Abstract
Background
Non-small-cell lung cancer (NSCLC) patients with epidermal growth factor receptor (EGFR) mutations might develop primary and secondary resistance to tyrosine kinase inhibitors (TKIs). The proapoptotic protein Bcl-2-like 11 (BIM) is a key modulator of apoptosis triggered by EGFR-TKIs. The recent studies have indicated that some patients with positive EGFR mutations were refractory to EGFR-TKIs if they harbored a BIM deletion polymorphism. The purpose of this study was to investigate whether BIM polymorphism predicts treatment efficacy of EGFR-TKIs in Chinese NSCLC patients.
Patients and methods
A cohort of advanced NSCLC patients with EGFR mutations and treated with EGFR-TKIs (gefitinib or erlotinib) were recruited. We drew peripheral blood to determinate BIM deletion status and then compared patients’ clinical outcomes according to the BIM deletion status. Additionally, we electronically searched eligible cohort studies and conducted a meta-analysis to pool event risk.
Results
The exploratory cohort study included 140 patients. Patients with and without the BIM deletion polymorphism had similar objective response rates (ORRs, 48.5 vs 63.0%, P=0.16), disease control rate (DCR, 93.9 vs 97.0%, P=0.60) and adverse reactions. Similar progression-free survival (PFS) and overall survival (OS) were noted in overall population (P=0.27 for PFS and P=0.61 for OS) and prespecified patient subgroups. The meta-analysis included 10 eligible cohort studies involving 1,317 NSCLC patients. It showed the positive BIM deletion was associated with shorter PFS (hazard ratio =1.45; P=0.02). Nonsignificant differences existed for ORR, DCR and OS.
Conclusion
The expanded meta-analysis results demonstrated the positive BIM deletion predicts shorter PFS in NSCLC patients after treatment with EGFR-TKIs while other clinical measures do not. A large multicenter well-designed cohort study involving other concurrent genetic alterations is warranted.
Keywords:
Introduction
Non-small-cell lung cancer (NSCLC) is the leading cause of cancer-related death.Citation1 Like other cancers, NSCLC develops when cells initiate to uncontrollably drive mutations due to changes in their genes. Using targeted therapies could specifically attack these changes and block the growth of cancer cells without damaging the normal cells like cytotoxic chemotherapy.Citation2 The representative targeted therapy – tyrosine kinase inhibitor (TKI), which targets mutated epidermal growth factor receptors (EGFRs) – has turned into a better alternative for treating advanced NSCLC.Citation3 Consequently, it has surprisingly changed the treatment of advanced NSCLC.Citation4–Citation6 NSCLC patients with EGFR mutations who receive first-line therapy with an EGFR-TKI, such as gefitinib or erlotinib, have longer progression-free survival (PFS) than those who are treated with platinum-based chemotherapy.Citation4,Citation7–Citation9 However, about 30% of these patients show primary resistance to EGFR-TKIs even if they have EGFR mutations; meanwhile, most patients who respond initially might acquire drug resistance after approximately 1 year of treatment.Citation4,Citation5,Citation9–Citation11 Mechanisms of acquired resistance to EGFR-TKI include T790M secondary mutation, or subsequently C797S mutation responsible for resistance to T790M-targeting EGFR inhibitors, and MET amplification.Citation12–Citation14
BIM, also known as Bcl-2-like 11 (BCL2L11), is a member of the Bcl-2 family of genes and encodes the protein BIM. By binding to all members of the prosurvival Bcl-2 subfamily with high affinity, BIM serves as a key element in promoting apoptosis. BIM deletion polymorphism is a 2,903-bp deletion located in exon 2 of the BCL2L11 gene that leads to alternative splicing of the mRNA of BIM, which results in expression of BIM isoforms lacking the pro-apoptotic BCL2-homology domain 3 (BH3).Citation15 It is hypothesized that BIM might be involved in the apoptotic signaling following EGFR disruption by TKIs.Citation2 The intrinsic resistance and incomplete response may be due, in part, to downregulation of BIM expression.Citation11,Citation12 A recent study has suggested that the BIM germline alteration would prevent apoptosis induced by EGFR-TKIs, which poses a potential mechanism conferring resistance.Citation16 Another study has showed that BIM deletion polymorphism is associated with primary drug resistance to EGFR-TKIs.Citation17 As shown by induction of apoptosis, the EGFR-mutant NSCLC cells with the BIM deletion polymorphism are much less sensitive to gefitinib than those with wild-type BIM.Citation15 Thus, therapies that upregulate BIM expression, such as histone deacetylase inhibitor, vorinostat, may resensitize some low BIM-expressing oncogene-addicted cancers to targeted therapies.Citation17
Given that EGFR-mutated lung tumors occur more frequently in East Asians and the BIM polymorphism is also prevalent in East Asian population and seldom found in Caucasian counterparts,Citation16 we carried out this exploratory cohort study in the People’s Republic of China to investigate the predictive role of BIM deletion polymorphism in advanced EGFR-mutant NSCLC patients treated with EGFR-TKIs. Besides, we sought to perform a meta-analysis incorporating all currently available evidences from cohort studies to compare the clinical outcomes according to the BIM polymorphism status in NSCLC patients with EGFR mutations after the treatment with EGFR-TKIs.
