 ?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.
?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.Abstract
Background
MicroRNAs (miRNAs) have been shown to be involved in the initiation and progression of cancers in the literature. In this study, we aimed to evaluate the clinicopathological role of miR-17 in breast cancer.
Materials and methods
The expression of miR-17 was measured in 132 breast cancer tissues and paired adjacent normal tissues by using real-time quantitative polymerase chain reaction. The association between miR-17 expression levels and clinicopathological parameters was also analyzed. 3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide and flow cytometry assays were used to investigate the role of miR-17 in the regulation of breast cancer cells.
Results
The expression of miR-17 was remarkably increased in breast cancer tissues and cell lines. Clinical association analysis revealed that a high expression of miR-17 was prominently associated with poor survival time in breast cancer. Overexpression of miR-17 promoted cell proliferation and induced tumor growth.
Conclusion
Our findings clarified that the upregulation of miR-17 played a vital role in breast cancer progression and suggested that miR-17 could be used as a prognostic biomarker for breast cancer.
Introduction
Breast cancer is one of the most common cancers worldwide,Citation1 with differences among tumors that are driven by multiple genetic/epigenetic alterations and molecular events.Citation2 Despite the improved prognosis of breast cancer patients due to early diagnosis, radical surgery and the development of adjuvant therapy, breast cancer remains one of the most common type of cancers among women.Citation3,Citation4 Prognostic factors that are frequently used for making clinical decisions in breast cancer are age, tumor size, status of lymph nodes, histological types of the tumor, pathological grade, and hormone receptor status.Citation5,Citation6 However, more biomarkers are needed for therapy and prediction of outcomes because human breast cancers are diverse in their genetic nature and their response to therapy.
MicroRNAs (miRNAs) are small noncoding endogenous RNA of 20–25 nucleotides that negatively regulate expression of target genes at the posttranscriptional level through mRNA degradation and translational repression.Citation7 Numerous studies have demonstrated that aberrantly expressed miRNAs are involved in diverse diseases,Citation8–Citation10 including cancer. Some miRNAs act as tumor suppressors or oncogenes and play critical roles in many aspects of breast cancer carcinogenesis, including cell proliferation, differentiation, metastasis, and angiogenesis.Citation11–Citation17 Given the high stability of miRNAs in formalin-fixed, paraffin-embedded (FFPE) tissue and circulation, there is growing recognition of the possibility of miRNAs as biomarkers for cancer diagnosis, prognosis, and prediction of treatment response.Citation18–Citation21
Recently, miR-17 has been shown to play a significant role in the pathogenesis of various cancers. The miR-17-5p was overexpressed in ovarian cancer cells, which activated AKT by downregulation of phosphatase and tensin homolog (PTEN) in ovarian cancer cells.Citation22 Overexpression of miR-17 significantly increases cell motility and migration of melanoma cells by inhibiting the translation of ETV1.Citation23 High levels of the oncogenic miRNA (oncomiR) guide strand called miR-17-5p is overexpressed in triple negative breast cancer (TNBC) and can inhibit ribosomal translation of tumor suppressor gene mRNAs, such as programmed cell death 4 (PDCD4) or PTEN.Citation24 miR-17 may help detecting early recurrence of colon cancer after radical surgery and adjuvant chemotherapy with high accuracy in clinical practice.Citation25 miR-17-92 plays an oncogenic role and provides chemoresistance to cisplatin in human prostate cancer cells.Citation26 However, little is known about the association between the level of miR-17 expression and the tumorigenesis and development.
In our study, we examined miR-17 expression in breast cancer tissues and cell lines. We also assessed the association between miR-17 expression and clinicopathological characteristics and overall survival of breast cancer patients. Moreover, we investigated the role of miR-17 in the regulation of breast cancer cells.
Materials and methods
Tissue specimens
A total of 132 breast cancer tissue specimens were obtained from patients who had undergone breast cancer surgery at the Affiliated Hospital of Nanjing Medical University, Changzhou No 2 People’s Hospital, Changzhou, People’s Republic of China. All the samples were shown to be correctly labeled clinically and pathologically and immediately frozen at −80°C until use. The ethical committees of Changzhou No 2 People’s Hospital affiliated to Nanjing Medical University approved this study, and written informed consent was obtained from all patients.
