Abstract
Purpose
This meta-analysis study aimed to reveal the prognostic relevance of ubiquitin-specific protease 22 (USP22) expression in patients with cancers.
Methods
PubMed, Embase, and the Cochrane Library electronic databases were searched for relevant studies published up to April 2017. The prognostic value of USP22 expression was evaluated by hazard ratio with 95% confidence intervals (CIs). Relative risk (RR) with 95% CIs assessed the effects of USP22 expression on clinicopathological parameters. A total of 16 studies of 2,233 Chinese patients were included in the final meta-analysis.
Results
A significant association was found between USP22 overexpression and survival in patients with cancers. The pooled RR indicated that USP22 overexpression was related to histological grade, advanced tumor–node–metastasis stage, positive lymph node metastasis, and distant metastasis.
Conclusion
This meta-analysis demonstrated that USP22 could be a novel biomarker for predicting prognosis in patients with cancers in the Chinese population.
Introduction
Cancer is a group of diseases involving abnormal cell growth with the potential to invade or spread to other parts of the body. It has a high mortality rate,Citation1 but the number of survivors has slightly increased because of improvements in treatment modalities and screening programs that identify the grade, stage, and size of the tumor in the last 10 years; however, these clinicopathological factors cannot fully predict individual outcomes.Citation2,Citation3 Therefore, to improve clinical care, it is imperative to explore effective biological prognostic markers.
Ubiquitin-specific protease 22 (USP22) is a subunit of the human SAGA transcriptional cofactor acetylation complex that functions to deubiquitinate histones H2B and H2A.Citation4 USP22 was identified as one of the 11-gene “death-from-cancer” signatures that predict rapid disease recurrence, distant metastatic sites, and poor response to therapy across multiple solid tumors.Citation5,Citation6 Its expression correlates with a stem cell-like expression profile and appears to be driven by the BMI-1 oncogene. Previous studies showed that USP22 enhances cancer cell proliferation through interaction with Rb and p53.Citation7–Citation9 USP22 is recruited by the Myc oncoprotein or nuclear receptors and counteracts heterochromatin silencing, thereby transactivating specific target genes in cancer cells.Citation10,Citation11 USP22 activation markedly contributes to aberrant cell cycle control and anoikis resistance, and its aberrant expression is associated with cancer progression and poor prognosis.Citation8,Citation12,Citation13
Many studies have evaluated the prognostic value of USP22 overexpression in patients with cancer; some other conflicting conclusions were arrived.Citation14,Citation15 Therefore, the impact of USP22 expression on cancer prognosis remains controversial. To better understand the relationship between cancer pathogenesis and USP22 overexpression, we performed a meta-analysis to evaluate the effects of USP22 on survival and clinicopathological parameters in patients with cancer.
Methods
Search strategy
A systematic literature search of the databases PubMed, Embase, and the Cochrane Library was performed to identify relevant English language studies published up to April 2017 using the following keywords: “USP22” AND “prognosis” OR “prognostic” OR “survival” OR “outcome” AND “cancer” OR “carcinoma” OR “neoplasm” OR “tumor” OR “malignant”. We also inspected the references listed in identified studies to find additional relevant studies. Eligible reports were identified independently by two investigators (NA and LW).
Inclusion criteria
Eligible studies had to meet the following inclusion criteria: 1) evaluating USP22 expression in human cancer tissues; 2) evaluating the association between USP22 expression and cancer prognosis; and 3) providing sufficient information to estimate hazard ratios (HRs) with 95% confidence intervals (CIs). The exclusion criteria were as follows: 1) reviews, case reports, letters, and conference abstracts and 2) overlapping data. A flow diagram of the study selection process is shown in . Because immunohistochemistry (IHC) is the most common method used to detect USP22 expression in tumors, and as mRNA expression levels were analyzed by reverse transcription polymerase chain reaction in cancer tissues than in paired adjacent normal tissues,Citation16–Citation23 we did not include studies reporting relationship of USP22 mRNA expression and clinicopathological parameters of patients in our current meta-analysis.
