Abstract
Objective
Non-melanoma skin cancer (NMSC) is the most common malignancy with annually rising incidence. The aim of this study was to estimate the association between three coding polymorphisms (Arg399Gln, Arg194Trp, and Arg280His) of the DNA repair gene X-ray repair cross-complementing group 1 (XRCC1) and NMSC susceptibility.
Methods
Online databases were searched to retrieve case–control studies published between January 2000 and November 2016. Pooled odds ratio (OR) and 95% confidence interval (CI) were employed to assess the strength of association. Overall, 10 relevant studies were finally included for analysis, including 3,143 NMSC patients and 3,540 controls. For each polymorphism of XRCC1 gene, there were 3,050 cases and 3,463 controls for Arg399Gln, 914 cases and 1,182 controls for Arg194Trp, and 279 cases and 413 controls for Arg280His.
Results
Our results showed that these three polymorphisms in the XRCC1 coding region were not associated with increased risk of NMSC in the total studied population. However, subgroup analysis by ethnicities demonstrated that Gln/Arg genotype of Arg399Gln polymorphism was associated with increased risk of NMSC under the heterogeneous model in Asian populations (Gln/Arg vs Arg/Arg: OR =1.39, 95% CI =1.04–1.87, P=0.03); subgroup analysis by tumor types showed that Trp/Trp genotype of Arg194Trp was positively associated with decreased cancer risk in squamous-cell skin cancer (SCC) type under the homogeneous model (Trp/Trp vs Arg/Arg: OR =0.38, 95% CI =0.16–0.92, P=0.03).
Conclusion
Our results suggested that Arg399Gln variant of XRCC1 gene might be a risk factor for NMSC in Asian populations, and Arg194Trp variant of XRCC1 gene might be a protective factor for patients with SCC. In addition, future case–control studies are still needed to further evaluate the effect of XRCC1 polymorphisms in NMSC risk.
Introduction
Skin cancer is one of the most frequent malignant diseases that arise from the skin in humans.Citation1 It is categorized into three main types: basal-cell skin cancer (BCC), squamous-cell skin cancer (SCC), and melanoma.Citation2 Non-melanoma skin cancer (NMSC) is the most commonly diagnosed cancer in white-skinned individuals with a worldwide increasing incidence.Citation3 It comprises BCC and SCC, as well as a host of rare tumors.Citation4 BCC and SCC together account for 95%–98% of all NMSC cases: the former is characterized by local invasiveness with rare metastasis; the latter has a higher metastatic potential and is responsible for the majority of deaths from NMSC.Citation5 The incidence of NMSC has been rising by 3%–8% per year since 1960, with as much as 300% increase in the past 2 decades.Citation6 Moreover, recent estimates demonstrate that more cases of NMSC have occurred in the past 30 years than all other forms of cancer combined.Citation7 The rising incidence rates of NMSC are probably caused by a combination of increased exposure to ultraviolet (UV) or sun light, ozone depletion, genetics, and immune suppression.Citation8 There are many approaches to the management of NMSC,Citation9 but the current treatment is limited due to low complete clearance rates.Citation10 In addition, treatment of NMSC is substantial and increasing, posing a considerable burden on the health care system.Citation11 Therefore, there is an urgent need to identify particular biomarkers that can predict this disease and guide the treatment options.