Patients and methods
Patients
In this exploratory cohort study, a total of 140 NSCLC patients harboring EGFR mutation who were treated with EGFR-TKIs were recruited from June 2009 through May 2013. This study was approved by the Ethics Committees of Shanghai Cancer Center, Fudan University, and was carried out in accordance with the World Medical Association’s Declaration of Helsinki (1964) and its later amendments. Informed consent was obtained from each participating patient before any study-related procedure was performed.
Patients received either oral gefitinib (250 mg per day) or oral erlotinib (150 mg per day). Every 2 months, patients were assessed for response using Response Evaluation Criteria in Solid Tumors (RECIST) version 1.1.Citation18 According to this criteria, overall response rate (ORR) was defined as the proportion of patients who had complete response and partial response, while disease control rate (DCR) was defined as the proportion of patients who had a best response rating of complete response, partial response or stable disease. PFS was calculated from the date EGFR-TKIs therapy was initiated to the date of either tumor progression or death from any cause. Overall survival (OS) was defined as the time from the initiation of EGFR-TKIs therapy to death from any cause. Adverse events related to EGFR-TKIs treatment were evaluated using the National Cancer Institute Common Terminology Criteria for Adverse Events (NCI CTCAE) Version 4.0 (2009).
EGFR mutations and BIM deletion polymorphism
We used direct sequencing to determinate EGFR (exons 18–21) mutations in polymerase chain reaction (PCR) fragments amplified with genomic DNA from formalin-fixed paraffin-embedded tissue.Citation19,Citation20 BIM deletion polymorphism analysis (the presence of wild-type or deletion alleles) was performed on genomic DNA extracted (QIAamp DNA blood mini kit; Qiagen NV, Venlo, the Netherlands) from peripheral blood samples using PCR amplification and agarose gel electrophoresis. The primer sequences were as follows: wild-type BIM forward primer, 5′-ACTGTAAAACGACGGCCAGTCCTCATGATGAAGGCTAACTCAA-3′; and reverse primer, 5′-ACCAGGAAACAGCTATGACCAACCTCTGACAAGTGACCACCA-3′. For the BIM deletion polymorphism, the forward primer sequence was the same as that used for wild-type BIM, and the reverse sequence was 5′-ACCAGGAAACAGCTATGACCGGCACAGCCTCTATGGAGAACA-3′. The PCR conditions were 95°C for 3 minutes, and then 40 cycles of 94°C for 30 seconds, 58°C for 30 seconds and 72°C for 30 seconds. The final elongation step was performed at 72°C for 5 minutes. The PCR products were subjected to electrophoresis in 2% agarose gel stained with ethidium bromide and visualized using an ultraviolet illuminator.
Statistical analysis
R version 3.1.2 and SAS®9.2 software were used for all statistical analyses, including those in the meta-analysis. Two-sided P-values of less than 0.05 were considered statistically significant.
In the exploratory study, demographic and clinicopathological characteristics and adverse reactions were summarized by BIM deletion polymorphism status using descriptive statistics. ORR and DCR between patients with and without BIM deletion polymorphism were compared using Pearson’s Chi-square test. Survival curves were drawn by the Kaplan–Meier method, and statistical test was performed using log-rank test. To calculate hazard ratios (HRs) and 95% confidence intervals (CIs), Cox regression analysis was applied among both overall population and those prespecified subgroups according to the following prognostic factors: age, gender, smoking status, type of EGFR mutation, chemotherapy history and EGFR-TKIs treatment line.
