Abstract
Renal cell carcinoma (RCC) is a common type of kidney cancer. Normally, surgical treatment can prolong life, but only for patients with early stage tumors. However, it is difficult for early detection strategies to distinguish between benign and malignant kidney tumors. Therefore, potential biomarkers for early diagnosis and prognosis of RCC are needed. Intriguingly, mounting evidence has demonstrated that many long noncoding RNAs (lncRNAs) are strongly linked to cancers. Indeed, promising RCC-associated lncRNA biomarkers have also been identified. However, the functional and prognostic roles of the antisense (AS) serum deprivation response (SDPR) lncRNA (SDPR-AS) in RCC remain largely unknown. The aims of this study were to investigate the expression and prognostic relevance of SDPR-AS in RCC. We uncovered the downregulated expressions of both lncRNA SDPR-AS and its protein-coding gene, SDPR, in RCC tissues compared to the matched normal tissues. Furthermore, SDPR-AS and SDPR expressions were positively correlated. Overexpression and knockdown experiments suggested that SDPR-AS and SDPR were coregulated in RCC cell lines. In addition, overexpression of SDPR-AS suppressed cell migration and invasion, but not cell growth. Furthermore, expression of SDPR-AS was associated with tumor differentiation and lymphatic metastasis. Kaplan–Meier survival and log-rank tests demonstrated the association of elevated expression of SDPR-AS with increased overall survival. In conclusion, our results suggest that the SDPR-AS may serve as a prognostic biomarker and therapeutic target of RCC.
Introduction
Renal cell carcinoma (RCC) accounts for approximately 2%–4% of all human malignancies and is one of the most common cancers in urology.Citation1,Citation2 Because kidney diseases are mostly asymptomatic during the early stages, many RCC patients are diagnosed with advanced stage metastatic diseases that are often resistant to chemotherapy, and therefore this is associated with high mortality.Citation3 Despite the great advances made in the diagnosis and treatment of RCC, therapeutic options are limited and prognosis remains unfavorable.Citation4 Therefore, there is a need to identify effective biomarkers for predicting the progression and prognosis of RCC and to advance the development of new targeted therapies.
During the past decades, numerous studies have revealed that a large portion of the human genome is universally transcribed into noncoding RNAs (ncRNAs).Citation5 Long ncRNAs (lncRNAs) are RNA molecules that are larger than 200 nucleotides in length and with no protein coding capabilities.Citation6,Citation7 Recently, many studies have shown that lncRNAs are involved in carcinogenesis, including invasion and metastasis of kidney, breast, and colorectal cancers.Citation8–Citation10 Particularly in RCC, it has been reported that high expression of lncRNA ATB promotes cell migration and invasion, and correlates with metastases,Citation11 while low expression of lncRNA NBAT-1 increases cell proliferation, migration, and invasion, and is also associated with poor prognosis.Citation12 In addition, many other lncRNAs, including TUG1,Citation13 BX357664,Citation14 and MEG3,Citation15 have been demonstrated to participate in the development and progression of RCC. Despite the identification of numerous lncRNAs in RCC, the majority remain unexplored, and this is therefore the reason for conducting this study.
Through in vitro experiments, Burgener et alCitation16 in 1990 discovered serum deprivation response (SDPR), also named cavin-2, located on chromosome 2q32–q33 as a substrate for protein kinase C isoforms. SDPR has been confirmed to be involved in inducing membrane curvature and caveolae formation, and it is strongly linked to gut electrophysiological pacing functions, cell proliferation, and migration.Citation17–Citation19 Importantly, previous studies have indicated that SDPR is down-regulated in kidney, prostate, and breast cancers.Citation20,Citation21 SDPR has also been reported to be a possible biomarker for early detection and discrimination of malignant kidney tumors.Citation22 Through bioinformatic analysis, we uncovered an lncRNA, termed AC098617.1, which is located at chromosome 2q32.3, ranging from 192,711,265 to 192,901,485, with the full length up to about 190,221 bp (data from UCSC website). AC098617.1 (Ensembl Gene ID: ENSG00000233766) with 13 transcripts is also known as LOC105373813 Gene or Uncharacterized LOC105373813. Of particular note is that the lncRNA is the antisense (AS) transcript of SDPR (SDPR-AS) and is located on the upstream of the gene. The expression and functions of lncRNAs are often associated with their adjacent protein coding genes.Citation23 It is therefore tempting to speculate that the expression and functions of SDPR-AS may also be associated with the progression and prognosis of RCC. In this study, we first investigated the expression profiles of SDPR-AS and SDPR in RCC. Subsequently, the functional roles of SDPR-AS in RCC cells were evaluated. Additionally, the correlations between SDPR-AS expression and clinical variables, as well as prognosis, were examined.
