferation, migration, and invasion.
EVADR
EVADR is located on chromosome 6q13 in humans and has a length of 0.39 kb. Recent evidence shows that EVADR is overexpressed in ADC tissues, including lung ADC, and is correlated with decreased patient survival. Among nine MER48-associated lncRNAs, EVADR is the only one that is consistently expressed in ADC tissues. EVADR expression is regulated via an active promoter provided by the MER48 endogenous retrovirus element.Citation87
Tumor suppressor lncRNAs
MEG3
MEG3 is located on chromosome 14q32.3 in humans and has a length of 1.6 kb. Previous evidence indicates that MEG3 is a tumor suppressor because of its role in modulating angiogenesis.Citation88 MEG3 can act in both p53-dependent and p53-independent manner during different processes. However, overexpression of MEG3 decreases NSCLC cell proliferation and induces apoptosis via the activation of p53.Citation89 In addition, MEG3 expression is deregulated in NSCLC tissues, and a low expression level is significantly related with higher TNM stage, increased tumor size, and poor patient survival.Citation90,Citation91
SPRY4-IT1
SPRY4-IT1 is located on chromosome 5q31 in humans and has a length of 0.69 kb. SPRY4-IT1 is upregulated in melanoma, esophageal SCC, and clear cell renal cell carcinoma, suggesting a common oncogenic role.Citation92–Citation94 Sun et alCitation95 showed that SPRY4-IT1 is significantly downregulated in 94.2% of NSCLC cancerous tissues compared with normal tissues, which suggests an anti-oncogenic role. Ectopic expression of SPRY4-IT1 is associated with tumor size, advanced pathological stage, lymph node metastasis, and overall survival time in NSCLC patients. Reduced SPRY4-IT1 expression is an independent prognostic marker for NSCLC. EZH2, a methyltransferase and a catalytic subunit of PRC2, is overexpressed in NSCLC,Citation96 and its downregulation prevents it from binding to the SPRY4-IT1 promoter region. This decreases the H3K27me3 modification, resulting in the inhibition of SPRY4-IT1 expression. SPRY4-IT1 has also been shown to promote NSCLC cell proliferation and metastasis by modulating the process of EMT.Citation95
PANDAR
PANDAR is located on chromosome 6p21.2 in humans and has a length of 1.5 kb.Citation97 Although PANDAR is transcribed in antisense to CDKN1A, it is not a linked transcript of CDKN1A.Citation98 PANDAR is a direct transcriptional target of p53 in NSCLC cells, and can modulate Bcl-2 expression by binding to NF-YA, thus affecting NSCLC cell apoptosis.Citation97 PANDAR interacts with NF-YA (NF-YA is related to tumorigenesis) to decrease proapoptotic gene expression in a p53-dependent manner in normal human fetal lung fibroblasts. In NSCLC tissues, PANDAR interacts with NF-YA and is deregulated. PANDAR downregulation is associated with increased tumor size and advanced TNM stage. PANDAR expression is an independent prognostic predictor for NSCLC. Moreover, PANDAR, a transcriptional target of p53, affects NSCLC cell apoptosis partly by modulating Bcl-2 transcription through binding to NF-YA, thus affecting the proliferation of NSCLC cells in vitro and in vivo.Citation97
GAS5
Characteristics of GAS5
GAS5, comprising 12 exons, is located on chromosome 1q25 in humans and has a length of 0.65 kb. With respect to this locus, there are ten C/D box snoRNAs transcribed from its intronic regions. In addition, these snoRNAs have increasingly been linked to the functions of GAS5.Citation99 GAS5 plays an essential role in cell apoptosis and growth. Evidence to date indicates that GAS5 modulates the activity of glucocorticoid-responsive genes. In this process, GAS5 represents a clear example of a decoy lncRNA, which competitively binds to the glucocorticoid receptor and prevents it from binding to glucocorticoid response elements.Citation100
GAS5 in lung cancer
Multiple studies have indicated that GAS5 acts as a tumor suppressor in various cancers, such as gastric cancer,Citation101 hepatocellular cancer,Citation102 and colorectal cancer,Citation103 because of its role in the inhibition of proliferation and promotion of apoptosis. In NSCLC patient samples, decreased expression of GAS5 is linked to advanced TNM stage and increased tumor size.Citation104 Increased expression of GAS5 deregulates E2F1 and drives the expression of p21 p53 in NSCLC cells, indicating that GAS5 has a regulatory effect on NSCLC cell proliferation.Citation105 The activity of GAS5 can be regulated by miR-21, which has a putative binding site in GAS5. In turn, GAS5 suppresses miR-21 expression in a feedback loop between them.Citation106 Dong et alCitation107 showed that overexpression of GAS5 in the lung ADC A549 cell line reverts gefitinib resistance, suggesting its tumor-suppressive function. Moreover, overexpression of GAS5 reverses the resistance to EGFR-tyrosine kinase inhibitors in ADC in vitro and in vivo. Generally, increased EGFR is related to poor prognosis in NSCLC patients.Citation108 This study further indicated that GAS5 has an anti-oncogenic role.