Data extraction
Data from eligible studies were extracted independently by two investigators (NA and LW). The following items were collected: first author’s name, year of publication, nationality, number of cases, cancer type, method of detection, cutoff value of USP22 positivity, HRs with corresponding 95% CIs for disease-free survival (DFS) and overall survival (OS) analyses, and clinicopathological parameters. Any disagreements were settled by a third investigator (NA).
Quality assessment
The quality of included studies was assessed by two independent investigators (NA and LW) on the basis of the Newcastle–Ottawa scale (NOS) 9-point scoring system.Citation24 Studies with an NOS score of ≥6 were regarded as high quality.
Statistical analysis
HRs with their 95% CIs were used to evaluate the association between USP22 expression levels and survival. If available, we obtained the data directly from the article. Alternatively, we reviewed the data from Kaplan–Meier survival curves using the Engauge Digitizer version 4.1 software. All included HRs derived from univariable models. Then, we stratified the variables by clinicopathological parameters of patients shown to contribute to poor survival, including lymph invasion, M stage, tumor–node–metastasis (TNM) stage, and histodifferentiation. The impact of USP22 expression on clinicopathological parameters was assessed by relative risks (RRs) with their 95% CIs. Heterogeneity among the included studies was checked by the chi-squared Q test.Citation25 If the studies were heterogeneous (P<0.10 or I2>50%), the random effects model was used; otherwise, the fixed effects model was used.Citation26 A sensitivity analysis was carried out to evaluate whether any single study affected the pooled univariable HRs by sequentially discarding individual studies. Egger’s test and Begg’s funnel plots were applied to assess publication bias.Citation27 All statistical analyses were performed using the R 3.2.0 meta-analysis software. P-values <0.05 were considered statistically significant.
Results
Study characteristics
A total of 86 studies were selected following an initial search, of which 16 eligible studies from 2011 to 2015 met the inclusion and exclusion criteria and were included in this meta-analysis. The articles from January 2016 to April 2017 do not meet the inclusion criteria that we mentioned. The main characteristics of the eligible studies are summarized in . All studies were performed exclusively in patients from China. Seven of the studies were of patients with digestive system cancers (colorectal cancer, hepatocellular cancer, esophageal cancer, gastric cancer, and pancreatic cancer),Citation16,Citation18,Citation23,Citation28–Citation31 two of the studies were of patients with salivary carcinoma,Citation20,Citation22 and seven of the studies were of patients with other cancer types (oral squamous cell carcinoma, lung cancer, glioma cancer, breast cancer, epithelial ovarian cancer, and papillary thyroid carcinoma).Citation17,Citation19,Citation21,Citation32–Citation35 IHC was the main method used to detect USP22 expression in these studies.
Table 1 Main characteristics of the studies included in this meta-analysis
Meta-analysis results
USP22 expression and survival
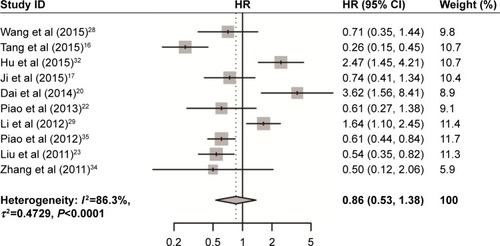
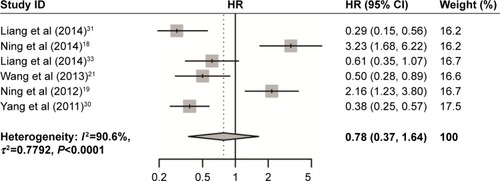
As shown in and , USP22 overexpression was significantly associated with poor DFS (HR =0.86, 95% CI: 0.53–1.38) and OS (HR =0.78, 95% CI: 0.37–1.64) in cancers. Owing to the heterogeneity detected in these studies, the random effects model was applied.