Various types of DNA damage could promote the development of human diseases.Citation12 DNA repair is a complicated biological process that maintains genome stability.Citation13 Epidemiologic research has found that genetic variants of DNA repair genes might be involved in cancer development.Citation14 X-ray repair cross-complementing group 1 (XRCC1) gene, located at chromosome 19q13.2, is a key component of base excision repair (BER) and is required for genetic stability.Citation15 It is required for repair in both DNA single-strand break repair and BER pathways and acts primarily as a scaffold protein that facilitates the assembly of multi-protein complexes and coordinating steps during damage.Citation16 XRCC1 is thought to play the repair role through protein–protein interactions or by direct contact with DNA.Citation17 Evidence has shown that polymorphisms in DNA repair genes could influence individual DNA repair capacity.Citation18 Genetic polymorphisms of XRCC1 gene might contribute to impair DNA repair, thus playing a role in heightening the risk of NMSC. Three main single nucleotide polymorphisms (SNPs) in the coding region of XRCC1 gene have been identified: amino acid substitution between arginine and glutamine at codon 399 (Arg399Gln, G to A base change, SNP number: rs25487) in exon 10, arginine and tryptophan at codon 194 (Arg194Trp, C to T base change, SNP number: rs1799782) in exon 6, and arginine and histidine at codon 280 (Arg280His, G to A base change, SNP number: rs25489) in exon 9. The 399Gln allele, located in the C-terminal functional domain, was shown to be associated with altering the phenotype of the XRCC1 protein;Citation19 the 194Trp variant, located in the linker region of the N-terminal functional domain, appeared to protect against genotoxic effects;Citation20 the 280His, located in the proliferating cell nuclear antigen-binding region, was suggested to impair and decrease DNA repair ability.Citation21 All these changes in conserved protein sites may alter the BER capacity and affect protein function, thus increasing the chances of DNA damage.Citation22,Citation23
Although several studies have identified these three SNPs in the XRCC1 gene and NMSC risk, the results still remain inconclusive. In addition, the incidence of NMSC varies widely worldwide based on geographical distribution with the highest rates in Australia and the lowest rates in parts of Africa.Citation24 Therefore, we conducted the present meta-analysis to systematically review all the published articles on this issue and to obtain a relatively reliable result.
Materials and methods
Search strategy
We searched the online databases of PubMed, Medline, Web of Science, and Embase to retrieve relevant articles published between January 2000 and November 2016. The Medical Subject Heading (MeSH) terms were “skin cancer or non-melanoma skin cancer”, “basal-cell skin cancer or squamous-cell skin cancer”, “DNA repair gene or X-ray repair cross-complementing group 1 or XRCC1”, and “polymorphism or single nucleotide polymorphism or variant”. We manually checked the references of retrieved articles to obtain more sources. Our study included articles written only in the English language. If the same authors had published more than one article on the same subject among the same participants, then only the most complete study was included in this meta-analysis.
Criteria for article screening
The articles that met the following criteria were included in the study: 1) case–control studies evaluating the relationship between XRCC1 polymorphisms and NMSC susceptibility, 2) studies in which NMSC was confirmed in patients by experienced pathologists, and controls were age-matched unrelated participants without any type of cancer, 3) studies in which the results were expressed as odds ratio (OR) with corresponding 95% confidence interval (CI), and 4) studies in which the genotype distribution of controls for a certain polymorphism was in Hardy–Weinberg Equilibrium (HWE). Studies whose data could not be extracted, those with duplicated data, and reviews or conference papers were excluded.
Qualification assessment and data extraction
According to the Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) statement,Citation25 two authors independently estimated the quality of extracted articles. Any disagreement was resolved by discussing with the third author. Each item reached a final consensus. The following information was retrieved: the name of first author, publication year, country, ethnicity, mean age of patients and mean age of controls, total number of cases and controls, tumor types, and genotyping methods.
Statistical analyses
Statistical analyses were carried out using Review Manager (Revman, version 5.3) software.Citation26 The association between XRCC1 polymorphisms and NMSC risk was evaluated by OR with 95% CI. The significance of the pooled OR was determined by Z test (P<0.05 was considered significant). For each genetic variant, allelic model, homogeneous model, heterogeneous model, domain model, and recessive model were calculated. Cochran’s Q test and I2 test were employed to evaluate the heterogeneity of the included articles;Citation27,Citation28 random-effect model was used when the P-value of Cochran’s Q test was <0.10 and I2 >50%; otherwise, the fixed-effect model was used. To assess whether our results were substantially influenced by the presence of any individual study, we conducted a sensitivity analysis by systematically removing each study and recalculating the significance of the results. The funnel plot was performed to examine the publication bias.