TUG1
TUG1 is located on chromosome 22q12.2 in humans and has a length of 7.1 kb. The TUG1 gene displays a high level of conservation in the human, mouse, rat, dog, and cow genomes.Citation109 TUG1 is involved in photoreceptor development and is deregulated in many kinds of human cancers. In NSCLC, TUG1 possesses tumor suppressor features such as inhibition of cell proliferation and promotion of apoptosis. TUG1, which is induced by p53, is found binding to PRC2 and epigenetically regulates the expression of HOXB7.Citation110 Thus, it is plausible that TUG1 modulates NSCLC cell growth via the AKT and MAPK signaling pathways because HOXB7 participates in these pathways. Moreover, patients with low level of TUG1 expression display a higher TNM stage, increased tumor size, and relatively poor overall survival.Citation110 TUG1 has an oncogenic role in NSCLC, but it is a bona fide ncRNA in other cancer entities, including urothelial carcinoma of the bladderCitation111 and osteosarcoma.Citation112
BANCR
BANCR is located on chromosome 9q21.11 and has a length of 0.69 kb. BANCR is upregulated in malignant melanoma, colorectal carcinoma, and papillary thyroid carcinoma tissues, suggesting a common oncogenic role.Citation113–Citation115 However, Sun et al showed that BANCR is significantly downregulated in NSCLC cancerous tissues compared with normal tissues.Citation116 In addition, deregulated expression of BANCR is associated with increased tumor size, advanced pathological stage, lymph node metastasis, and poor survival in NSCLC patients. Reduced BANCR expression is an independent prognostic marker for NSCLC. Furthermore, knockdown of BANCR expression leads to the promotion of cell migration and invasion but inhibition of metastasis. It is plausible that downregulated BANCR promotes cell proliferation by downregulating p21 expression.Citation117 Subsequent studies further indicate that BANCR has a critical role in EMT via modulation of E-cadherin, N-cadherin, and Vimentin expression. In sum, BANCR is proposed to modulate NSCLC cell-invasive and metastatic ability partially by modulating the EMT process.Citation116
Discussion
Lung cancer is responsible for the largest number of cancer-related deaths around the world. One of the main barriers to the success of lung cancer therapy is the lack of tumor biomarkers for early diagnosis. In the previous studies, evidences have mainly focused on elucidation of lncRNAs in cellular and mouse models. As shown in , the expression of MALAT1, HOTAIR, SOX2-OT, HNF1A-AS1, ANRIL, MVIH, and PVT1 is associated positively with tumor size; reciprocally, the expression of MEG3, SPRY4-IT1, GAS5, and TUG1 is negatively correlated. Besides, the expression of MALAT1, HOTAIR, HNF1A-AS1, ANRIL, and PVT1 is associated positively with lymph node metastases, whereas the expression of SPRY4-IT1 and BANCR is negatively correlated. Indeed, to accurately and comprehensively understand the role of lncRNAs in human, large clinical cases of pathological characteristics as well as the prognosis are needed. The previous studies have indicated that overexpression of MALAT1, SOX2-OT, ANRIL, CARLO-5, MVIH, and PVT1 is a negative prognostic marker for patient survival. Indeed, the clinical integration of lncRNAs with respect to prognostic and predictive biomarker signatures will increase the therapeutic benefit. Here, we summarize the recent research and regulatory networks of lncRNAs () in lung cancer. Interestingly, the key role of PRC2 in modulating proliferation and cell cycle of