Materials and methods
Statement of ethics
The tissue specimens and clinical materials used in this study were collected after each participant gave written informed consent, which was in accordance with our institutional ethical guidelines. The ethical committee of the Chinese People’s Liberation Army (PLA) General Hospital approved the utilization of tumor tissues for this study.
Patients and samples
In total, 56 RCC patients who underwent initial surgery at the Chinese PLA General Hospital between 2005 and 2007 were enrolled into this research. Tumor and adjacent normal tissues were immediately snap-frozen in liquid nitrogen after resection, then maintained at −80°C until use.
Cell culture
The human RCC cell lines, OS-RC-2, 786-O, 769-P, Caki-1, Caki-2, and ACHN, as well as normal renal cell line HKC were obtained from American Type Culture Collection (ATCC, Manassas, VA, USA). All cells were cultured in Dulbecco’s Modified Eagle’s Medium (Hyclone, UT, USA), containing 10% heat-inactivated fetal bovine serum, penicillin (100 U/mL), and streptomycin (100 μg/mL) in a 5% CO2 and humidified atmosphere at 37°C.
SDPR-AS overexpression experiments
The universally available pcDNA3.1 vector was used for cDNA-SDPR-AS plasmid construction. Following the manufacturer’s instructions, the SDPR-AS low-expressing OS-RC-2 cells were transfected with pcDNA-SDPR-AS using Lipofectamine 2000 (Invitrogen). The transfected cells were harvested for RNA isolation and functional assays.
SDPR-AS knockdown experiments
Small interfering RNAs (siRNAs) for the target SDPR-AS (SDPR-AS-si) and negative-control siRNAs (NC-si) were obtained from GenePharma (Shanghai, People’s Republic of China), and the sequences are listed in . The SDPR-AS high-expressing 769-P cell line was selected for transfection with SDPR-AS-si. Prior to transfection, 12-well plates were seeded with approximately 50% of the cells and cultured for 24 h. Then, siRNA transfections were carried out with X-treme GENE transfection reagents (Roche) according to the manufacturer’s instructions. RNA was isolated after 48 h of transfection and used to perform functional assays.
Table 1 List of primers and their sequences used in this study
RNA isolation and reverse transcription
Total RNA was isolated from cells and tissues using Trizol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s protocol. NanoDrop ND2000 (Thermo Scientific Inc., Santa Clara, CA, USA) was used to determine the purity and quantify the concentration of RNA. A First Strand cDNA Synthesis Kit (TakaRa, Dalian, People’s Republic of China) was used to perform reverse transcription.
Real-time quantitative polymerase chain reaction
Primers used for real-time quantitative polymerase chain reaction (RT-qPCR) were obtained from GenePharma (Shanghai, People’s Republic of China), and the sequences are listed in . According to the manufacturer’s instructions, RT-qPCR was performed using the SYBR Prime-Script RT-PCR kit (Takara) in an Applied Biosystems 7500 fluorescent quantitative PCR system (Applied Biosystems, Foster City, CA, USA). The reaction started at 95°C for 5 min, followed by 40 cycles of 95°C for 30 s, 59°C for 30 s, and 72°C for 30 s. GAPDH was used as the internal reference to normalize qPCR results. Relative gene expression levels were measured using cycle threshold (CT) in the ΔΔCT calculation.