Results
Characteristics of eligible studies
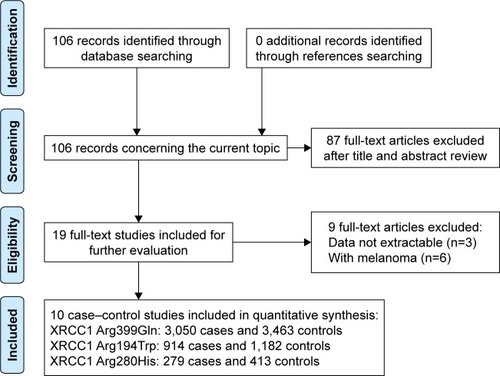
We firstly identified 106 relevant articles. After applying the inclusion and exclusion criteria, a total of 10 articles were finally selected, including 6,683 participants (3,143 NMSC patients and 3,540 controls). shows the selection process of this meta-analysis. The 10 articles included were conducted in 10 different countries: UK,Citation29 Denmark,Citation30,Citation31 USA,Citation32 Sweden/Finland,Citation33 Sweden,Citation34 Korea,Citation35 Japan,Citation36 Germany,Citation37 and Taiwan (China).Citation38 Three were conducted in Asian populations and seven in Caucasian populations. The sample size ranged from 40 to 1,176. The genotypes of XRCC1 polymorphisms were determined using three methods. The genotype distributions of the controls of all studies were in agreement with HWE (P>0.05). The main characteristics of the studies included are presented in .
Table 1 The detailed characteristics of all studies included in this meta-analysis
Association between XRCC1 Arg399Gln polymorphism and NMSC susceptibility
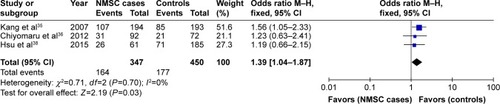
The summary of the test for association and test for heterogeneity of this meta-analysis is shown in . All these 10 articles concerned the effect of XRCC1 Arg399Gln polymorphism in NMSC risk, including 3,050 cases and 3,463 controls. No significant between-study heterogeneity was observed, and the fixed-effect model was employed. Our statistical analyses found that XRCC1 Arg399Gln polymorphism was not associated with increased risk of NMSC under the allelic model (Gln vs Arg: OR =0.97, 95% CI =0.90–1.05, P=0.42) as shown in . This insignificance was detected in other genetic models as well (Gln/Gln vs Arg/Arg: OR =0.90, 95% CI =0.77–1.06, P=0.20; Gln/Arg vs Arg/Arg: OR =1.02, 95% CI =0.92–1.14, P=0.69; Gln/Gln + Gln/Arg vs Arg/Arg: OR =0.99, 95% CI =0.90–1.10, P=0.89; Gln/Gln vs Gln/Arg + Arg/Arg: OR =0.90, 95% CI =0.77–1.04, P=0.16).
Figure 2 Forest plot of the association between XRCC1 Arg399Gln polymorphism and non-melanoma skin cancer risk under allelic model.
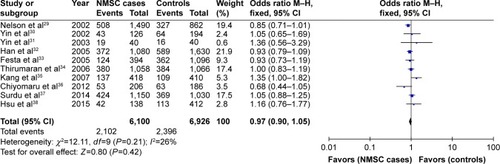
Table 2 Meta-analysis and stratified analyses of Arg399Gln, Arg194Trp, and Arg280His in the coding region of XRCC1 gene with NMSC risk under each genetic model
Subgroup analysis by ethnicities showed that only Gln/Arg genotype of this variant was significantly correlated with increased risk of NMSC under the heterogeneous model in Asian populations (Gln/Arg vs Arg/Arg: OR =1.39, 95% CI =1.04–1.87, P=0.03) as shown in . This positive relationship was not found in other genetic models. Moreover, no significant relationship was found between XRCC1 Arg399Gln variant and NMSC risk under any genetic models in Caucasian populations (P>0.05). Subgroup analysis by NMSC types showed that XRCC1 Arg399Gln variant was not associated with NMSC risk in both BCC and SCC types.