lung cancer cells has been emerging. PCR2 indirectly functions as a double-edged sword through ROB1, ROCK1, and E2F1. First, MALAT1, ANAIL, SOX2-OT, and HOTAIR promote proliferation and cell cycle by regulating PRC2. TUG1 inhibits proliferation by regulating PRC2. Meanwhile, PRC2 is able to inhibit the expression of SPRY4-IT1. Several growth-related genes including p21 and p53 have been shown in the hinge of network. BANCE, GAS5, and MEG3 enhance activity of these tumor suppressor genes. CARLO-5 and HOTAIR have been proposed to highlight oncogenic feature by inhibiting the expression of p21 and p53. Generally, MMPs play a critical role in tumor cell growth and metastasis by altering the environments in which the cells grow.Citation118 We have shown that HOTAIR and MVIH possibly regulate proliferation and metastatic ability of lung cancer cells. Noteworthy is that the altering expression of E-cadherin, Vimentin, and N-cadherin is a fundamental event in EMT.Citation119 It is plausible that MALAT1, BANCE, SPRY4-IT1, and H19 act in concert with them to regulate the progress of EMT during lung tumorigenesis. Additionally, miRNAs as high-potential biomarkers have critical position in regulatory network of lung cancer; for example, miR-196a represses HOX5 expression, thereby promoting metastasis. It is very likely that lncRNAs harbor miRNA seed regions and enrich target RNA-binding motifs. Thus, miRNAs are proposed to modulate lung tumorigenesis by lncRNAs.
Although an impressive number of studies in the last decade focused on the characteristics and functions of ncRNAs, research is still in its infancy and presents great challenges. First, as the sequence and structure of lncRNAs are only poorly conserved, the canonical knockdown and knockout methods may have no effect. In addition, ectopic expression of lncRNAs may not show obvious phenotypes as with protein-coding transcripts. Second, reference value is not always high between different researches, owing to different and multiple functions of lncRNAs in different tissues and cells. Therefore, elucidating the biological functions of lncRNAs is not easy. Third, the limited bioinformatical resources are another reason. Current lncRNA annotation is lacking compared with other RNA databases. Similarly, bioinformatic tools, such as lncRNA secondary structure prediction, remain to be developed. Unraveling the functions and regulatory mechanisms of lncRNAs in lung cancer might be a future breakthrough to improve our understanding of this network. The integration of miRNA and lncRNA signature profiling in lung cancer may be a useful tool for clinical applications.
Conclusion
lncRNAs are increasingly being recognized as critical molecules in various biological processes. In addition to the various types, there is a large number of lncRNAs, and they show numerous modes of interaction. Based on the location concerning the nearest protein-coding gene, lncRNAs can be classified into four subclasses, namely exonic, intronic, overlapping, and intergenic lncRNAs. According to their function, lncRNAs can be categorized as signal, decoy, sponge, guide, and scaffold molecules. It has become increasingly clear that lncRNAs are involved in tumorigenesis in many cancers. This network should serve as a guide for “navigating” through the lncRNAs research in the literature.
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (Nos 30871189, 81171841, 81200366, 81372515, 81401901, and 81572281).
Disclosure
The authors report no conflicts of interest in this work.