Cell proliferation assay
The CCK8 assay was used to examine the proliferation of RCC cells. After 48 h of transfection, cells were seeded into 96-well plates together with the CCK8 reagent and incubated for 1 h at 37°C. The Tecan infinite M200 multimode microplate reader (Tecan, Mechelen, Belgium) was then used to detect cell density at 450 nm absorption wavelength. All experiments were performed in triplicate.
Scratch wound-healing assay
The scratch wound-healing assay was used to evaluate the migratory properties of RCC cells. Cells were seeded into 12-well plates to achieve a confluent monolayer after 48 h of transfection. Then, uniform wounds were scraped with sterile 200 μL pipette tips. To remove floating cells, each well was washed thrice with PBS. The initial distance (0 h) and the distances traveled by cells after 24, 48, and 72 h of scratching were detected microscopically.
Cell invasion assay
Transwell assays were conducted with 8 mm pore size Matrigel Invasion Chambers (Millipore Corporation, Billerica, MA, USA) to evaluate the invasive properties of RCC cells. After 48 h of transfection, single-cell suspensions were seeded into the upper chamber, with the lower chamber filled with 600 μL Dulbecco’s Modified Eagle’s Medium containing 10% fetal bovine serum. After 24–48 h of incubation, the noninvading cells were wiped off from the upper surface of the membranes, whereas cells that had invaded were stained with 0.5% crystal violet solution after cell fixation in 95% methyl aldehyde. The images of migrated cells per well were captured and counted in random fields using a microscope. Each experiment was conducted in triplicate.
Statistical analyses
Statistical analyses were performed with SPSS version 18.0 (SPSS, Chicago, IL, USA). All experiments mentioned above were repeated three times, and data were presented as mean ± standard deviation. Two-tailed t-test and χ2 test were used for comparisons between different groups. Kaplan–Meier estimator was used to perform survival analysis. P<0.05 was considered statistically significant.
Results
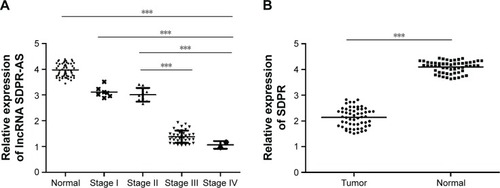
Downregulation of both SDPR-AS and SDPR expression in RCC tissue samples
The expression levels of SDPR-AS and its protein coding gene SDPR were examined by RT-qPCR in panel-paired specimens obtained from the 56 patients with RCC. Compared to the matched normal tissues, the expression levels of SDPR-AS (, ***P<0.001) and SDPR (, ***P<0.001) in tumor tissues were significantly downregulated.
Figure 1 Downregulation of both SDPR-AS and SDPR expression in RCC tissue samples.
Abbreviations: SDPR, serum deprivation response; SDPR-AS, antisense (AS) transcript of SDPR; RCC, renal cell carcinoma; TNM, tumor node metastasis.
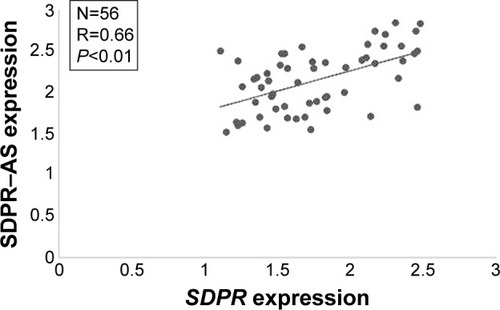
Expressions of SDPR-AS and SDPR are positively correlated
To investigate the relationship between SDPR-AS and SDPR, we assessed the correlation of their expression levels by RT-qPCR. The result showed a positive correlation in the levels of their expression (, N=56, R=0.66, P<0.01).
