Abstract
Aim
To explore the expression and clinical significance of long noncoding RNA (lncRNA) gastric cancer-associated transcript 3 (GACAT3) in human colorectal cancer (CRC).
Methods
Expression of GACAT3 in CRC tissues and cell lines was measured using quantitative real-time PCR. CCK-8 and colony formation assays were used to assess the effect of GACAT3 on CRC cell line proliferation. Transwell invasion and migration assays were performed to detect the effect of GACAT3 on CRC cell line invasion and migration. Bioinformatics prediction, luciferase reporter assay, and pull-down assay were used to determine if miR-149 was a target of GACAT3. In addition, we also conducted colony formation assays and invasion assays to verify that GACAT3 promotes tumor progression through miR-149. Finally, in vivo tumorigenesis studies were used to demonstrate subcutaneous tumor growth.
Results
In the present study, we found that GACAT3 was highly expressed in CRC tissues and cell lines. Si-GACAT3 significantly decreased cell proliferation, motility, and invasiveness both in vitro and in vivo. We confirmed that downregulated GACAT3 significantly increased the expression of miR-149, and miR-149 binds to GACAT3 in a sequence-specific manner using luciferase reporter assays and pull-down assay. Further functional experiments indicated that GACAT3 could directly upregulate SP1 and STAT3 expressions by functioning as a competing endogenous RNA for miR-149, and consequentially, promoting CRC cell proliferation and invasion in vitro.
Conclusion
This study demonstrated that GACAT3 promotes tumor progression through competitive binding to miR-149 and suggests a promising new strategy for anti-CRC therapy.
Introduction
Worldwide, colorectal cancer (CRC) is the third most common cancer and the fourth most common cause of cancer-related death.Citation1 Despite the diagnosis and therapeutic advances in CRC, a significant proportion of CRC patients experience recurrence and die within 5 years after surgical treatment of their primary tumor.Citation2 Thus, it is necessary to elucidate the molecular mechanisms underlying CRC proliferation. In the past decades, intensive investigations identified a variety of molecular markers for CRC characterization and prognosis.Citation3,Citation4 Recently, evidences have suggested that various long noncoding RNAs (lncRNAs) also act as modulators in carcinogenesis and progression of human CRC and may serve as novel therapeutic targets.Citation5–Citation7
As a novel class of regulatory genes, lncRNAs (>200 nucleotides in length) have limited or no protein-coding capacity.Citation8,Citation9 Evidences suggest that lncRNAs play important roles in a wide range of cellular processes, such as epithelial–mesenchymal transition (EMT)Citation10 and cell proliferation and migration.Citation11
Studies indicate that lncRNAs are frequently aberrantly expressed in diverse human diseases, including various cancers.Citation12,Citation13 Several studies have revealed that lncRNAs can regulate gene expression at the transcriptional, posttranscriptional, and epigenetic levels.Citation14,Citation15 A large number of lncRNAs may function by competing with endogenous RNA (ceRNA) for miRNA targets, thereby preventing a single miRNA or multiple miRNAs from binding to their proper regulatory targets.Citation16,Citation17 A growing body of evidences strongly suggest that several lncRNAs, such as linc-NeD12,5,Citation18 HOTAIR,Citation19 and MALAT1,Citation20 may function as ceRNAs, exerting essential roles in many biological processes.
Gastric cancer-associated transcript 3 (GACAT3), previously named as AC130710, has been officially named as GACAT3 now by the HUGO Gene Nomenclature Committee (HGNC).Citation21 GACAT3 acts as a potential prognosis marker of GC.Citation22 In GC cells, the expression of GACAT3 was suppressed by miR-129-5p, a tumor suppressor downregulated in the GC cells.Citation23 However, the functions of GACAT3 in CRC and the underlying mechanisms remain unknown. In the present study, we found that GACAT3 was highly expressed in CRC. Knockdown of GACAT3 inhibited the malignancy of CRC cells, including proliferation, motility, and invasiveness. Meanwhile, we found that downregulated GACAT3 significantly increased the expression of miR-149. Bioinformatics, luciferase reporter assays, and pull-down assay confirmed that miR-149 binds to GACAT3 in a sequence-specific manner. This suggests that GACAT3-mediated derepression of miR-149 is a potential mechanism by which this lncRNA promotes cell proliferation in CRC.
Methods
Patient samples
Twenty-six human CRC and their adjacent noncancerous tissues were collected from patients who underwent surgery for CRC at the Tumor Tissue Bank of Central Hospital of Cangzhou. There was no radiotherapy or chemotherapy prior to the operation. The adjacent noncancerous tissues were 5 cm from the edge of tumor. The diagnoses of these CRC and noncancerous tissues were verified by pathologists. All of the experimental protocols were conducted in accordance with the approved guidelines, and the study was approved by the Institutional Research Committee of Cangzhou Medical College. Written informed consent was provided by all the patients.