Association between XRCC1 Arg194Trp polymorphism and NMSC susceptibility
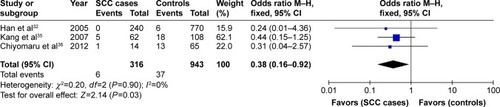
Four articles focused on this genetic variant, containing 914 cases and 1,182 controls. The frequency of Trp allele was a little higher in the patient group than that in the control group (14.6% vs 12.6%), but statistical analyses did not detect significant association between XRCC1 Arg194Trp variant and NMSC risk under the five genetic models as shown in . Subgroup analysis by ethnicities showed that XRCC1 Arg194-Trp polymorphism was not associated with NMSC risk among Asian and Caucasian populations (P>0.05). Subgroup analysis by NMSC types showed that Trp/Trp genotype was positively associated with decreased cancer risk in SCC type under the homogeneous model (Trp/Trp vs Arg/Arg: OR =0.38, 95% CI =0.16–0.92, P=0.03) as shown in . However, this correlation was not found in patients with BCC type.
Association between XRCC1 Arg280His polymorphism and NMSC susceptibility
Two articles included 279 cases and 413 controls. Our results showed that XRCC1 Arg280His polymorphism was not associated with NMSC risk under each genetic model (His vs Arg: OR =0.88, 95% CI =0.38–2.05, P=0.78; His/His vs Arg/Arg: OR =0.87, 95% CI =0.21–3.50, P=0.84; His/Arg vs Arg/Arg: OR =0.97, 95% CI =0.48–1.95, P=0.93; His/His + His/Arg vs Arg/Arg: OR =0.92, 95% CI =0.41–2.07, P=0.84; His/His vs His/Arg + Arg/Arg: OR =0.87, 95% CI =0.21–3.53, P=0.84).
Sensitivity analysis and publication bias
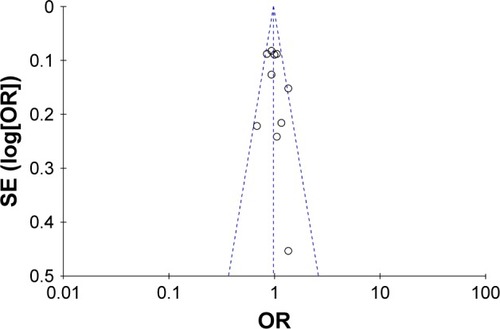
We systematically omitted each single article one at a time to verify whether our results were influenced by each included study. Our results showed that the pooled ORs were not significantly changed. The funnel plot was used to assess the publication bias of the literature. All genetic comparisons in our study showed no evidence of publication bias as shown in , which indicates that there was no publication bias in the present meta-analysis.
Discussion
In this meta-analysis, we screened 10 relevant articles in total. We found that Arg399Gln, Arg194Trp, and Arg280His polymorphisms of the XRCC1 gene were not associated with NMSC risk in the total studied populations. However, subgroup analysis showed that only Gln/Arg genotype of XRCC1 Arg399Gln variant was significantly correlated with increased risk of NMSC in Asian populations, and Trp/Trp genotype of XRCC1 Arg194Trp variant was positively associated with decreased risk of SCC type. Our results were not consistent with the previous meta-analysis, which studied the association between polymorphisms of the XRCC1 gene and skin cancer risk, and showed that no significant association was observed in stratified analyses of Arg399Gln and Arg194Trp polymorphisms and tumor type (SCC, BCC, and melanoma).Citation39
Harmful effects of solar UV radiation on human health is one of the causes of NMSC.Citation40 Exposure to UV radiation initiates ~90% of NMSC, causing malignant transformation of keratinocytes and suppression of inflammatory response. A reduced capacity to repair UV-induced DNA damage is one of the underlying molecular mechanisms for sunlight-induced skin carcinogenesis in the general population.Citation41 Evidence showed that a low DNA repair capacity was a susceptibility factor for NMSC.Citation42–Citation44 The XRCC1 protein, involved in the DNA repair pathway, is essential for the maintenance of genomic stability.Citation45 It serves to orchestrate BER via its role as a central scaffolding protein physically associated with DNA ligase III and via its function in recognizing and binding to single-strand breaks.Citation46,Citation47 XRCC1 is also involved in the organization of BER into multi-protein complexes of different sizes to DNA damage sites,Citation48 and defects in BER are shown to be linked with immunodeficiency, neurodegenerative disorders, and cancer predisposition.Citation49