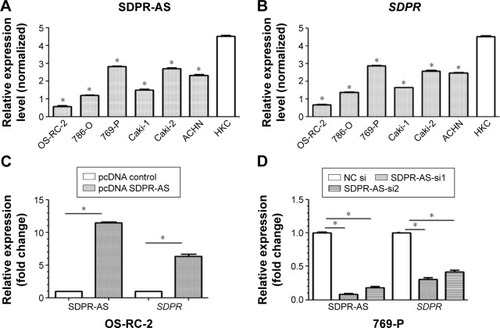
SDPR expression was coregulated with SDPR-AS overexpression or knockdown in RCC cell lines
RT-qPCR was performed to determine the expression levels of SDPR-AS and SDPR in six RCC cell lines (OS-RC-2, 786-O, 769-P, Caki-1, Caki-2, and ACHN) and the normal cell line (HKC). The results indicated that the expression levels of SDPR-AS and SDPR were significantly lower in the six RCC cell lines compared to the normal HKC cell line (, *P<0.05). In addition, as shown in , among the six RCC cell lines, the expression levels of SDPR-AS and SDPR were highest in the 769-P and lowest in the OS-RC-2 cell lines. After being transfected with pcDNA-SDPR-AS plasmid, SDPR-AS was overexpressed in the OS-RC-2 cell line, while the expression level of SDPR was apparently upregulated (, *P<0.05). On the contrary, SDPR and SDPR-AS expressions were significantly decreased in the 769-P cell line transfected with SDPR-AS-si (, *P<0.05). The data indicated that SDPR and SDPR-AS were coregulated in the RCC cell lines.
Figure 3 SDPR expression was co-regulated with SDPR-AS overexpression or knockdown in RCC cell lines.
Abbreviations: SDPR, serum deprivation response; SDPR-AS, antisense (AS) transcript of SDPR; NC, negative control; pc, plasmid complementary; HKC, renal tubular cell.
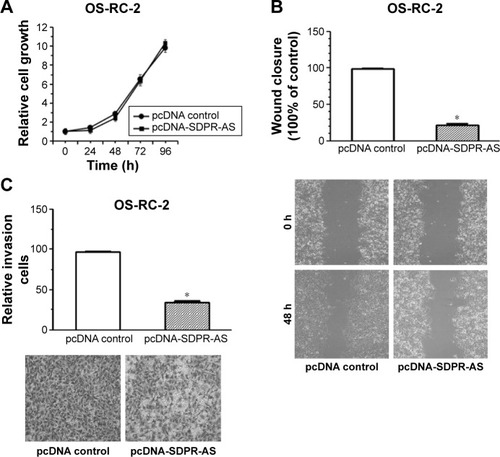
Overexpression of SDPR-AS suppressed the migration and invasion of OS-RC-2 cell line but had no evident association with proliferation
To further investigate the functional role of SDPR-AS, we examined its effects on OS-RC-2 cells transfected with pcDNA-SDPR-AS or the pcDNA control. There was no significant difference in the growth of OS-RC-2 cells transfected with either pcDNA-SDPR-AS or pcDNA control (). However, the wound healing assay showed significant inhibition of cell migration by pcDNA-SDPR-AS compared to pcDNA control (, *P<0.05). The matrigel invasion assay also revealed remarkable suppression of cell invasion by pcDNA-SDPR-AS compared to pcDNA control (, *P<0.05).
Figure 4 Overexpression of SDPR-AS suppressed the migration and invasion of OS-RC-2 cell line but had no evident association with proliferation.
Abbreviations: pc, plasmid complementary; SDPR, serum deprivation response; SDPR-AS, antisense (AS) transcript of SDPR.
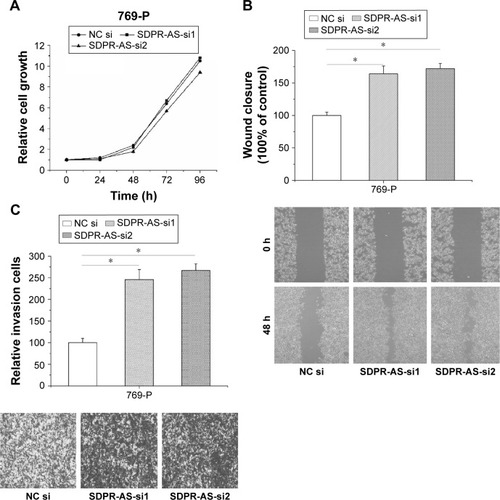
Knockdown of SDPR-AS promoted the migration and invasion of 769-P cell line but showed no evident association with proliferation
To further validate the functional role of SDPR-AS in RCC cells, we performed knockdown experiments by transfecting the 769-P cell line with SDPR-AS-si or NC si and examined their effects on cellular activity. There was no evident difference in proliferation of 769-P cells transfected with either SDPR-AS-si or NC si (). The wound healing and matrigel invasion assays, however, revealed significantly enhanced migration and invasion abilities of 769-P cells treated with SDPR-AS-si compared with those treated with NC si (, *P<0.05).