References
- WuYLiuHShiXYaoYYangWSongYThe long non-coding RNA HNF1A-AS1 regulates proliferation and metastasis in lung adenocarcinomaOncotarget20156119160917225863539
- ChenZFillmoreCMHammermanPSKimCFWongKKNon-small-cell lung cancers: a heterogeneous set of diseasesNat Rev Cancer201414853554625056707
- DavidsonMRGazdarAFClarkeBEThe pivotal role of pathology in the management of lung cancerJ Thorac Dis20135Suppl 5S463S47824163740
- LangerCJBesseBGualbertoABrambillaESoriaJCThe evolving role of histology in the management of advanced non-small-cell lung cancerJ Clin Oncol201028365311532021079145
- Cancer Genome Atlas Research NetworkComprehensive genomic characterization of squamous cell lung cancersNature2012489741751952522960745
- IyerMKNiknafsYSMalikRThe landscape of long noncoding RNAs in the human transcriptomeNat Genet201547319920825599403
- DerrienTJohnsonRBussottiGThe GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expressionGenome Res20122291775178922955988
- KapustaAKronenbergZLynchVJTransposable elements are major contributors to the origin, diversification, and regulation of vertebrate long noncoding RNAsPLoS Genet201394e100347023637635
- JohnsonRGuigoRThe RIDL hypothesis: transposable elements as functional domains of long noncoding RNAsRNA201420795997624850885
- GuttmanMAmitIGarberMChromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammalsNature2009458723522322719182780
- DiederichsSThe four dimensions of noncoding RNA conservationTrends Genet201430412112324613441
- GutschnerTHammerleMEissmannMThe noncoding RNA MALAT1 is a critical regulator of the metastasis phenotype of lung cancer cellsCancer Res20137331180118923243023
- ChuCQuKZhongFLArtandiSEChangHYGenomic maps of long noncoding RNA occupancy reveal principles of RNA-chromatin interactionsMol Cell201144466767821963238
- ShiXSunMLiuHYaoYSongYLong non-coding RNAs: a new frontier in the study of human diseasesCancer Lett2013339215916623791884
- WuLMuratPMatak-VinkovicDMurrellABalasubramanianSBinding interactions between long noncoding RNA HOTAIR and PRC2 proteinsBiochemistry201352529519952724320048
- BetancurJGTomariYCryptic RNA-binding by PRC2 components EZH2 and SUZ12RNA Biol201512995996526177152
- KhalilAMGuttmanMHuarteMMany human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expressionProc Natl Acad Sci U S A200910628116671167219571010
- PandeyRRMondalTMohammadFKcnq1ot1 antisense non-coding RNA mediates lineage-specific transcriptional silencing through chromatin-level regulationMol Cell200832223224618951091
- WangKCChangHYMolecular mechanisms of long noncoding RNAsMol Cell201143690491421925379
- HuarteMGuttmanMFeldserDA large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 responseCell2010142340941920673990
- PolisenoLSalmenaLZhangJCarverBHavemanWJPandolfiPPA coding-independent function of gene and pseudogene mRNAs regulates tumour biologyNature201046573011033103820577206
- HuYWangJQianJLong noncoding RNA GAPLINC regulates CD44-dependent cell invasiveness and associates with poor prognosis of gastric cancerCancer Res201474236890690225277524
- WangHZhangXLiuYDownregulated miR-31 level associates with poor prognosis of gastric cancer and its restoration suppresses tumor cell malignant phenotypes by inhibiting E2F2Oncotarget201653657736589
- HansenTBJensenTIClausenBHNatural RNA circles function as efficient microRNA spongesNature2013495744138438823446346
- LoewenGJayawickramarajahJZhuoYShanBFunctions of lncRNA HOTAIR in lung cancerJ Hematol Oncol2014719025491133
- LiWNotaniDMaQFunctional roles of enhancer RNAs for oestrogen-dependent transcriptional activationNature2013498745551652023728302
- ZhaoJSunBKErwinJASongJJLeeJTPolycomb proteins targeted by a short repeat RNA to the mouse X chromosomeScience2008322590275075618974356