Some studies have shown that polymorphisms in DNA repair genes might change the risk of cancer development.Citation50–Citation52 Variant genotypes of XRCC1 gene that cause amino acid substitutions may affect protein structure and gene expression, and impair the interaction of XRCC1 with the other enzymatic proteins, thus altering the BER function. For example, XRCC1 polymorphisms could influence cell responses to X-rays, and Arg399Gln of XRCC1 was shown to be associated with higher levels of DNA breaks;Citation53 the polymorphic variant Arg280His exhibited a slightly shorter retention time at DNA breaks.Citation54 XRCC1 polymorphisms might be associated with human cancer and could function as biomarkers of cancer susceptibility: Arg399Gln polymorphism was associated with lung cancerCitation55 and breast cancer;Citation56 Arg194Trp polymorphism was found to be associated with gastric cancerCitation57 and breast cancer;Citation58 and Arg280His was associated with invasive cervical cancerCitation59 and pancreatic cancer.Citation60 A previous review showed that the homozygous variant (Gln/Gln) of Arg399Gln variant can have 3–4-fold diminished capacity to remove DNA adducts and oxidized DNA damage; Arg280His variant appeared to decrease repair function, but Arg194Trp variant appears to protect against genotoxic effects.Citation20 This was consistent with our result. Several studies have identified the role of XRCC1 variants in skin cancer risk. Previous meta-analysis suggested that XRCC1 Arg399Gln polymorphism was a risk factor for cutaneous melanoma in a population-based subgroup.Citation61 Only the 10 studies included in the present meta-analysis analysed the effect of XRCC1 polymorphism in NMSC risk. For instance, a study that included ~300 Koreans showed that Arg/Gln and Gln/Gln genotypes had an ~2-fold increased risk of BCC compared to Arg/Arg individuals,Citation35 whereas another study that included ~1,000 Caucasians showed that Arg/Arg genotype had significantly reduced risk of both BCC and SCC.Citation29 In addition, genes of DNA BER pathway might be evolving as biomarkers for personalized therapies in cancer.Citation62 Polymorphisms in DNA repair genes might exert a role in survival, might facilitate response to chemotherapy, and might predict prognosis. Figl et al showed that variants in potential regulatory regions of DNA repair genes might influence disease outcome in melanoma patients and have potentially significant implications for patient surveillance and tailored treatment.Citation63 XRCC1 risk allele may result in resistance to therapy and shorter survival in bladder cancer patients treated with chemotherapy.Citation64 XRCC1 Gln allele genotype showed significant prognostic associations in hepatocellular carcinoma.Citation65 Bhandaru et al demonstrated that loss of XRCC1 expression was correlated with the progression of melanoma from dysplastic nevi to primary melanoma and to metastatic melanoma, and with worse overall and disease-specific 5-year and 10-year survival of patients with melanoma.Citation66
Several limitations were presented in this meta-analysis. 1) The included studies were conducted only in Asian and Caucasian populations, and other ethnicities were not considered. 2) The number of included studies for a certain SNP such as Arg280His variants was small, which may result in possible bias related to the selection of individual study participants. 3) Other risk factors such as age and gender should be analyzed. 4) The interaction of gene–gene and gene–environment should be included in future research because the combined effect of multiple SNPs in several genes of DNA repair pathways could have a large impact on pathological phenotypes.
Conclusion
Our results found that Arg399Gln polymorphism of XRCC1 gene might be a risk factor for NMSC in Asian populations and Arg194Trp variant of XRCC1 gene might be a protective factor for SCC patients. However, future large-scale, well-designed studies with more ethnicities are still needed to further investigate the role of XRCC1 polymorphisms in NMSC risk.
Disclosure
The authors report no conflicts of interest in this work.