Cell lines and cell culture
Human breast cancer cell lines (MCF-7, MDA-MB-231, and MDA-MB-435S) were obtained from the Central Lab of the Affiliated Hospital of Nanjing Medical University, Changzhou No 2 People’s Hospital, Changzhou, People’s Republic of China. The cell lines were cultured in Roswell Park Memorial Institute 1640 medium and Dulbecco’s Modified Eagle’s Medium (DMEM; Thermo Fisher Scientific, Waltham, MA, USA) containing 10% fetal bovine serum (FBS; Invitrogen) and were grown in a humidified 5% CO2 incubator at 37°C.
RNA extraction and expression analysis
Total RNA was extracted using TRIzol reagent (Invitrogen) and a mirVana miRNA Isolation Kit (Ambion, Austin, TX, USA) according to the manufacturer’s instructions. cDNA was synthesized with the RevertAid™ First Strand cDNA Synthesis Kit (Thermo Fisher Scientific). Real-time polymerase chain reaction (PCR) was performed using SYBR Green PCR Master Mix (Vazyme, Nanjing, People’s Republic of China) on ABI VII7 Real-time RT-PCR system (Bio-Rad Laboratories Inc., Hercules, CA, USA). For miR-17 detection, U6 snRNA was used as an internal control. The relative levels of miRNA expression were calculated from the relevant signals by normalization with the signal of U6 snRNA expression. RNA expression levels were obtained using the comparative cycle threshold
method and analyzed as mean ± SD.
RNA oligoribonucleotides and cell transfection
Both miR-17 and all RNA oligoribonucleotides for in vitro studies were purchased from GenePharma (Shanghai, People’s Republic of China). Their sequences were listed as follows: miR-17 scramble F1-5′-GUCCUGAGAAGGCUAGCAUAGAU-3′, F2-5′-CUAUGCUAGCCUUCUCAGGACUU-3′; miR-17 mimics F1-5′-CAAAGUGCUUACAGUGCAGGUAG-3′, F2-5′-ACCUGCACUGUAAGCACUUUGUU-3′; and miR-17 inhibitor F1-5′-CUACCUGCACUGUAAGCACUUUG. The transfection of RNA oligoribonucleotides was performed using Lipofectamine 2000 (Invitrogen). Unless otherwise indicated, 100 nM of RNA duplex or 80 nM of miRNA inhibitor was used for each transfection, and all the experiments were repeated in triplicate.
Cell proliferation and cell cycle analyses
Cell viability was analyzed using 3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT; Sigma) assays as previously described. Briefly, 5×103 cells per well were seeded into a 96-well plate. After miRNA transfection, the cells were maintained for 72 hours and cell viabilities were determined using a Benchmark Plus™ microplate spectrometer (Bio-Rad Laboratories Inc.). For cell cycle analysis, the cells were harvested 48 h following transfection, washed with PBS, and fixed in 75% ethanol at −20°C. After overnight fixation, the cells were washed with PBS and stained with propidium iodide (Beckman Coulter, Fullerton, CA, USA) for 30 min. Cell cycle analysis was performed using the BD Flow Cytometry System with FACSDiva software (BD, Franklin Lakes, NJ, USA). The cell cycle distribution is presented as the percentage of cells in G1, S, and G2 phases. The data were analyzed with FlowJo v5.7.2 (BD).
Stable transfection and in vivo animal studies
All animal experiments in this study were approved by the ethics committee of Nanjing Medical University, and the guidelines of National Animal Care and Use Committee were followed. Twenty mice were randomly divided into two groups, and each mouse was injected subcutaneously with 1×108 cells. The agomiR-17 expression constructs were generated by GenePharma, and miR-17 expression was confirmed by quantitative reverse transcription (qRT)-PCR. The tumor volume (mm3) was measured every 7 days and was calculated using the following formula: volume = width × width × height/2. The animals were sacrificed 42 days after seeding the tumor cells. All tumor grafts were excised, weighed, and harvested.