- BaoXWuHZhuXThe p53-induced lincRNA-p21 derails somatic cell reprogramming by sustaining H3K9me3 and CpG methylation at pluripotency gene promotersCell Res2015251809225512341
- YangLLinCJinClncRNA-dependent mechanisms of androgen-receptor-regulated gene activation programsNature2013500746459860223945587
- PanQShaiOLeeLJFreyBJBlencoweBJDeep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencingNat Genet200840121413141518978789
- BarashYCalarcoJAGaoWDeciphering the splicing codeNature20104657294535920445623
- Saint-AndreVBatscheERachezCMuchardtCHistone H3 lysine 9 trimethylation and HP1gamma favor inclusion of alternative exonsNat Struct Mol Biol201118333734421358630
- GonzalezIMunitaRAgirreEA lncRNA regulates alternative splicing via establishment of a splicing-specific chromatin signatureNat Struct Mol Biol201522537037625849144
- TripathiVEllisJDShenZThe nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylationMol Cell201039692593820797886
- JiPDiederichsSWangWMALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancerOncogene200322398031804112970751
- WiluszJEJnBaptisteCKLuLYA triple helix stabilizes the 3′ ends of long noncoding RNAs that lack poly(A) tailsGenes Dev201226212392240723073843
- BrownJAValensteinMLYarioTATycowskiKTSteitzJAFormation of triple-helical structures by the 3′-end sequences of MALAT1 and MENbeta noncoding RNAsProc Natl Acad Sci U S A201210947192021920723129630
- FriedelCCDolkenLRuzsicsZKoszinowskiUHZimmerRConserved principles of mammalian transcriptional regulation revealed by RNA half-lifeNucleic Acids Res20093717e11519561200
- WiluszJEFreierSMSpectorDL3′ end processing of a long nuclear-retained noncoding RNA yields a tRNA-like cytoplasmic RNACell2008135591993219041754
- BernsteinEDuncanEMMasuiOGilJHeardEAllisCDMouse polycomb proteins bind differentially to methylated histone H3 and RNA and are enriched in facultative heterochromatinMol Cell Biol20062672560256916537902
- HutchinsonJNEnsmingerAWClemsonCMLynchCRLawrenceJBChessAA screen for nuclear transcripts identifies two linked noncoding RNAs associated with SC35 splicing domainsBMC Genomics200783917270048
- YoshimotoRMayedaAYoshidaMMALAT1 long non-coding RNA in cancerBiochimica et biophysica acta20161859119219926434412
- GutschnerTBaasMDiederichsSNoncoding RNA gene silencing through genomic integration of RNA destabilizing elements using zinc finger nucleasesGenome Res201121111944195421844124
- GutschnerTHammerleMDiederichsSMALAT1 – a paradigm for long noncoding RNA function in cancerJ Mol Med (Berl)201391779180123529762
- TanoKMizunoROkadaTMALAT-1 enhances cell motility of lung adenocarcinoma cells by influencing the expression of motility-related genesFEBS Lett2010584224575458020937273
- ShenLChenLWangYJiangXXiaHZhuangZLong non-coding RNA MALAT1 promotes brain metastasis by inducing epithelial-mesenchymal transition in lung cancerJ Neurooncol2015121110110825217850
- YangFYiFHanXDuQLiangZMALAT-1 interacts with hnRNP C in cell cycle regulationFEBS Lett2013587193175318123973260
- SchmidtLHGorlichDSpiekerTPrognostic impact of Bcl-2 depends on tumor histology and expression of MALAT-1 lncRNA in non-small-cell lung cancerJ Thorac Oncol2014991294130425036876
- ZhangJZhangBWangTWangHLncRNA MALAT1 overexpression is an unfavorable prognostic factor in human cancer: evidence from a meta-analysisInt J Clin Exp Med2015845499550526131129
- FaticaABozzoniILong non-coding RNAs: new players in cell differentiation and developmentNat Rev Genet201315172124296535
- SomarowthuSLegiewiczMChillonIMarciaMLiuFPyleAMHOTAIR forms an intricate and modular secondary structureMol Cell201558235336125866246