USP22 expression and clinicopathological parameters
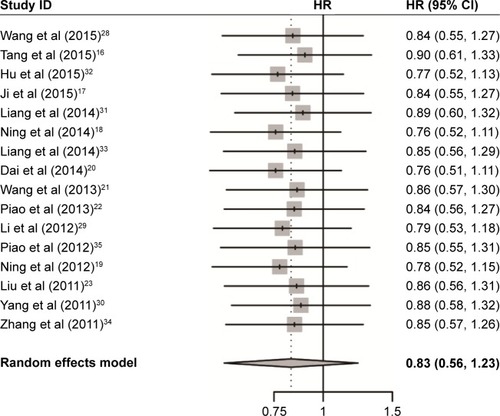
As shown in and , increased USP22 was significantly associated with histological grade (RR =0.72, 95% CI: 0.66–0.78, P=0.002), advanced TNM stage (RR =0.55, 95% CI: 0.49–0.61, P=0.000), positive lymph node metastasis (RR =0.69, 95% CI: 0.64–0.75, P=0.000), and positive distant metastasis (RR =0.66, 95% CI: 0.61–0.72, P=0.017) but not with gender (P=0.748). Owing to insufficient data, we did not detect a relationship between USP22 overexpression and other clinicopathological parameters.
Figure 4 Forest plot of the relationship between high USP22 expression and patient clinicopathological parameters.
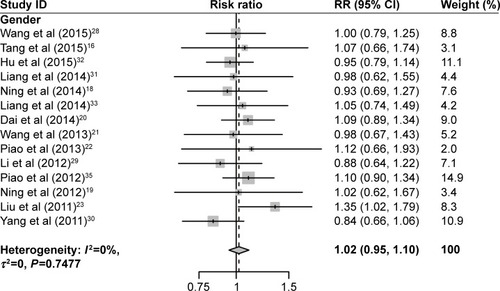
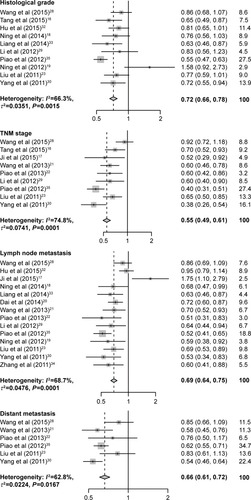
Table 2 Meta-analysis: RR value and 95% CI of clinicopathological parameters
Sensitivity analysis
Sensitivity analysis revealed no significant changes in HRs when any individual study was discarded (). The investigation of the impact of USP22 overexpression on survival gave a Begg’s test score of P=0.533 and an Egger’s test score of P=0.368, reflecting no evidence of publication bias in the analysis.
Discussion
The USP22 gene is located on human chromosome 17 and consists of 14 exons. Its 1,578 bp open reading frame encodes a 525 amino acid polypeptide with a molecular weight of ~66 kDa, containing a C-terminal peptidase domain of the C19 family and an N-terminal zinc finger motif that mediates the association between the enzyme and other proteins.Citation8,Citation12 USP22 was previously shown to be weakly expressed or absent in normal tissues but highly expressed in various cancers.Citation36 Although the focus area of much USP22 research is digestive system cancers, the role of USP22 in other malignant tumors has received increasing attention in recent years. Several studies reported that USP22 could be considered as an important prognostic marker for malignant cancers.Citation19,Citation23,Citation29,Citation32,Citation33,Citation37,Citation38 However, because only some reports correlated USP22 expression with prognosis, no pooled analysis has previously been performed. Our study conducted a pooled analysis of the prognostic value of USP22 expression relevant to 2,233 patients with cancers in China. Some independent prognostic factors are indicated via pooled analysis in general.
In the present meta-analysis, high USP22 expression was associated with worse survival in patients with cancer. Schrecengost et alCitation39 previously showed that USP22 depletion dramatically downregulated androgen receptor protein levels and abolished androgen receptor activity in both androgen deprivation therapy and castration-resistant prostate adenocarcinoma cells. Xu et alCitation7 speculated that the observed increase in USP22 activity in CRC might stabilize major vault protein by rescuing it from proteolysis and thus contributing to drug resistance. Therefore, the potential use of USP22 as a target for cancer therapy warrants further study.