Statistical analyses
The significance of differences between groups was estimated by the Wilcoxon test for two-group comparisons and the Kruskal–Wallis test for k-group comparisons (with k>2). Cases were divided into two groups, high or low, using the median value of miR-17 expression as a cutoff. Fisher’s exact test was used to evaluate the relationship between miR-17 expression and clinical features. Overall survival was calculated as the time from the date of surgery resection to the date of last contact or death. Survival analyses were performed using the Kaplan–Meier method and the log-rank test. P-value <0.05 was considered statistically significant. All statistical analyses were performed using the SPSS version 19.0 (IBM Corp, Armonk, NY, USA).
Results
MiR-17 is frequently upregulated in breast cancer
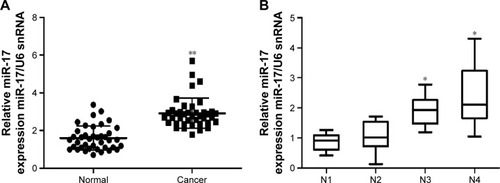
In order to explore the role of miR-17 in breast carcinogenesis, we used qRT-PCR to measure miR-17 expression in breast cancer and adjacent noncancerous tissues. The expression of miR-17 was significantly increased in cancer tissues compared with adjacent normal tissues (P<0.0001, n=40; ). In addition, the expression of miR-17 was higher in tumors with pathological stages (N1 and N2, P=0.5612; N1 and N3, P<0.05; N1 and N4, P<0.05; ). These data suggested that upregulated miR-17 was a frequent event in human breast cancer and could be involved in cancer progression.
Figure 1 The expression of miR-17 in breast cancer tissues.
Abbreviation: qRT-PCR, quantitative reverse transcription polymerase chain reaction.
We also investigated the association between the expression of miR-17 and clinicopathological characteristics. Specimens were divided into two groups according to the expression of miR-17. The data showed that the expression of miR-17 differed significantly according to human epidermal growth factor receptor 2 status (P<0.05) and estrogen receptor status (P<0.05). As shown in , no difference was found between miR-17 expression and other clinical features, such as age, cyclooxygenase-2, CD44, epidermal growth factor receptor, progesterone receptor, P-glycoprotein status, and lymph node metastasis.
Table 1 The relationship between miR-17 expression and clinicopathological parameters
MiR-17 promotes proliferation of breast cancer cells in vitro
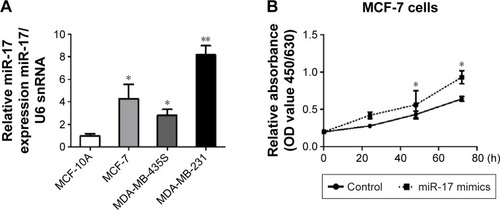
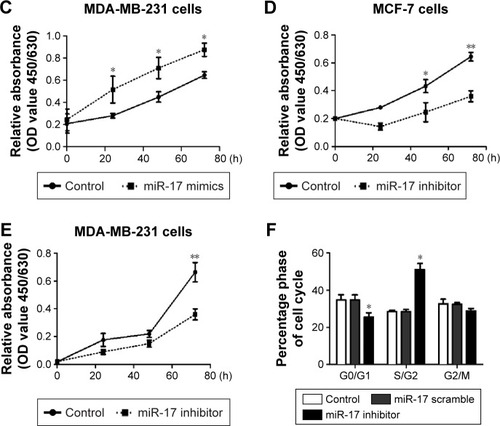
To determine the role of miR-17 in breast cancer cells, we detected miR-17 expression in the breast cancer cell lines MCF-7, MDA-MB-435S, and MDA-MB-231. We found that the expression of miR-17 was significantly higher in cancer cell lines compared with MCF10A cells (). Then, we transfected MCF-7 and MDA-MB-231 cells with miR-17 mimics or an inhibitor. We found that miR-17 mimics promoted miR-17 expression and miR-17 inhibitor inhibited miR-17 expression in breast cancer cells (Figure S1). MTT assays indicated that miR-17 mimics induced breast cancer cell proliferation (). In contrast, miR-17 inhibitor suppressed cell proliferation in breast cancer cells (). In addition to cell proliferation, we also investigated the role of miR-17 on cell cycle progression. Cell cycle profile was examined by flow cytometry with propidium iodide staining, and the cell numbers were counted according to the DNA content of G0/G1, S, and G2/M phases. The data showed that miR-17 inhibitor arrested cell cycle in S phases (). These data suggested that miR-17 increased cell proliferation and miR-17 inhibitor inhibited cell proliferation and arrested cell cycle in the S-phase.