Figure 5 Knockdown of SDPR-AS promoted the migration and invasion of 769-P cell line but showed no evident association with proliferation.
Abbreviations: NC, negative control; SDPR, serum deprivation response; SDPR-AS, antisense (AS) transcript of SDPR.
Correlation analyses of SDPR-AS expression and clinical variables of RCC patients
Studies were carried out to assess the correlation between SDPR-AS and clinical characteristics of RCC patients. The statistical analysis revealed an evident association of SDPR-AS expression levels with differentiation grade, tumor node metastasis (TNM) stage, and lymph node metastasis (). However, no statistically significant associations were found in other clinicopathological features, including age, tumor size, and tumor location.
Table 2 Correlation between SDPR-AS expression and clinicopathologic features of RCC patients
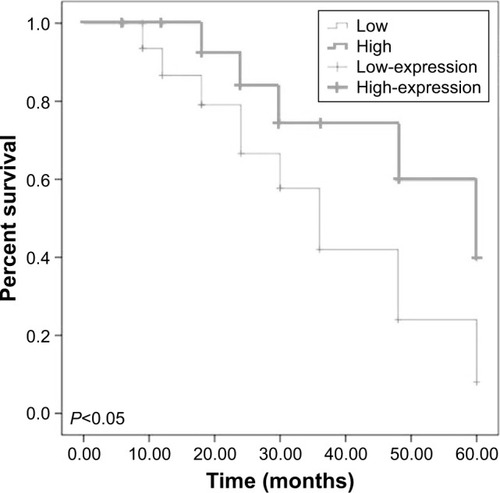
The association between SDPR-AS expression and prognosis of RCC patients
Kaplan–Meier survival and log-rank tests were performed to determine the association between SDPR-AS expression and prognosis. The overall expression levels of SDPR-AS were classified based on the mean relative expression ratio in RCC tissue specimens as high (SDPR-AS expression ratio ≥ median ratio, n=21) and low (SDPR-AS expression ratio ≤ median ratio, n=35). The data suggested that high expression of SDPR-AS was significantly associated with increased overall survival, whereas low expression indicated poor overall survival outcomes. Thus, SDPR-AS expression may be associated with prognosis of RCC patients (, *P<0.05).
Discussion
Dozens of lncRNAs have recently been discovered through RNA sequencing and are annotated by the GENCODE project.Citation24 Interest has been generated as a result of the fact that numerous lncRNAs are strongly linked with various cancers. For example, Chen et alCitation9 reported that long intergenic non-coding RNA ROR (lincRNA-ROR) was involved in breast cancer invasion through participating in the epithelial–mesenchymal transition process. Huang et alCitation25 demonstrated that low expression of DGCR5 was an important negative prognostic factor for hepatocellular carcinoma. In RCC, a number of lncRNAs, including CRNDE,Citation26 TCL6,Citation27 and MALAT-1Citation28 have been reported to be aberrantly expressed. Although alterations of lncRNAs in RCC have already been recognized, the functional roles of many remain to be understood. In the present study, we analyzed the functional roles of the lncRNA SDPR-AS and its prognostic relevance in RCC.
Recently, a number of studies have demonstrated that AS lncRNAs can regulate their sense mRNA partners in a discordant or concordant fashion.Citation29,Citation30 Numerous AS lncRNAs have been identified to be associated with a variety of malignant tumors.Citation31,Citation32 Generally, they modulate their sense partners by epigenetic regulation at the promoter regions or through other mechanisms.Citation33,Citation34 lncRNA SDPR-AS is the AS partner of the mRNA of SDPR. It has also been demonstrated that SDPR is an important tumor suppressor in certain cancers, including prostate cancer,Citation20,Citation35 breast cancer,Citation21,Citation36 and hepatocellular carcinoma.Citation37 In particular, a previous study reported that SDPR was remarkably downregulated in RCC cells,Citation22 which sparked our interest in conducting this study to investigate the functional and prognostic roles of its AS lncRNA in RCC.