We stratified the variables by clinicopathological parameters of patients shown to contribute to poor survival, including lymph invasion, M stage, TNM stage, and histodifferentiation. The random effects model was applied because of the significant heterogeneity observed for the association between evaluated USP22 expression and histological grade, TNM stage, and distant metastasis. Our meta-analysis has some limitations that should be acknowledged. First, most of the included studies focused on digestive system cancers and the small number of studies focusing on other cancer types might weaken our conclusions; thus, additional studies of different cancers could be included in a future meta-analysis. Second, all the included studies were of patients from China; hence, our conclusion might not be reliably extrapolated to non-Asian populations. Third, our extraction of data from Kaplan–Meier curves to derive HRs with 95% CIs might not have been reliable. Finally, although all studies detected USP22 expression by IHC, the use of different antibody concentrations and variable cutoff values might have influenced the results.
Conclusion
This meta-analysis indicated that USP22 overexpression was significantly associated with poor prognosis in cancer patients and that it could be used as a biomarker to predict clinical outcomes, especially in patients with cancers of the digestive system. Further clinical research is needed to support our conclusions and to confirm the precise prognostic value of USP22 in cancers.
Author contributions
All authors contributed toward data analysis, drafting and critically revising the paper and agree to be accountable for all aspects of the work.
Acknowledgments
The authors acknowledge the previous authors researched on USP22 and PubMed, Embase, and the Cochrane Library electronic databases.
Disclosure
The authors report no conflicts of interest in this work.
References
- SchneiderGSchmidt-SupprianMRadRSaurDTissue-specific tumorigenesis: context mattersNat Rev Cancer201717423925328256574
- AllardBLonghiMSRobsonSCStaggJThe ectonucleotidases CD39 and CD73: novel checkpoint inhibitor targetsImmunol Rev2017276112114428258700
- AndrewsLPMarciscanoAEDrakeCGVignaliDALAG3 (CD223) as a cancer immunotherapy targetImmunol Rev20172761809628258692
- ZhangXYPfeifferHKThorneAWMcMahonSBUSP22, an hSAGA subunit and potential cancer stem cell marker, reverses the polycomb-catalyzed ubiquitylation of histone H2ACell Cycle20087111522152418469533
- GlinskyGVBerezovskaOGlinskiiABMicroarray analysis identifies a death-from-cancer signature predicting therapy failure in patients with multiple types of cancerJ Clin Invest200511561503152115931389
- GlinskyGVGenomic models of metastatic cancer: functional analysis of death-from-cancer signature genes reveals aneuploid, anoikis-resistant, metastasis-enabling phenotype with altered cell cycle control and activated Polycomb Group (PcG) protein chromatin silencing pathwayCell Cycle20065111208121616760651
- XuHLiuYLYangYMDongXSKnock-down of ubiquitin-specific protease 22 by micro-RNA interference inhibits colorectal cancer growthInt J Colorectal Dis2012271213021773699
- CaoJYanQHistone ubiquitination and deubiquitination in transcription, DNA damage response, and cancerFront Oncol201222622649782
- DingFBaoCTianYUSP22 promotes NSCLC tumorigenesis via MDMX up-regulation and subsequent p53 inhibitionInt J Mol Sci201416130732025547493
- PijnappelWWTimmersHTDubbing SAGA unveils new epigenetic crosstalkMol Cell200829215215418243109
- ZhaoYLangGItoSA TFTC/STAGA module mediates histone H2A and H2B deubiquitination, coactivates nuclear receptors, and counteracts heterochromatin silencingMol Cell20082919210118206972
- ColeAJClifton-BlighRMarshDJHistone H2B monoubiquitination: roles to play in human malignancyEndocr Relat Cancer2015221T19T3324891457
- ZhangXYVarthiMSykesSMThe putative cancer stem cell marker USP22 is a subunit of the human SAGA complex required for activated transcription and cell-cycle progressionMol Cell200829110211118206973