In the meta-analysis, risk ratios (RRs) for binary data (ORR and DCR) as well as HRs for survival time (PFS and OS) were pooled along with 95% CIs using fixed-effect model and additionally displayed using forest plots. Statistical heterogeneity was considered significant when P-value was less than 0.10 for the Q-test. Publication bias was evaluated using funnel plot and Begg’s and Egger’s tests.Citation21,Citation22
Results
Demographic and clinicopathological characteristics
The relevant characteristics of the study patients at the initiation of EGFR-TKI treatment are summarized in . The median age of all the included patients was 58.5 years, 94 (67.1%) patients were female and 117 (83.6%) did not report family history of lung cancer. Approximately three-fourths of patients were nonsmokers and could not undergo radical surgery as well. These patients had previously received a median number of four treatment cycles. The vast majority of patients had an Eastern Cooperative Oncology Group (ECOG) performance status of 1 (86.4%) and the pathological diagnosis of adenocarcinoma (91.4%). In addition, there were 54 (38.6%) patients receiving EGFR-TKIs as first-line treatment. The most common EGFR mutation was seen in exon 19, accounting for 52.1% of mutations, followed by exon 21 mutation (42.1%). In this cohort, 37 (26.4%) patients were identified with heterozygous (eg, positive) BIM deletion polymorphism.
Table 1 Demographic and clinicopathological characteristics of the patients included in cohort study
Clinical responses and survival
We analyzed the association between the BIM deletion polymorphism status and clinical outcomes. In total, 133 patients were eligible for response assessment. The ORR and DCR in patients with heterozygous BIM deletion and treated with an EGFR-TKI were 48.5% (95% CI: 30.8%–66.5%) and 93.9% (95% CI: 79.8%–99.3%), respectively, which were not significantly different from those (63.0% [95% CI: 52.8%–72.4%] and 97.0% [95% CI: 91.5%–99.4%], respectively) observed in patients without the BIM deletion (P=0.16 and P=0.60, respectively) ().
Table 2 Clinical response and adverse reactions after EGFR-TKIs therapy in cohort study
The median follow-up duration was 29 months (range 2–61) for the entire patient cohort. At the time of the data analysis, 125 patients developed disease progression, including 32 (86.5%) in the heterozygous BIM deletion group and 93 (90.3%) in the wild-type group. The median PFS was 21 months (95% CI: 12–22) for patients with heterozygous BIM deletion polymorphism and 17 months (95% CI: 12–19) for the wild-type population. The Kaplan–Meier curve for PFS showed no significant difference between the heterozygous and wild type population after EGFR-TKIs therapy (P=0.27; ). The possible predictive factors of EGFR-TKIs treatment efficacy in terms of PFS were further investigated using prespecified subgroups (). Each subgroup analysis showed patients with or without the deletion polymorphism did not differ on PFS. Seventy-eight patients (55.7%) died, including 24 (64.9%) in the heterozygous BIM deletion group and 54 (52.4%) in the wild-type group. The median OS was 34 months for patients with the BIM deletion and 33 months for those without the BIM deletion (P=0.61; ). The median OS was also not significantly different between the heterozygous BIM and wild-type groups (P=0.61; ). Furthermore, no significant differences in OS were found between patients with or without the deletion polymorphism with respect to selected patient subgroups ().
Figure 1 Kaplan–Meier curves for (A) progression-free survival and (B) overall survival according to BIM deletion status.
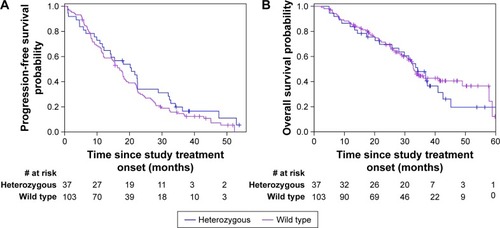
Table 3 Progression-free survival analysis in patient subgroups according to BIM deletion status
Table 4 Overall survival analysis in patient subgroups according to BIM deletion status
Adverse reactions
The study patients after EGFR-TKI treatment had similar adverse reactions of all types regardless of their BIM deletion polymorphism status (48.6% [heterozygous] vs 53.4% [wild type]). Rash and diarrhea were the most reported adverse reactions ().