- BhanAMandalSSLncRNA HOTAIR: A master regulator of chromatin dynamics and cancerBiochim Biophys Acta20151856115116426208723
- WuYZhangLZhangLLong non-coding RNA HOTAIR promotes tumor cell invasion and metastasis by recruiting EZH2 and repressing E-cadherin in oral squamous cell carcinomaInt J Oncol20154662586259425901533
- TsaiMCManorOWanYLong noncoding RNA as modular scaffold of histone modification complexesScience2010329599268969320616235
- HangHXingZManiSKKRNA helicase DDX5 regulates PRC2/HOTAIR function in Hepatitis B Virus infection and hepatocarcinogenesisHepatology201615327331377
- ChiyomaruTYamamuraSFukuharaSGenistein inhibits prostate cancer cell growth by targeting miR-34a and oncogenic HOTAIRPLoS One201388e7037223936419
- YoonJHAbdelmohsenKKimJScaffold function of long non-coding RNA HOTAIR in protein ubiquitinationNat Commun20134293924326307
- ZhaoWAnYLiangYXieXWRole of HOTAIR long noncoding RNA in metastatic progression of lung cancerEur Rev Med Pharmacol Sci201418131930193625010625
- OnoHMotoiNNaganoHLong noncoding RNA HOTAIR is relevant to cellular proliferation, invasiveness, and clinical relapse in small-cell lung cancerCancer Med20143363264224591352
- NakagawaTEndoHYokoyamaMLarge noncoding RNA HOTAIR enhances aggressive biological behavior and is associated with short disease-free survival in human non-small cell lung cancerBiochem Biophys Res Commun2013436231932423743197
- LiuXHLuKHWangKMMicroRNA-196a promotes non-small cell lung cancer cell proliferation and invasion through targeting HOXA5BMC Cancer20121234822876840
- ZhuangYWangXNguyenHTInduction of long intergenic non-coding RNA HOTAIR in lung cancer cells by type I collagenJ Hematol Oncol201363523668363
- LiuZSunMLuKThe long noncoding RNA HOTAIR contributes to cisplatin resistance of human lung adenocarcinoma cells via downregualtion of p21 (WAF1/CIP1) expressionPLoS One2013810e7729324155936
- ZhouCYeLJiangCBaiJChiYZhangHLong noncoding RNA HOTAIR, a hypoxia-inducible factor-1alpha activated driver of malignancy, enhances hypoxic cancer cell proliferation, migration, and invasion in non-small cell lung cancerTumour Biol201536129179918826088446
- ShahryariAJaziMSSamaeiNMMowlaSJLong non-coding RNA SOX2OT: expression signature, splicing patterns, and emerging roles in pluripotency and tumorigenesisFront Genet2015619626136768
- HouZZhaoWZhouJA long noncoding RNA Sox2ot regulates lung cancer cell proliferation and is a prognostic indicator of poor survivalInt J Biochem Cell Biol20145338038824927902
- AmaralPPNeytCWilkinsSJComplex architecture and regulated expression of the Sox2ot locus during vertebrate developmentRNA200915112013202719767420
- BassAJWatanabeHMermelCHSOX2 is an amplified lineage-survival oncogene in lung and esophageal squamous cell carcinomasNat Genet200941111238124219801978
- PasmantELaurendeauIHeronDVidaudMVidaudDBiècheICharacterization of a germ-line deletion, including the entire INK4/ARF locus, in a melanoma-neural system tumor family: identification of ANRIL, an antisense noncoding RNA whose expression coclusters with ARFCancer Res20076783963396917440112
- NieFQSunMYangJSLong noncoding RNA ANRIL promotes non-small cell lung cancer cell proliferation and inhibits apoptosis by silencing KLF2 and P21 expressionMol Cancer Ther201514126827725504755
- Fernandez-ZapicoMELomberkGATsujiSA functional family-wide screening of SP/KLF proteins identifies a subset of suppressors of KRAS-mediated cell growthBiochem J2011435252953721171965
- ZhangEBKongRYinDDLong noncoding RNA ANRIL indicates a poor prognosis of gastric cancer and promotes tumor growth by epigenetically silencing of miR-99a/miR-449aOncotarget2014582276229224810364
- RenXSYinMHZhangXTumor-suppressive microRNA-449a induces growth arrest and senescence by targeting E2F3 in human lung cancer cellsCancer Lett2014344219520324211326