References
- LeeDSkin cancerParkJWJungDiIntegumentary Physical TherapySpringerBerlin Heidelberg2016165191
- FranceschiSLeviFRandimbisonLLa VecchiaCSite distribution of different types of skin cancer: new aetiological cluesInt J Cancer199667124288690520
- FindlayMAllyMSRossDFarhadiehNWBulstrodeNCWiley Online Library. Non-melanoma skin cancersPlastic and Reconstructive Surgery: Approaches and Techniques2015100
- GriffinLLAliFRLearJTNon-melanoma skin cancerClin Med (Lond)2016161626526833519
- RaczJJoshuaAMLipaJESunAWrightFCNon-melanoma Skin CancerSpringer International PublishingBerlin Heidelberg2016225232
- EisemannNWaldmannAGellerACNon-melanoma skin cancer incidence and impact of skin cancer screening on incidenceJ Invest Dermatol20141341435023877569
- National cancer instituteSkin Cancer Prevention (PDQ®)National Cancer Institute2013 Available from: https://www.cancer.gov/types/skin/patient/skin-prevention-pdqAccessed June 29, 2017
- LeiterUEigentlerTGarbeCEpidemiology of skin cancerAdv Exp Med Biol201481012014025207363
- MicaliGLacarrubbaFBhattKNascaMRMedical approaches to non-melanoma skin cancerExpert Rev Anticancer Ther201313121409142124236820
- BuchananMALevinBVenessMNon-melanoma skin cancer: primary non-surgical therapies and prevention strategiesRiffatFPalmeCEVenessMNon-Melanoma Skin Cancer of the Head And NeckIndiaSpringer20153751
- GuyGPJrMachlinSREkwuemeDUYabroffKRPrevalence and costs of skin cancer treatment in the U.S., 2002–2006 and 2007–2011Am J Prev Med201448218318725442229
- WangL-EWeiQNucleotide Excision Repair: DNA Repair Capacity Variability and Cancer SusceptibilityDNA Repair and Cancer: From Bench to Clinic2013288 Available from: https://books.google.co.jp/books?hl=zh-CN&lr=&id=hmPOBQAAQBAJ&oi=fnd&pg=PA288&ots=Pq1v9bUU2m&sig=YwI2P2hJkGloxN5bT5q-JvWoR_Q&redir_esc=y#v=onepage&q&f=falseAccessed June 29, 2017
- BernsteinCAnilRNfonsamVBernsteiHDNA Damage, DNA Repair and Cancer2013
- RicceriFMatulloGVineisPIs there evidence of involvement of DNA repair polymorphisms in human cancer?Mutat Res20127361–211712121864546
- TaylorRMThistlethwaiteACaldecottKWCentral role for the XRCC1 BRCT I domain in mammalian DNA single-strand break repairMol Cell Biol20022282556256311909950
- LondonREThe structural basis of XRCC1-mediated DNA repairDNA Repair (Amst)2015309010325795425
- MokMCharacterization of Protein and DNA Interactions of the Human DNA Repair Protein XRCC1 [doctoral dissertation]McMaster UniversityHamilton, Ontario, Canada2016
- CornettaTFestaFTestaACozziRDNA damage repair and genetic polymorphisms: assessment of individual sensitivity and repair capacityInt J Radiat Oncol Biol Phys200666253754516965996
- LunnRMLangloisRGHsiehLLThompsonCLBellDAXRCC1 polymorphisms: effects on aflatoxin B1-DNA adducts and glycophorin A variant frequencyCancer Res199959112557256110363972
- GinsbergGAngleKGuytonKSonawaneBPolymorphism in the DNA repair enzyme XRCC1: utility of current database and implications for human health risk assessmentMutat Res20117271–211521352951
- TakanamiTNakamuraJKubotaYHoriuchiSThe Arg280His polymorphism in X-ray repair cross-complementing gene 1 impairs DNA repair abilityMutat Res20055821–213514515781218
- HungRJHallJBrennanPBoffettaPGenetic polymorphisms in the base excision repair pathway and cancer risk: a HuGE reviewAm J Epidemiol20051621092594216221808
- AuWWNavasumritPRuchirawatMUse of biomarkers to characterize functions of polymorphic DNA repair genotypesInt J Hyg Environ Health2004207430131315471094
- LomasALeonardi-BeeJBath-HextallFA systematic review of worldwide incidence of nonmelanoma skin cancerBr J Dermatol201216651069108022251204