Meta-analysis of BIM deletion status and clinical outcomes
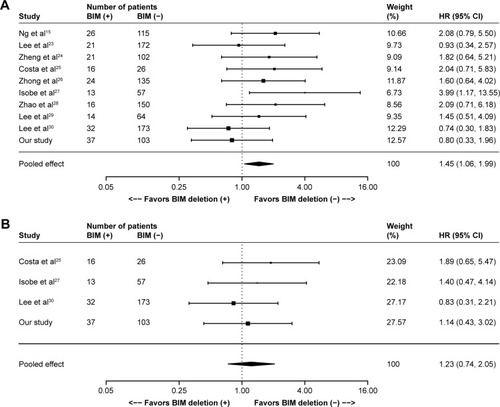
One hundred sixty-nine records were identified in PubMed (from 1965 to November 2015), Embase (from 1965 to November 2015) and Cochrane Library databases according to the search strategy that used key words associated with “Lung cancer”, “BIM or (BCL2L11 deletion) or (Bcl-2-like protein 11 deletion)” and “EGFR-mutant or (epidermal growth factor receptor mutation) or EGFR” without language limit. Finally, nine eligible previous cohort studies,Citation15,Citation23–Citation30 together with our present cohort study, were included for the meta-analysis, which involved a total of 1,317 NSCLC patients with EGFR mutations that referred to the efficacy of EGFR-TKIs (gefitinib, erlotinib or afatinib) stratified by BIM polymorphism status. The flow chart of study selection is summarized in Figure S1, and the characteristics of all the studies included in the meta-analysis are presented in . All of the ten studies presented HR of PFS data for pooling; nonetheless, data of ORR, DCR and OS were not available in several distinct studies, and so they were excluded from their respective pooling. Study quality was assessed by using the Newcastle–Ottawa Scale.Citation31 In general, the overall quality of included cohort studies could be rated as good (data not shown). Funnel plots (Figure S2), Egger’s tests and Begg’s test with regard to PFS indicated potential publication bias (Egger’s P=0.02; Begg’s P=0.02). Given the absence of heterogeneity (Q [df=9] =8.78; P=0.46) in the ten included studies, the results of fixed-effects models were used to draw study conclusions.
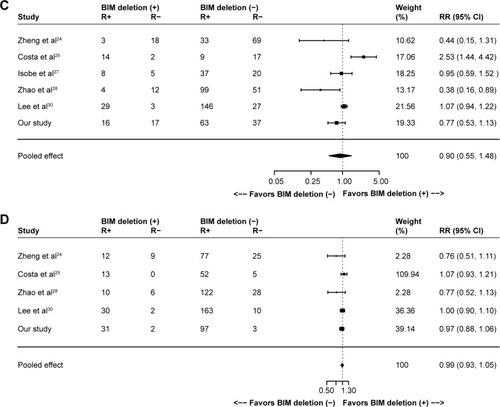
In the ten included studies, the positive BIM polymorphism did not show a completely consistent effect on PFS (). With a large sample size after pooling, however, a significant difference was then found between patients with or without the deletion polymorphism (HR =1.45, 95% CI: 1.06–1.99; P=0.02; ). However, such difference was not observed in terms of OS (HR =1.23, 95% CI: 0.74–2.05; P=0.43; ), ORR (RR =0.90, 95% CI: 0.55–1.48; P=0.69; ) and DCR (RR =0.99, 95% CI: 0.93–1.05; P=0.64; ).
Figure 2 Meta-analyses of (A) PFS, (B) OS, (C) ORR and (D) DCR according to BIM deletion status in EGFR-mutant non-small-cell lung cancer patients receiving EGFR-TKIs. (C and D) R+ represents responders and R- represents nonresponders.