Cell culture and transfection
The following cell lines were obtained from the Cell Bank of the Chinese Academy of science: FHC, HT29, HCT116, SW480, LoVo, and 293T. Cells were cultured in Dulbecco’s Modified Eagle’s Medium, Iscove’s modified Dulbecco’s medium, or F-12K medium supplemented with 10% fetal bovine serum (FBS), 100 U/mL penicillin, and 100 mg/mL streptomycin (GIBCO, Carlsbad, CA, USA) in humidified air at 37°C with 5% CO2. Cell transfections were performed using the Lipofectamine 3000 kit (Thermo Fisher Scientific, San Jose, CA, USA) according to the manufacturer’s instructions.
Vector construction
GACAT3 gene inhibitor plasmid and the respective empty vector plasmids were purchased from GeneCopoeia™ (Rockville, MD, USA). The target sequence of si-GACAT3 is 5′-GGAUGAGAUGAUGAGGAUUTT-3′. The target GACAT3-wild and GACAT3-mutant gene expression plasmids (p-GACAT3′-UTR-wt and p-GACAT3′ UTR-mut) were purchased from GeneCopoeia (Rockville, MD, USA). MiR-149 gene inhibitor plasmid was purchased from GeneCopoeia (Rockville, MD, USA) and its target sequence is 5′-AATTCAAAAAAGGGAGGGACGGGGGCTGTGC-3′.
RNA extraction and quantitative real-time PCR analysis
Samples were immediately frozen in liquid nitrogen after surgical dissection. Total RNA was extracted using TRIzol Reagent (Thermo Fisher Scientific, San Jose, CA, USA) according to the manufacturer’s protocol. RNA quantity was measured by the Nano Drop ND-2000 spectrophotometer (optical density 260 nm, Nano Drop, Wilmington, DE, USA), and RNA integrity was assessed using standard denaturing agarose gel electrophoresis. The expression of GACAT3 lncRNA from tissue samples or the cultured cells was quantified according to the manufacturer’s instructions using SYBR Premix EX Taq™ II kit (TaKaRa, RR820A) on the ABI Prism 7500 (Applied Biosystems, Foster City, CA, USA). MiRNA was harvested using the PureLink™ miRNA Isolation Kit (Thermo Fisher Scientific), and miRNA expression was quantified by TaqMan MicroRNA Assay Kit. The relative expression levels of mRNA/lncRNA and miRNA were determined using the 2−ΔΔCT method with human glyceraldehyde 3-phosphate dehydrogenase (GAPDH) and U6 snRNA as internal controls, respectively. The sequences of the primers used in the study are shown in . Each experiment was performed in triplicate.
Western blot assay
Tissue samples and the treated cells were lysed in Radio Immunoprecipitation Assay (RIPA) buffer (Thermo Scientific, Rockford, IL, USA) and Protease Inhibitor (Thermo Fisher Scientific, Waltham, MA, USA). The total protein (30 μg) in equal concentration was separated by 10% SDS/PAGE gels and transferred to a polyvinylidene fluoride (PVDF) membrane (Millipore Corporation, Billerica, MA, USA). The PVDF membranes were blocked and incubated with the primary antibodies at 4°C overnight. The membranes were then incubated with horseradish peroxidase-conjugated secondary antibody for 1 h at room temperature. All blots were visualized using an enhanced chemiluminescence (ECL) substrate kit (Amersham Biosciences, Little Chalfont, United Kingdom) and an ECL detection system (Amersham Biosciences). The antibodies used were rabbit anti-GAPDH (1:1,000; Santa Cruz Biotechnology, Santa Cruz, CA, USA) and rabbit anti-STAT3 (1:500; Abcam, Cambridge, UK).
Bioinformatics prediction and luciferase reporter assay
The potential miRNA binding sites of GACAT3 predicted by computer-aided algorithms were obtained from microRNA.org target program (www.microRNA.org). Two luciferase reporters containing the wild-type (WT) GACAT3 (psiCHECK2-GACAT3-WT) or mutant GACAT3 were generated to analyze the interaction between GACAT3 and miR-149. Mutant GACAT3 contained a mutation site (psiCHECK2-GACAT3-MU) abolishing targeting by miR-149. CRC cells were cotransfected with 500 ng of the luciferase construct along with miR-149 and miR-149 control. At 48 h posttransfection, luciferase activity assays were performed with the Secrete-Pair™ Dual Luminescence Assay Kits (GeneCopoeia). The results were obtained from three independent experiments performed in duplicate.