Figure 2 miR-17 regulated breast cancer cell proliferation and cell cycle arrest in vivo.
Abbreviations: qRT-PCR, quantitative reverse transcription polymerase chain reaction; MTT, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide; OD, optical density; SD, standard deviation.
MiR-17 induces tumor growth in vivo
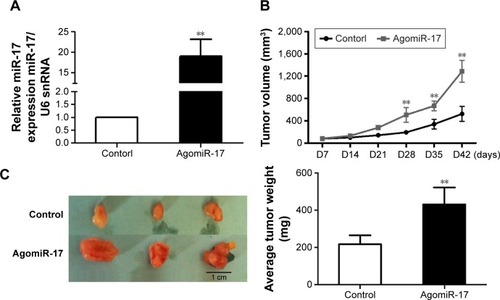
To measure the effect of miR-17 on tumor growth in vivo, we transfected MCF-7 cells with agomiR-17 (a modified miR-17 mimics used for in vivo studies). miR-17 was highly expressed in MCF-7 cells by qPCR (). The three batches of cells were injected subcutaneously into nude mice. After 42 days, we observed faster tumor growth in the agomiR-17 group compared with the control group (). The average weight of tumors from the two groups was significantly different (). These results showed that overexpression of miR-17 induced tumor growth in vivo.
Figure 3 miR-17 induced xenograft tumor growth in vivo.
Abbreviations: qPCR, quantitative polymerase chain reaction; SD, standard deviation.
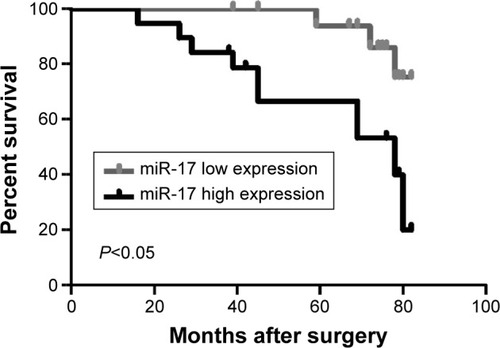
Overexpression of miR-17 is associated with poor prognosis of breast cancer patients
To further investigate the association of miR-17 expression with survival of breast cancer patients, Kaplan–Meier curve with long rank analysis was performed. Breast cancer patients with a low expression of miR-17 had a significantly longer survival time compared with those with a high expression of miR-17 (P=0.0044, P<0.05, n=36; ). These data suggested that miR-17 expression was a prognostic factor in breast cancer.
Figure 4 Relative miR-17 expression associated with overall survival of breast cancer patients.
Discussion
There is mounting evidence demonstrating that miRNAs play an important role in the development of human malignancies,Citation27–Citation29 which exhibit oncogenic or tumor suppressive role by directly regulating oncogenes or tumor-suppressor genes.Citation30,Citation31
It would be conducive to expand our view to better understand cancer carcinogenesis by analyzing mRNA target association and miRNA-mediated pathways.
Here, we focused on miR-17, which had been shown to regulate growth, proliferation, apoptosis, migration, and invasion of tumor cells in certain cancers. The expression of miR-17 was significantly increased in prostate cancer,Citation25,Citation32 gastric cancer,Citation33–Citation35 and colon cancer cells,Citation36–Citation38 which suggests a potential tumor promoter function of miR-17. In this study, we observed that upregulation of miR-17 was a frequent event in breast cancer tissues compared with that in adjacent tissues. In addition, the expression level of miR-17 was higher in tumors with the pathological stage. We also found that miR-17 could induce cell proliferation in breast cancer cells and arrest cell cycle in the S-phase.