Discussion
To the best of our knowledge, the predictive role of BIM deletion polymorphism in efficacy of EGFR-TKIs among NSCLC patients with EGFR mutations remains elusive. BIM deletion polymorphism is only found in East Asian descent.Citation15 A recent study randomly selected a wide range of 6,858 participants and used real-time PCR assay with high-resolution melting to detect BIM and EGFR mutation. The results showed that there were four outcomes of BIM: non-detection of 2,903 bp BIM (NA), non-deletion of 2,903 bp BIM (homozygous non-deletion-type DNA, II), 2,903 bp deletion BIM (homozygous deletion-type DNA, DD) and heterozygote (ID).Citation32 We conducted our present study in the People’s Republic of China to investigate whether the BIM polymorphism status would affect clinical efficacy of EFGR-TKIs and prognosis of NSCLC patients with EGFR mutations treated with EFGR-TKIs. Furthermore, we included all of the eligible cohort studies or appropriate subgroups of cohort studies in our meta-analysis to achieve an adequate sample size to draw a reliable conclusion. The present exploratory cohort study did not show positive BIM deletion was associated with poorer clinical outcomes in advanced and metastatic NSCLC patients after EGFR-TKIs treatment. With a substantially expanded sample size (n=1,317) in the current meta-analysis, however, the positive BIM deletion displayed significant predictive effects on shorter PFS (P=0.02), while it failed to demonstrate significant difference regarding the other three common clinical outcome measures OS (P=0.43), ORR (P=0.69) and DCR (P=0.64).
In our cohort, the positive BIM deletion polymorphism occurred in 37 (26.4%) patients, which is relatively higher than the rate (9.6%–20%) reported in other cohort studies included for meta-analysis,Citation15,Citation23,Citation24,Citation26–Citation30 except for one study which has also quantitatively reported low/intermediate BIM mRNA expression.Citation25 The characteristics of the patients at baseline indicated that our cohort patients with heterozygous BIM deletion polymorphism were likely associated with marginally better prognosis factors in terms of younger age, less smoking, better performance status, less metastasis and earlier disease stage as of TKIs treatment onset. These slight inequalities of distribution may partially contribute to the estimated HR of <1 observed for PFS (HR =0.80; with vs without BIM deletion polymorphism). Even so, similar observation of variants was reported in two published studies conducted in Korea (HR =0.74, 95% CI: 0.30–1.83; HR =0.93, 95% CI: 0.34–2.57, respectively).Citation23,Citation30 The authors of these studies and Chinese counterpartsCitation9 discussed these findings using the following potential arguments: (1) uncertainties may be by chance due to small size of included study patients; (2) they did not consider other proapoptotic Bcl-2 family members such as BAX, BAK, and other BH3-only proteins including BAD and PUMA which might be key players in the apoptotic response in oncogene-addicted cancer; (3) unconsidered concomitant genetic alterations beyond EGFR mutations could conceivably accelerate or delay cancer progression; and (4) BIM RNA levels in treatment-naïve tissue were not measured; these measurements could be helpful for better understanding of the meaning of BIM deletion in patients with EGFR-mutant NSCLC. All of these highlighted points were echoed in our current study indeed. Subject to limited number of included studies and data availability, our study did not further analyze the role of EGFR subtypes with BIM polymorphism in predicting efficacy of EGFR-TKIs, either. Nevertheless, we analyzed toxic effects and obtained similar findings, with rash and diarrhea being the most common adverse reactions as in the EGFR-TKI group of the randomized control trials.Citation4,Citation5,Citation7,Citation8
Several prior studiesCitation15,Citation24–Citation29 reported that patients with BIM deletion polymorphism had significantly shorter PFS after EGFR-TKI treatment than did patients without BIM deletion polymorphism. As a result, our meta-analysis of these studies also reflected this finding. Although several previous meta-analysesCitation9,Citation33–Citation36 mentioned similar findings regarding PFS, the current pooled analysis containing more eligible studies further provided a possibility to analyze other clinical outcome measures. As a comprehensive meta-analysis of PFS, OS, ORR and DCR with the largest sample size to date, our study provided a more reliable answer regarding the impact of BIM polymorphism status on treatment efficacy of EGFR-TKIs in advanced and metastatic NSCLC patients with EGFR mutations.