In the present study, we first demonstrated the significant downregulation of SDPR-AS in RCC tissues in comparison to their matched nontumorous tissues. By loss-of-function and gain-of-function approaches, we confirmed that SDPR-AS played a critical role in cell migration and invasion, but had no evident effect on cell proliferation. In addition, the results also demonstrated a positive correlation between SDPR and SDPR-AS expression. Based on clinical data analysis, we determined that low expression of SDPR-AS was associated with tumor differentiation grade, TNM stage and lymph node metastasis. However, no statistically significant associations were found in other clinicopathological features, including age, tumor size, and tumor location. Furthermore, a multivariate analysis suggested that SDPR-AS expression level could serve as an independent prognostic predictor for RCC. Taken together, these results revealed a key role of SDPR-AS in RCC metastasis, possibly through modulation of the levels and functions of its sense partner. To our knowledge, our data may provide the first evidence for clarifying the functional and prognostic roles of SDPR-AS in RCC.
Conclusion
This study revealed the molecular mechanistic insights of SDPR-AS in RCC. In addition, this study provides some evidence that SDPR-AS may act as an independent prognostic predictor of RCC patients.
Acknowledgments
This work was supported by Medical Scientific Research Foundation of Heilongjiang Province, People’s Republic of China (2016–214) and China Postdoctoral Science Foundation.
Disclosure
The authors report no conflicts of interest in this work.
References
- LjungbergBCowanNCHanburyDCEAU guidelines on renal cell carcinoma: the 2010 updateEur Urol201058339840620633979
- LjungbergBCampbellSCChoiHYThe epidemiology of renal cell carcinomaEur Urol201160461562121741761
- ZhengTYangCGTargeting SPOP with small molecules provides a novel strategy for kidney cancer therapySci China Life Sci2017601919327888385
- PantuckAJZismanABelldegrunASThe changing natural history of renal cell carcinomaJ Urol200116651611162311586189
- GuttmanMAmitIGarberMChromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammalsNature2009458723522322719182780
- WashietlSKellisMGarberMEvolutionary dynamics and tissue specificity of human long noncoding RNAs in six mammalsGenome Res201424461662824429298
- GuttmanMRinnJLModular regulatory principles of large non-coding RNAsNature2012482738533934622337053
- RenXLanTChenYShaoZYangCPengJLncRNA uc009yby.1 promotes renal cell proliferation and is associated with poor survival in patients with clear cell renal cell carcinomasOncol Lett20161231929193427602119
- ChenYMLiuYWeiHYLvKZFuPLinc-ROR induces epithelial-mesenchymal transition and contributes to drug resistance and invasion of breast cancer cellsTumour Biol2016378108611087026883251
- IguchiTUchiRNambaraSA long noncoding RNA, lncRNA-ATB, is involved in the progression and prognosis of colorectal cancerAnticancer Res20153531385138825750289
- XiongJLiuYJiangLZengYTangWHigh expression of long non-coding RNA lncRNA-ATB is correlated with metastases and promotes cell migration and invasion in renal cell carcinomaJpn J Clin Oncol201646437838426848077
- XueSLiQWCheJPGuoYYangFQZhengJHDecreased expression of long non-coding RNA NBAT-1 is associated with poor prognosis in patients with clear cell renal cell carcinomaInt J Clin Exp Pathol2015843765377426097558
- ZhangMLuWHuangYDownregulation of the long noncoding RNA TUG1 inhibits the proliferation, migration, invasion and promotes apoptosis of renal cell carcinomaJ Mol Histol201647442142827323757