- MoherDLiberatiATetzlaffJAltmanDGPRISMA GroupPreferred reporting items for systematic reviews and meta-analyses: the PRISMA statementPLoS Med200967e100009719621072
- Review Manager (RevMan) [Computer program]. Version 5.3CopenhagenThe Nordic Cochrane Centre, The Cochrane Collaboration2011Accessed January 5, 2016
- PatilKDCochran’s Q test: exact distributionJ Am Stat Assoc197570349186189
- DeeksJJAltmanDGBradburnMJMatthiasEGeorgeDSDouglasGAStatistical methods for examining heterogeneity and combining results from several studies in meta-analysisSystematic Reviews in Health Care: Meta-analysis in Context2nd edLondon, UKWiley Online Library, BMJ Publishing Group2001285312
- NelsonHHKelseyKTMottLAKaragasMRThe XRCC1 Arg399Gln polymorphism, sunburn, and non-melanoma skin cancer: evidence of gene-environment interactionCancer Res200262115215511782372
- YinJRockenbauerEHedayatiMMultiple single nucleotide polymorphisms on human chromosome 19q13.2-3 associate with risk of Basal cell carcinomaCancer Epidemiol Biomarkers Prev200211111449145312433725
- YinJVogelUGerdesLUDybdahlMBolundLNexoBATwelve single nucleotide polymorphisms on chromosome 19q13.2-13.3: linkage disequilibria and associations with basal cell carcinoma in Danish psoriatic patientsBiochem Genet2003411–2273712645871
- HanJColditzGALiuJSHunterDJGenetic variation in XPD, sun exposure, and risk of skin cancerCancer Epidemiol Biomarkers Prev20051461539154415941969
- FestaFKumarRSanyalSBasal cell carcinoma and variants in genes coding for immune response, DNA repair, folate and iron metabolismMutat Res20055741–210511115914210
- ThirumaranRKBermejoJLRudnaiPSingle nucleotide polymorphisms in DNA repair genes and basal cell carcinoma of skinCarcinogenesis20062781676168116501254
- KangSYLeeKGLeeWPolymorphisms in the DNA repair gene XRCC1 associated with basal cell carcinoma and squamous cell carcinoma of the skin in a Korean populationCancer Sci200798571672017355263
- ChiyomaruKNaganoTNishigoriCXRCC1 Arg194Trp polymorphism, risk of nonmelanoma skin cancer and extramammary Paget’s disease in a Japanese populationArch Dermatol Res2012304536337022639094
- SurduSFitzgeraldEFBloomMSPolymorphisms in DNA repair genes XRCC1 and XRCC3, occupational exposure to arsenic and sunlight, and the risk of non-melanoma skin cancer in a European case-control studyEnviron Res201413438238925218703
- HsuLIWuMMWangYHAssociation of environmental arsenic exposure, genetic polymorphisms of susceptible genes, and skin cancers in TaiwanBioMed Res Int2015201589257926295053
- ZhangHLiWFranklinMJDudekAZPolymorphisms in DNA repair gene XRCC1 and skin cancer risk: a meta-analysisAnticancer Res201131113945395222110224
- KrickerAArmstrongBKEnglishDRSun exposure and non-melanocytic skin cancerCancer Causes Control1994543673928080949
- WeiQMatanoskiGMFarmerERHedayatiMAGrossmanLDNA repair capacity for ultraviolet light-induced damage is reduced in peripheral lymphocytes from patients with basal cell carcinomaJ Invest Dermatol199510469339367769261
- MattaJLVillaJLRamosJMDNA repair and nonmelanoma skin cancer in Puerto Rican populationsJ Am Acad Dermatol200349343343912963906
- WangLELiCStromSSRepair capacity for UV light induced DNA damage associated with risk of nonmelanoma skin cancer and tumor progressionClin Cancer Res200713216532653917975167
- BendeskyAMichelASordoMDNA damage, oxidative mutagen sensitivity, and repair of oxidative DNA damage in nonmelanoma skin cancer patientsEnviron Mol Mutag2006477509517
- CampalansAMarsinSNakabeppuYO’ConnorTRBoiteuxSRadicellaJPXRCC1 interactions with multiple DNA glycosylases: a model for its recruitment to base excision repairDNA Repair (Amst)20054782683515927541