Despite the comprehensive findings, there exist several limitations in our cohort study and meta-analysis. First, unlike randomized controlled trials, this present observational study, especially of small sample size, predisposes to imbalanced distribution of baseline characteristics. In this case, we did a serial of subgroup analyses to ascertain possibly consistent effect. In addition, the data of the cohort study are from a single hospital in the People’s Republic of China, which would potentially limit any extrapolations of the study conclusions. For the part of meta-analysis, reported aggregate data from several cohort studies were used rather than individual patient data, which may not provide robust estimation for the comparative efficacy. Publication bias might exist, although we did citations search without any language limit. Moreover, the quality of meta-analysis was subject to the quality of individual studies included. Second, although the prevalence of BIM deletion polymorphism was examined carefully in this study, we did not consider other coexisting genetic alterations beyond that of BIM deletion polymorphism. The underlying biology of EGFR-mutant NSCLC and tumor prognosis should be complex enough.Citation3,Citation9,Citation37,Citation38 Therefore, more efforts should be made to investigate the potential mechanisms of the primary and secondary resistance to EGFR-TKIs induced by BIM polymorphism and other ones in order to find oriented solutions and develop new therapies.
In summary, our meta-analysis of studies demonstrated that BIM deletion polymorphism is associated with shorter PFS after EGFR-TKIs treatment in advanced NSCLC EGFR-mutant patients than those without BIM polymorphism. Even so, additional large multicenter well-designed cohort studies comprising essential BIM gene alteration and other concurrent genetic alterations are warranted to uncover more underlying biology of EGFR-mutant NSCLC used for predicting clinical prognosis in the future. This further clarification will provide benefits for new drug development in the relevant therapeutic area.
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (81401892), Shanghai Health Bureau Foundation (201440423) and the Program for Cooperation among Industry, Academy, and Research, the Committee of Science and Technology, Baoshan District, Shanghai (No bkw2014122). The authors thank Sam Zhong for his generous assistance with this study.
Supplementary materials
Figure S1 PRISMA flow diagram of the studies search and selection process.
Abbreviation: PRISMA, preferred reporting items for systematic reviews and meta-analyses.
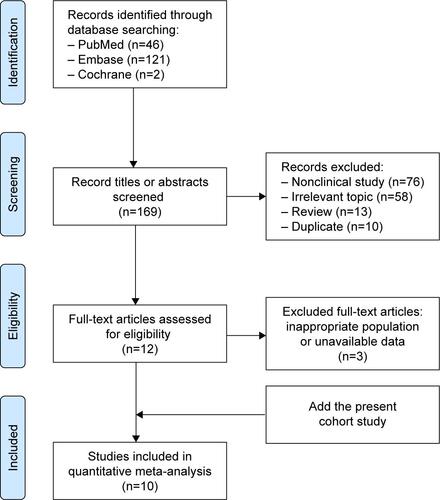
Figure S2 Funnel plot of hazard ratio for PFS and standard error of hazard ratio.
Abbreviation: PFS, progression-free survival.
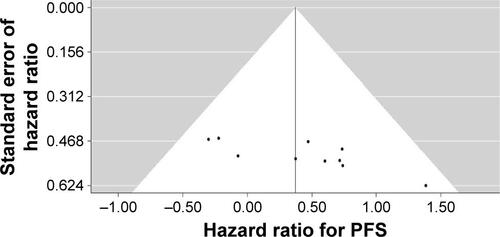
Table S1 Characteristics of cohort studies included in meta-analyses
References
- NgKPHillmerAMChuahCTA common BIM deletion polymorphism mediates intrinsic resistance and inferior responses to tyrosine kinase inhibitors in cancerNat Med201218452152822426421
- LeeJKShinJYKimSPrimary resistance to epidermal growth factor receptor (EGFR) tyrosine kinase inhibitors (TKIs) in patients with non-small-cell lung cancer harboring TKI-sensitive EGFR mutations: an exploratory studyAnn Oncol20132482080208723559152
- ZhengLLinBSongZRelationship between BIM gene polymorphism and therapeutic efficacy in the retreatment of advanced non-small cell lung cancer with tyrosine kinase inhibitorZhongguo Fei Ai Za Zhi20131612632638 Chinese [with English abstract]24345487
- CostaCMolinaMADrozdowskyjAThe impact of EGFR T790M mutations and BIM mRNA expression on outcome in patients with EGFR-mutant NSCLC treated with erlotinib or chemotherapy in the randomized phase III EURTAC trialClin Cancer Res20142072001201024493829
- ZhongJLiZXZhaoJAnalysis of BIM (BCL-2 like 11 gene) deletion polymorphism in Chinese non-small cell lung cancer patientsThorac Cancer20145650951626767045
- IsobeKHataYTochigiNClinical significance of BIM deletion polymorphism in non-small-cell lung cancer with epidermal growth factor receptor mutationJ Thorac Oncol20149448348724736070
- ZhaoMZhangYCaiWThe Bim deletion polymorphism clinical profile and its relation with tyrosine kinase inhibitor resistance in Chinese patients with non-small cell lung cancerCancer2014120152299230724737648
- LeeJHLinYLHsuWHBcl-2-like protein 11 deletion polymorphism predicts survival in advanced non-small-cell lung cancerJ Thorac Oncol2014991385139225057939
- LeeJYKuBMLimSHThe BIM deletion polymorphism and its clinical implication in patients with EGFR-mutant non-small-cell lung cancer treated with EGFR tyrosine kinase inhibitorsJ Thorac Oncol201510690390926001141
Disclosure
The authors report no conflicts of interest in this work.