- LiuYQianJLiXLong noncoding RNA BX357664 regulates cell proliferation and epithelial-to-mesenchymal transition via inhibition of TGF-beta1/p38/HSP27 signaling in renal cell carcinomaOncotarget2016749814108142227806310
- WangMHuangTLuoGLong non-coding RNA MEG3 induces renal cell carcinoma cells apoptosis by activating the mitochondrial pathwayJ Huazhong Univ Sci Technolog Med Sci201535454154526223924
- BurgenerRWolfMGanzTBaggioliniMPurification and characterization of a major phosphatidylserine-binding phosphoprotein from human plateletsBiochem J199026937297342390065
- HansenCGBrightNAHowardGNicholsBJSDPR induces membrane curvature and functions in the formation of caveolaeNat Cell Biol200911780781419525939
- ElliottMHAshpoleNEGuXCaveolin-1 modulates intraocular pressure: implications for caveolae mechanoprotection in glaucomaSci Rep201663712727841369
- DrabMVerkadePElgerMLoss of caveolae, vascular dysfunction, and pulmonary defects in caveolin-1 gene-disrupted miceScience200129355392449245211498544
- LiXJiaZShenYCoordinate suppression of Sdpr and Fhl1 expression in tumors of the breast, kidney, and prostateCancer Sci20089971326133318422756
- OzturkSPapageorgisPWongCKSDPR functions as a metastasis suppressor in breast cancer by promoting apoptosisProc Natl Acad Sci U S A2016113363864326739564
- GianazzaEChinelloCMaininiVAlterations of the serum peptidome in renal cell carcinoma discriminating benign and malignant kidney tumorsJ Proteomics20127612514022868251
- PrekerPAlmvigKChristensenMSPROMoter uPstream Transcripts share characteristics with mRNAs and are produced upstream of all three major types of mammalian promotersNucleic Acids Res201139167179719321596787
- HarrowJFrankishAGonzalezJMGENCODE: the reference human genome annotation for The ENCODE ProjectGenome Res20122291760177422955987
- HuangRWangXZhangWDown-regulation of LncRNA DGCR5 correlates with poor prognosis in hepatocellular carcinomaCell Physiol Biochem2016403–470771527898409
- ShaoKShiTYangYWangXXuDZhouPHighly expressed lncRNA CRNDE promotes cell proliferation through Wnt/beta-catenin signaling in renal cell carcinomaTumour Biol Epub2016106
- SuHSunTWangHDecreased TCL6 expression is associated with poor prognosis in patients with clear cell renal cell carcinomaOncotarget2017845789579927494890
- ChenSMaPZhaoYBiological function and mechanism of MALAT-1 in renal cell carcinoma proliferation and apoptosis: role of the MALAT-1-Livin protein interactionJ Physiol Sci Epub2016921
- FaghihiMAWahlestedtCRegulatory roles of natural antisense transcriptsNat Rev Mol Cell Biol200910963764319638999
- WahlestedtCNatural antisense and noncoding RNA transcripts as potential drug targetsDrug Discov Today20061111–1250350816713901
- YangXSongJHChengYLong non-coding RNA HNF1A-AS1 regulates proliferation and migration in oesophageal adenocarcinoma cellsGut201463688189024000294
- TakayamaKHorie-InoueKKatayamaSAndrogen-responsive long noncoding RNA CTBP1-AS promotes prostate cancerEMBO J201332121665168023644382
- MussotterTKluweLHogelJNon-coding RNA ANRIL and the number of plexiform neurofibromas in patients with NF1 microdeletionsBMC Med Genet2012139823101500
- PasmantESabbaghAVidaudMBiecheIANRIL, a long, noncoding RNA, is an unexpected major hotspot in GWASFASEB J201125244444820956613
- TentaRKatopodisHChatziioannouAMicroarray analysis of survival pathways in human PC-3 prostate cancer cellsCancer Genomics Proteomics20074430931817878531
- TianYYuYHouLKSerum deprivation response inhibits breast cancer progression by blocking transforming growth factor-beta signalingCancer Sci2016107327428026749136
- JingWLuoPZhuMAiQChaiHTuJPrognostic and diagnostic significance of SDPR-Cavin-2 in hepatocellular carcinomaCell Physiol Biochem201639395096027513662