- OnyangoPFeinbergAPA nucleolar protein, H19 opposite tumor suppressor (HOTS), is a tumor growth inhibitor encoded by a human imprinted H19 antisense transcriptProc Natl Acad Sci U S A201110840167591676421940503
- MatoukIRavehEOhanaPThe increasing complexity of the oncofetal h19 gene locus: functional dissection and therapeutic interventionInt J Mol Sci20131424298431623429271
- SchoenherrCJLevorseJMTilghmanSMCTCF maintains differential methylation at the Igf2/H19 locusNat Genet2003331666912461525
- KallenANZhouXBXuJThe imprinted H19 lncRNA antagonizes let-7 microRNAsMol Cell201352110111224055342
- MatoukIJHalleDGilonMHochbergAThe non-coding RNAs of the H19-IGF2 imprinted loci: a focus on biological roles and therapeutic potential in lung cancerJ Transl Med20151311325884481
- MatoukIJRavehEAbu-lailROncofetal H19 RNA promotes tumor metastasisBiochim Biophys Acta2014184371414142624703882
- MatoukIJDeGrootNMezanSThe H19 non-coding RNA is essential for human tumor growthPLoS One200729e84517786216
- KaplanRLuettichKHeguyAHackettNRHarveyBGCrystalRGMonoallelic up-regulation of the imprinted H19 gene in airway epithelium of phenotypically normal cigarette smokersCancer Res20036371475148212670893
- LuoJTangLZhangJLong non-coding RNA CARLo-5 is a negative prognostic factor and exhibits tumor pro-oncogenic activity in non-small cell lung cancerTumour Biol20143511115411154925129441
- KimTCuiRJeonYJLong-range interaction and correlation between MYC enhancer and oncogenic long noncoding RNA CARLo-5Proc Natl Acad Sci U S A2014111114173417824594601
- NieFQZhuQXuTPLong non-coding RNA MVIH indicates a poor prognosis for non-small cell lung cancer and promotes cell proliferation and invasionTumour Biol20143587587759424793017
- BarsottiAMBeckermanRLaptenkoOHuppiKCaplenNJPrivesCp53-Dependent induction of PVT1 and miR-1204J Biol Chem201228742509251922110125
- YangYRZangSZZhongCLLiYXZhaoSSFengXJIncreased expression of the lncRNA PVT1 promotes tumorigenesis in non-small cell lung cancerInt J Clin Exp Pathol20147106929693525400777
- GibbEAWarrenRLWilsonGWActivation of an endogenous retrovirus-associated long non-coding RNA in human adenocarcinomaGenome Med2015712225821520
- HanahanDWeinbergRAHallmarks of cancer: the next generationCell2011144564667421376230
- LuKHLiWLiuXHLong non-coding RNA MEG3 inhibits NSCLC cells proliferation and induces apoptosis by affecting p53 expressionBMC Cancer20131346124098911
- ZhouYZhongYWangYActivation of p53 by MEG3 non-coding RNAJ Biol Chem200728234247312474217569660
- ZhangXRiceKWangYMaternally expressed gene 3 (MEG3) noncoding ribonucleic acid: isoform structure, expression, and functionsEndocrinology2010151393994720032057
- KhaitanDDingerMEMazarJThe melanoma-upregulated long noncoding RNA SPRY4-IT1 modulates apoptosis and invasionCancer Res201171113852386221558391
- XieHWWuQQZhuBLong noncoding RNA SPRY4-IT1 is upregulated in esophageal squamous cell carcinoma and associated with poor prognosisTumour Biol20143587743775424810925
- ZhangHMYangFQYanYCheJPZhengJHHigh expression of long non-coding RNA SPRY4-IT1 predicts poor prognosis of clear cell renal cell carcinomaInt J Clin Exp Pathol2014795801580925337221
- SunMLiuXHLuKHEZH2-mediated epigenetic suppression of long noncoding RNA SPRY4-IT1 promotes NSCLC cell proliferation and metastasis by affecting the epithelial-mesenchymal transitionCell Death Dis20145e129824967960
- BehrensCSolisLMLinHEZH2 protein expression associates with the early pathogenesis, tumor progression, and prognosis of non-small cell lung carcinomaClin Cancer Res201319236556656524097870