References
- ChenWZhengRBaadePDCancer statistics in China, 2015CA Cancer J Clin201666211513226808342
- CostaDBHalmosBKumarABIM mediates EGFR tyrosine kinase inhibitor-induced apoptosis in lung cancers with oncogenic EGFR mutationsPLoS Med200741016691679 discussion 168017973572
- NiJZhangLEvaluation of three small molecular drugs for targeted therapy to treat nonsmall cell lung cancerChin Med J (Engl)2016129333234026831237
- MaemondoMInoueAKobayashiKNorth-East Japan Study GroupGefitinib or chemotherapy for non-small-cell lung cancer with mutated EGFRN Engl J Med2010362252380238820573926
- MokTSWuYLThongprasertSGefitinib or carboplatin-paclitaxel in pulmonary adenocarcinomaN Engl J Med20093611094795719692680
- AokiTIgawaSFuruyaNImpacts of treatment lines and initiation timing of erlotinib for advanced non-small cell lung cancerAnticancer Res201232260160822287751
- MitsudomiTMoritaSYatabeYWest Japan Oncology GroupGefitinib versus cisplatin plus docetaxel in patients with non-small-cell lung cancer harboring mutations of the epidermal growth factor receptor (WJTOG3405): an open label, randomised phase 3 trialLancet Oncol201011212112820022809
- ZhouCWuYLChenGErlotinib versus chemotherapy as first-line treatment for patients with advanced EGFR mutation-positive non-small-cell lung cancer (OPTIMAL, CTONG-0802): a multicentre, open-label, randomised, phase 3 studyLancet Oncol201112873574221783417
- HuangWFLiuAHZhaoHJDongHMLiuLYCaiSXBIM gene polymorphism lowers the efficacy of EGFR-TKIs in advanced non small cell lung cancer with sensitive EGFR mutations: a systematic review and meta-analysisMedicine (Baltimore)20159433e126326287412
- PaoWChmieleckiJRational, biologically based treatment of EGFR-mutant non-small-cell lung cancerNat Rev Cancer2010101176077420966921
- FordePMEttingerDSManaging acquired resistance in EGFR-mutated non-small cell lung cancerClin Adv Hematol Oncol201513852853226351816
- WangSCangSLiuDThird-generation inhibitors targeting EGFR T790M mutation in advanced non-small cell lung cancerJ Hematol Oncol201693427071706
- WangSTsuiSTLiuCSongYLiuDEGFR C797S mutation mediates resistance to third-generation inhibitors in T790M-positive non-small cell lung cancerJ Hematol Oncol2016915927448564
- WangSSongYYanFLiuDMechanisms of resistance to third-generation EGFR tyrosine kinase inhibitorsFront Med201610438338827770386
- NgKPHillmerAMChuahCTA common BIM deletion polymorphism mediates intrinsic resistance and inferior responses to tyrosine kinase inhibitors in cancerNat Med201218452152822426421
- O’ConnorLStrasserAO’ReillyLABim: a novel member of the Bcl-2 family that promotes apoptosisEMBO J19981723843959430630
- NakagawaTTakeuchiSYamadaTEGFR-TKI resistance due to BIM polymorphism can be circumvented in combination with HDAC inhibitionCancer Res20137382428243423382048
- EisenhauerEATherassePBogaertsJNew response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1)Eur J Cancer200945222824719097774
- PaezJGJännePALeeJCEGFR mutations in lung cancer: correlation with clinical response to gefitinib therapyScience200430456761497150015118125
- YiSZhuangYZhouJA comparison of epidermal growth factor receptor mutation testing methods in different tissue types in non-small cell lung cancerInt J Mol Med201434246447424891042
- BeggCBMazumdarMOperating characteristics of a rank correlation test for publication biasBiometrics1994504108811017786990