- VidalAEBoiteuxSHicksonIDRadicellaJPXRCCI coordinates the initial and late stages of DNA abasic site repair through protein-protein interactionsEMBO J200120226530653911707423
- CaldecottKWAoufouchiSJohnsonPShallSXRCC1 polypeptide interacts with DNA polymerase beta and possibly poly (ADP-ribose) polymerase, and DNA ligase III is a novel molecular ‘nick-sensor’ in vitroNucleic Acids Res19962422438743948948628
- HanssenbauerASolvanggartenKSundheimOXRCC1 coordinates disparate responses and multiprotein repair complexes depending on the nature and context of the DNA damageEnviron Mol Mutagen201152862363521786338
- WilsonDM3rdKimDBerquistBRSigurdsonAJVariation in base excision repair capacityMutat Res20117111–210011221167187
- KarahalilBBohrVAWilsonDM3rdImpact of DNA polymorphisms in key DNA base excision repair proteins on cancer riskHum Exp Toxicol20123110981100523023028
- JeggoPAPearlLHCarrAMDNA repair, genome stability and cancer: a historical perspectiveNat Rev Cancer2016161354226667849
- ZhangKZhouBWangYRaoLZhangLThe XRCC1 Arg280His polymorphism contributes to cancer susceptibility: an update by meta-analysis of 53 individual studiesGene201251029310122975644
- GdowiczklosokAWidelMRzeszowskawolnyJThe influence of XPD, APE1, XRCC1, and NBS1 polymorphic variants on DNA repair in cells exposed to X-raysMutat Res20137551424823669291
- BerquistBRSinghDKFanJFunctional capacity of XRCC1 protein variants identified in DNA repair-deficient Chinese hamster ovary cell lines and the human populationNucleic Acids Res201038155023503520385586
- NatukulaKJamilKPingaliURAttiliVSMadireddyURThe codon 399 Arg/Gln XRCC1 polymorphism is associated with lung cancer in IndiansAsian Pac J Cancer Prev20131495275527924175813
- Przybylowska-SygutKStanczykMKusinskaRKordekRMajsterekIAssociation of the Arg194Trp and the Arg399Gln polymorphisms of the XRCC1 gene with risk occurrence and the response to adjuvant therapy among Polish women with breast cancerClin Breast Cancer2012131616823103366
- ChenBZhouYYangPWuXTPolymorphisms of XRCC1 and gastric cancer susceptibility: a meta-analysisMol Biol Rep20123921305131321604176
- Al MutairiFMAlanaziMShalabyMAlabdulkarimHAPathanAAParineNRAssociation of XRCC1 gene polymorphisms with breast cancer susceptibility in Saudi patientsAsian Pac J Cancer Prev20131463809381323886187
- BajpaiDBanerjeeAPathakSJainSSinghNAbstract 5368: Genetic susceptibility to cervical cancer: role of XRCC1Cancer Res20147419 Suppl5368
- HouBHJianZXCuiPLiSJTianRQOuJRAssociation and intragenic single-nucleotide polymorphism interactions of the XRCC1 polymorphisms for pancreatic cancer susceptibilityPancreas201645454655126418909
- JiangHXuWZhangFQuantitative assessment of the association between XRCC1 Arg399Gln and Arg194Trp polymorphisms and risk of cutaneous melanomaMelanoma Res201626329029926967970
- MohanVMadhusudanSDNA base excision repair: evolving bio-markers for personalized therapies in cancerChenCNew Research Directions in DNA RepairInTechOpenRijeka, Croatia2013529557
- FiglASchererDNagoreESingle nucleotide polymorphisms in DNA repair genes XRCC1 and APEX1 in progression and survival of primary cutaneous melanoma patientsMutation Res20096611–2788419073198
- SacerdoteCGuarreraSRicceriFPolymorphisms in the XRCC1 gene modify survival of bladder cancer patients treated with chemotherapyInt J Cancer201313382004200923553206
- LiQWLuCRYeMXiaoWHLiangJEvaluation of DNA repair gene XRCC1 polymorphism in prediction and prognosis of hepatocellular carcinoma riskAsian Pac J Cancer Prev201213119119422502666
- BhandaruMMartinkaMLiGRotteALoss of XRCC1 confers a metastatic phenotype to melanoma cells and is associated with poor survival in patients with melanomaPigment Cell Melanoma Res201427336637524410901