- HanLZhangEBYinDDLow expression of long noncoding RNA PANDAR predicts a poor prognosis of non-small cell lung cancer and affects cell apoptosis by regulating Bcl-2Cell Death Dis20156e166525719249
- HungTWangYLinMFExtensive and coordinated transcription of noncoding RNAs within cell-cycle promotersNat Genet201143762162921642992
- HiroseTSteitzJAPosition within the host intron is critical for efficient processing of box C/D snoRNAs in mammalian cellsProc Natl Acad Sci U S A20019823129141291911606788
- KinoTHurtDEIchijoTNaderNChrousosGPNoncoding RNA gas5 is a growth arrest- and starvation-associated repressor of the glucocorticoid receptorSci Signal20103107a8
- SunMJinFYXiaRDecreased expression of long noncoding RNA GAS5 indicates a poor prognosis and promotes cell proliferation in gastric cancerBMC Cancer20141431924884417
- TuZQLiRJMeiJZLiXHDown-regulation of long non-coding RNA GAS5 is associated with the prognosis of hepatocellular carcinomaInt J Clin Exp Pathol2014774303430925120813
- YinDHeXZhangEKongRDeWZhangZLong noncoding RNA GAS5 affects cell proliferation and predicts a poor prognosis in patients with colorectal cancerMed Oncol2014311125325326054
- ShiXSunMLiuHA critical role for the long non-coding RNA GAS5 in proliferation and apoptosis in non-small-cell lung cancerMol Carcinog201554Suppl 1E1E1224357161
- YuXLiZLong non-coding RNA growth arrest-specific transcript 5 in tumor biologyOncol Lett20151041953195826622780
- ZhangZZhuZWatabeKNegative regulation of lncRNA GAS5 by miR-21Cell Death Differ201320111558156823933812
- DongSQuXLiWThe long non-coding RNA, GAS5, enhances gefitinib-induced cell death in innate EGFR tyrosine kinase inhibitor-resistant lung adenocarcinoma cells with wide-type EGFR via downregulation of the IGF-1R expressionJ Hematol Oncol201584325925741
- SelvaggiGNovelloSTorriVEpidermal growth factor receptor overexpression correlates with a poor prognosis in completely resected non-small-cell lung cancerAnn Oncol2004151283214679115
- YoungTLMatsudaTCepkoCLThe noncoding RNA taurine upregulated gene 1 is required for differentiation of the murine retinaCurr Biol200515650151215797018
- ZhangEBYinDDSunMP53-regulated long non-coding RNA TUG1 affects cell proliferation in human non-small cell lung cancer, partly through epigenetically regulating HOXB7 expressionCell Death Dis20145e124324853421
- HanYLiuYGuiYCaiZLong intergenic non-coding RNA TUG1 is overexpressed in urothelial carcinoma of the bladderJ Surg Oncol2013107555555922961206
- ZhangQGengPLYinPWangXLJiaJPYaoJDown-regulation of long non-coding RNA TUG1 inhibits osteosarcoma cell proliferation and promotes apoptosisAsian Pac J Cancer Prev20131442311231523725133
- WangYChenWChenJPanQPanJLncRNA expression EGFR exon 19 deletions in lung adenocarcinoma ascertained by using microarray analysisMed Oncol201431913725085781
- GuoQZhaoYChenJBRAF-activated long non-coding RNA contributes to colorectal cancer migration by inducing epithelial-mesenchymal transitionOncol Lett20148286987525013510
- FlockhartRJWebsterDEQuKBRAFV600E remodels the melanocyte transcriptome and induces BANCR to regulate melanoma cell migrationGenome Res20122261006101422581800
- SunMLiuXHWangKMDownregulation of BRAF activated non-coding RNA is associated with poor prognosis for non-small cell lung cancer and promotes metastasis by affecting epithelial-mesenchymal transitionMol Cancer2014136824655544
- ShiYLiuYWangJDownregulated long noncoding RNA BANCR promotes the proliferation of colorectal cancer cells via downregulation of p21 expressionPLoS One2015104e122679
- van KempenLCLCoussensLMMMP9 potentiates pulmonary metastasis formationCancer Cell20022425125212398887
- SeanoGChiaverinaGGagliardiPAEndothelial podosome rosettes regulate vascular branching in tumour angiogenesisNat Cell Biol201416109319411825218639