- EggerMDavey SmithGSchneiderMMinderCBias in meta-analysis detected by a simple, graphical testBMJ199731571096296349310563
- LeeJKShinJYKimSPrimary resistance to epidermal growth factor receptor (EGFR) tyrosine kinase inhibitors (TKIs) in patients with non-small-cell lung cancer harboring TKI-sensitive EGFR mutations: an exploratory studyAnn Oncol20132482080208723559152
- ZhengLLinBSongZRelationship between BIM gene polymorphism and therapeutic efficacy in the retreatment of advanced non-small cell lung cancer with tyrosine kinase inhibitorZhongguo Fei Ai Za Zhi20131612632638 Chinese [with English abstract]24345487
- CostaCMolinaMADrozdowskyjAThe impact of EGFR T790M mutations and BIM mRNA expression on outcome in patients with EGFR-mutant NSCLC treated with erlotinib or chemotherapy in the randomized phase III EURTAC trialClin Cancer Res20142072001201024493829
- ZhongJLiZXZhaoJAnalysis of BIM (BCL-2 like 11 gene) deletion polymorphism in Chinese non-small cell lung cancer patientsThorac Cancer20145650951626767045
- IsobeKHataYTochigiNClinical significance of BIM deletion polymorphism in non-small-cell lung cancer with epidermal growth factor receptor mutationJ Thorac Oncol20149448348724736070
- ZhaoMZhangYCaiWThe Bim deletion polymorphism clinical profile and its relation with tyrosine kinase inhibitor resistance in Chinese patients with non-small cell lung cancerCancer2014120152299230724737648
- LeeJHLinYLHsuWHBcl-2-like protein 11 deletion polymorphism predicts survival in advanced non-small-cell lung cancerJ Thorac Oncol2014991385139225057939
- LeeJYKuBMLimSHThe BIM deletion polymorphism and its clinical implication in patients with EGFR-mutant non-small-cell lung cancer treated with EGFR tyrosine kinase inhibitorsJ Thorac Oncol201510690390926001141
- WellsGASheaBO’ConnellDThe Newcastle-Ottawa Scale (NOS) for assessing the quality of nonrandomized studies in meta-analysesOttawa, ONOttawa Hospital Research Institute2011 Available from: http://www.ohri.ca/programs/clinical_epidemiology/oxford.htmAccessed January 1, 2011
- XiaJJZhaoSFXiongLWReal-time PCR assay with high resolution melting for EGFR and BIM mutation of lung cancerEur Rev Med Pharmacol Sci201620132805281127424979
- NieWTaoXWeiHChenWSLiBThe BIM deletion polymorphism is a prognostic biomarker of EGFR-TKIs response in NSCLC: a systematic review and meta-analysisOncotarget2015628256962570026325082
- ZouQZhanPLvTSongYThe relationship between BIM deletion polymorphism and clinical significance of epidermal growth factor receptor-mutated non-small cell lung cancer patients with epidermal growth factor receptor-tyrosine kinase inhibitor therapy: a meta-analysisTransl Lung Cancer Res20154679279626798589
- YingHQChenJHeBSThe effect of BIM deletion polymorphism on intrinsic resistance and clinical outcome of cancer patient with kinase inhibitor therapySci Rep201551134826076815
- MaJYYanHLGuWAssociation between BIM deletion polymorphism and clinical outcome of EGFR-mutated NSCLC patient with EGFR-TKI therapy: a meta-analysisJ Cancer Res Ther201511239740226148607
- LeeJYLimSHKimMIs there any predictor for clinical outcome in EGFR mutant NSCLC patients treated with EGFR TKIs?Cancer Chemother Pharmacol20147351063107024663503
- HsiaoSHLiuHELeeHLDistinct clinical outcomes of non-small cell lung cancer patients with epidermal growth factor receptor (EGFR) mutations treated with EGFR tyrosine kinase inhibitors: non-responders versus respondersPLoS One2013812e8326624376677