- AoNLiuYBianXFengHLiuYUbiquitin-specific peptidase 22 inhibits colon cancer cell invasion by suppressing the signal transducer and activator of transcription 3/matrix metalloproteinase 9 pathwayMol Med Rep20151222107211325902005
- AoNLiuYFengHUbiquitin-specific peptidase USP22 negatively regulates the STAT signaling pathway by deubiquitinating SIRT1Cell Physiol Biochem20143361863187524969755
- TangBLiangXTangFExpression of USP22 and Survivin is an indicator of malignant behavior in hepatocellular carcinomaInt J Oncol20154762208221626497847
- JiMShiHXieYUbiquitin specific protease 22 promotes cell proliferation and tumor growth of epithelial ovarian cancer through synergy with transforming growth factor beta1Oncol Rep201533113314025369910
- NingZWangALiangJUSP22 promotes the G1/S phase transition by upregulating FoxM1 expression via beta-catenin nuclear localization and is associated with poor prognosis in stage II pancreatic ductal adenocarcinomaInt J Oncol20144541594160824993031
- NingJZhangJLiuWLangYXueYXuSOverexpression of ubiquitin-specific protease 22 predicts poor survival in patients with early-stage non-small cell lung cancerEur J Histochem2012564e4623361242
- DaiWYaoYZhouQSunCFUbiquitin-specific peptidase 22, a histone deubiquitinating enzyme, is a novel poor prognostic factor for salivary adenoid cystic carcinomaPLoS One201491e8714824466336
- WangHLiYPChenJHPrognostic significance of USP22 as an oncogene in papillary thyroid carcinomaTumour Biol20133431635163923412977
- PiaoSMaJWangWIncreased expression of USP22 is associated with disease progression and patient prognosis of salivary duct carcinomaOral Oncol201349879680123664741
- LiuYLYangYMXuHDongXSAberrant expression of USP22 is associated with liver metastasis and poor prognosis of colorectal cancerJ Surg Oncol2011103328328921337558
- StangACritical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analysesEur J Epidemiol201025960360520652370
- HigginsJPThompsonSGDeeksJJAltmanDGMeasuring inconsistency in meta-analysesBMJ2003327741455756012958120
- LauJIoannidisJPSchmidCHQuantitative synthesis in systematic reviewsAnn Intern Med199712798208269382404
- EggerMDavey SmithGSchneiderMMinderCBias in meta-analysis detected by a simple, graphical testBMJ199731571096296349310563
- WangZZhuLGuoTWangYYangJDecreased H2B monoubiquitination and overexpression of ubiquitin-specific protease enzyme 22 in malignant colon carcinomaHum Pathol20154671006101425971547
- LiJWangZLiYUSP22 nuclear expression is significantly associated with progression and unfavorable clinical outcome in human esophageal squamous cell carcinomaJ Cancer Res Clin Oncol201213881291129722447106
- YangDDCuiBBSunLYThe co-expression of USP22 and BMI-1 may promote cancer progression and predict therapy failure in gastric carcinomaCell Biochem Biophys201161370371021735131
- LiangJXNingZGaoWUbiquitin specific protease 22 induced autophagy is correlated with poor prognosis of pancreatic cancerOncol Rep20143262726273425241857
- HuJYangDZhangHUSP22 promotes tumor progression and induces epithelial-mesenchymal transition in lung adenocarcinomaLung Cancer201588323924525907317
- LiangJZhangXXieSUbiquitin-specific protease 22: a novel molecular biomarker in glioma prognosis and therapeuticsMed Oncol201431489924573640
- ZhangYYaoLZhangXElevated expression of USP22 in correlation with poor prognosis in patients with invasive breast cancerJ Cancer Res Clin Oncol201113781245125321691749
- PiaoSLiuYHuJUSP22 is useful as a novel molecular marker for predicting disease progression and patient prognosis of oral squamous cell carcinomaPLoS One201278e4254022880026
- Melo-CardenasJZhangYZhangDDFangDUbiquitin-specific peptidase 22 functions and its involvement in diseaseOncotarget2016728448484485627057639
- YangMLiuYDWangYYLiuTBGeTTLouGUbiquitin-specific protease 22: a novel molecular biomarker in cervical cancer prognosis and therapeuticsTumour Biol201435292993423979981
- HuJLiuYLPiaoSLYangDDYangYMCaiLExpression patterns of USP22 and potential targets BMI-1, PTEN, p-AKT in non-small-cell lung cancerLung Cancer201277359359922717106
- SchrecengostRSDeanJLGoodwinJFUSP22 regulates oncogenic signaling pathways to drive lethal cancer progressionCancer Res201474127228624197134