Conclusion
This study analyzed the association between miR-17 and the prognosis of breast cancer. We found that miR-17 was upregulated in breast cancer tissues and induced breast cancer cell proliferation. We provided evidence to verify that breast cancer patients with a low expression of miR-17 had a significantly longer survival time. These findings suggested that miR-17 might be a novel prognostic indicator and a potential target for diagnosis and gene therapy in breast cancer.
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (81272240 to LF).
Supplementary material
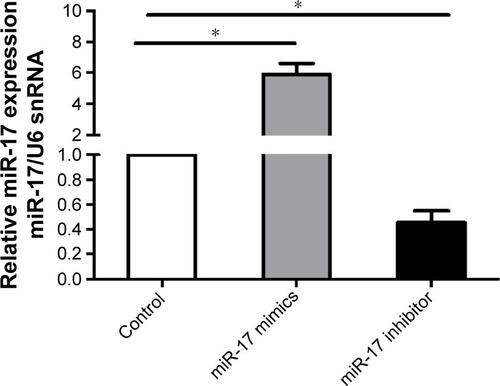
Figure S1 The expression of miR-17 in breast cancer cells.
Notes: MCF-7 cells with miR-17 mimics and miR-17 inhibitor treatment, and miR-17 expression was examined by qRT-PCR and normalized to U6 snRNA expression in breast cancer cells. The data were shown as mean ± SD from three independent experiments. *P<0.05.
Abbreviation: qRT-PCR, quantitative reverse transcription polymerase chain reaction.
Disclosure
The authors report no conflicts of interest in this work.
References
- DeSantisCEFedewaSAGoding SauerAKramerJLSmithRAJemalABreast cancer statistics, 2015: convergence of incidence rates between black and white womenCA Cancer J Clin2016661314226513636
- MancaGTardelliERubelloDSentinel lymph node biopsy in breast cancer: a technical and clinical appraisalNucl Med Commun201637657057626886421
- FanLGossPEStrasser-WeipplKCurrent status and future projections of breast cancer in AsiaBreast Care (Basel)201510637237826989355
- RussoAAndreanoAAnghinoniEA set of indicators to monitor the adherence to the guidelines for the diagnosis and treatment of breast cancerEpidemiol Prev20143811628 Italian [with English abstract]24736958
- CardosoFHarbeckNFallowfieldLKyriakidesSSenkusEGroupEGWLocally recurrent or metastatic breast cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-upAnn Oncol201223suppl 7vii11vii1922997442
- WangDDuanLTuZThe glasgow prognostic score predicts response to chemotherapy in patients with metastatic breast cancerChemotherapy201661421722226905743
- LuJGetzGMiskaEAMicroRNA expression profiles classify human cancersNature2005435704383483815944708
- LyonsPJPoitrasJJCourteauLAStoreyKBMorinPJrIdentification of differentially regulated microRNAs in cold-hardy insectsCryo Letters2013341838923435712
- SchneiderMRSamborskiABauersachsSZouboulisCCDifferentially regulated microRNAs during human sebaceous lipogenesisJ Dermatol Sci2013702889323452545
- WinbanksCEOoiJYNguyenSSMcMullenJRBernardoBCMicroRNAs differentially regulated in cardiac and skeletal muscle in health and disease: potential drug targets?Clin Exp Pharmacol Physiol201441972773725115402
- WangYChenJLinZRole of deregulated microRNAs in non-small cell lung cancer progression using fresh-frozen and formalin-fixed, paraffin-embedded samplesOncol Lett201611180180826870288
- VorvisCKoutsioumpaMIliopoulosDDevelopments in miRNA gene signaling pathways in pancreatic cancerFuture Oncol20161291135115026984178
- Meryet-FiguiereMLambertBGauduchonPAn overview of long non-coding RNAs in ovarian cancersOncotarget2016728447194473426992233
- LinCWLinPYYangPCNoncoding RNAs in tumor epithelial-to-mesenchymal transitionStem Cells Int20162016273270526989421
- KwanJYPsarianosPBruceJPYipKWLiuFFThe complexity of microRNAs in human cancerJ Radiat Res201657suppl 1i106i11126983984
- Fred Henry WalterRVollbrechtCWernerRmicroRNAs are differentially regulated between MDM2-positive and negative malignant pleural mesotheliomaOncotarget2016714187131872126918730
- FengTXuDTuCMiR-124 inhibits cell proliferation in breast cancer through downregulation of CDK4Tumour Biol20153685987599725731732
- TreeceALDuncanDLTangWGastric adenocarcinoma microRNA profiles in fixed tissue and in plasma reveal cancer-associated and Epstein-Barr virus-related expression patternsLab Invest201696666167126950485
- GomesBCSantosBRueffJRodriguesASMethods for studying MicroRNA expression and their targets in formalin-fixed, paraffin-embedded (FFPE) breast cancer tissuesMethods Mol Biol2016139518920526910075
- HanYXuGXLuHDysregulation of miRNA-21 and their potential as biomarkers for the diagnosis of cervical cancerInt J Clin Exp Pathol2015867131713926261606
- WenJYeFHeXDevelopment and validation of a prognostic nomogram based on the log odds of positive lymph nodes (LODDS) for breast cancerOncotarget2016715210462105326992235
- FangYXuCFuYMicroRNA-17-5p induces drug resistance and invasion of ovarian carcinoma cells by targeting PTEN signalingJ Biol Res (Thessalon)2015221226500892
- CohenRGreenbergENemlichYSchachterJMarkelGmiR-17 regulates melanoma cell motility by inhibiting the translation of ETV1Oncotarget2015622190061901626158900
- JinYYAndradeJWickstromENon-specific blocking of miR-17-5p guide strand in triple negative breast cancer cells by amplifying passenger strand activityPLoS One20151012e014257426629823
- ZhouPMaLZhouJmiR-17-92 plays an oncogenic role and conveys chemo-resistance to cisplatin in human prostate cancer cellsInt J Oncol20164841737174826891588
- ConevNVDonevISKonsoulova-KirovaAAChervenkovTGKashlovJKIvanovKDSerum expression levels of miR-17, miR-21, and miR-92 as potential biomarkers for recurrence after adjuvant chemotherapy in colon cancer patientsBiosci Trends20159639340126781797
- PalanichamyJKRaoDSmiRNA dysregulation in cancer: towards a mechanistic understandingFront Genet201455424672539
- MulraneLMcGeeSFGallagherWMO’ConnorDPmiRNA dysregulation in breast cancerCancer Res201373226554656224204025
- IliopoulosDPolytarchouCHatziapostolouMMicroRNAs differentially regulated by Akt isoforms control EMT and stem cell renewal in cancer cellsSci Signal2009292ra6219825827
- ErolesPTormoEPinedaBEspinELluchAMicroRNAs in breast cancer: one more turn in regulationCurr Drug Targets20161791083110025694121
- LiuHMicroRNAs in breast cancer initiation and progressionCell Mol Life Sci201269213587359922926415
- GongAYEischeidANXiaoJmiR-17-5p targets the p300/CBP-associated factor and modulates androgen receptor transcriptional activity in cultured prostate cancer cellsBMC Cancer20121249223095762
- BahariFEmadi-BaygiMNikpourPmiR-17-92 host gene, uderexpressed in gastric cancer and its expression was negatively correlated with the metastasisIndian J Cancer2015521222526837962
- ZhangXKongYXuXF-box protein FBXO31 is down-regulated in gastric cancer and negatively regulated by miR-17 and miR-20aOncotarget20145156178619025115392
- WuQLuoGYangZmiR-17-5p promotes proliferation by targeting SOCS6 in gastric cancer cellsFEBS Lett2014588122055206224801601
- ZhuJDongHZhangQZhangSCombined assays for serum carcinoembryonic antigen and microRNA-17-3p offer improved diagnostic potential for stage I/II colon cancerMol Clin Oncol2015361315131826807240
- KnudsenKNNielsenBSLindebjergJHansenTFHolstRSorensenFBmicroRNA-17 is the most up-regulated member of the miR-17-92 cluster during early colon cancer evolutionPLoS One20151010e014050326465597
- HuSLiuLChangEBWangJYRaufmanJPButyrate inhibits pro-proliferative miR-92a by diminishing c-Myc-induced miR-17-92a cluster transcription in human colon cancer cellsMol Cancer201514118026463716