Pull-down assay with biotinylated DNA probe
Pull-down assay was performed as reported.Citation24,Citation25 The GACAT3 pull-down probe sequence was 5′-AAGACTGAGGCCGACTGAGCCAGA-3′, and the random pull-down probe sequence used as negative control was 5′-AAGACTGAGGCCGACTGCAAATCA-3′.
Cell proliferation assays
We used the Cell proliferation Reagent Kit I (3-(4,5-dimethythiazol-2-yl)-2,5-diphenyl tetrazolium bromide [MTT]; Roche) to assess the ability of cell proliferation according to the manufacturer’s instructions. The colony formation assay was performed as reported.Citation26
Cell migration and invasion assays
In the migration assays, 1×104 cells at 48 h after transfection were placed into the upper chamber of an insert in serum-free medium (8-μm pore size, Millipore Corporation). For the invasion assays, 1×105 cells in serum-free medium were placed into the upper chamber coated with Matrigel (Sigma-Aldrich Co.). Medium containing 10% FBS was added to the lower chamber. The cells remaining on the upper membrane were removed after incubation for 24 h, and the cells that had migrated or invaded through the membrane were stained with 0.1% crystal violet, imaged, and counted using an IX71 inverted microscope (Olympus, Tokyo, Japan). All experiments were independently repeated three times.
Animal studies
For the subcutaneous xenograft model, 5×106 cells (stably transfected with p-GACAT3-inhibitor and control vectors) were suspended in 100 μL of PBS and then subcutaneously injected into the upper right flank region of each mouse. After 32 days, mice were sacrificed, and tumors were harvested and weighted. All of the experimental protocols were conducted in accordance with American Association for the Accreditation of Laboratory Animal Care guidelines and were approved by the Ethics Committee of Cangzhou Medical College.
Statistical analysis
All data were presented as the mean ± SD and evaluated using SPSS 22 (SPSS Inc., Chicago, IL, USA). All statistical analyses were performed using ANOVA or a two-tailed Student’s t-test to compare data. Differences were considered statistically significant at P<0.05.
Results
Long noncoding RNA GACAT3 is highly expressed in CRC tissues and cell lines
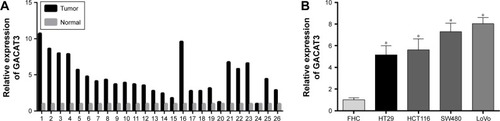
In order to investigate the role of GACAT3 in CRC tumorigenesis, we evaluated the expression of GACAT3 in 26 paired CRC tissues and adjacent normal tissues by quantitative real-time PCR (qRT-PCR). As shown in , compared with the adjacent normal mucosa tissues, the expression of GACAT3 was higher in 24 of 26 paired CRC specimens (P<0.001). Meanwhile, we detected the expression of GACAT3 in the CRC cell lines HT29, HCT116, SW480, LoVo, and normal colorectal epithelium cell line FHC. The expression of GACAT3 was also significantly upregulated in four CRC cell lines compared with the normal colorectal epithelium cell line ().
Figure 1 GACAT3 expression is elevated in CRC tissues and cell lines.
Abbreviations: CRC, colorectal cancer; GACAT3, gastric cancer-associated transcript 3; qRT-PCR, quantitative real-time PCR.
Knockdown of GACAT3 inhibited cell proliferation in vivo and in vitro
To evaluate the roles of GACAT3 in CRC, we knocked down GACAT3 with si-GACAT3 in HT29 and LoVo cells and convinced that GACAT3 expression was significantly reduced in CRC cell lines by qRT-PCR (). The effect of GACAT3 on the proliferation of CRC cells was examined using the MTT assay. The results revealed that the ability of proliferation was suppressed with si-GACAT3 in both HT29 and LoVo cells (). Similarly, GACAT3 knockdown significantly decreased the clone formation activity in cell lines ().
Figure 2 Knockdown of GACAT3 inhibited cell proliferation in vivo and in vitro.
Abbreviations: CRC, colorectal cancer; GACAT3, gastric cancer-associated transcript 3; MTT, 3-(4,5-dimethythiazol-2-yl)-2,5-diphenyl tetrazolium bromide.
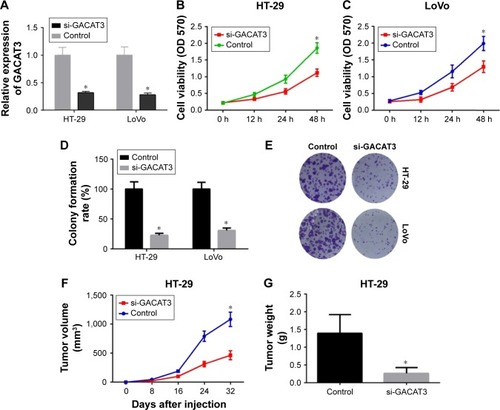
To validate the function of GACAT3 in vivo, HT29/sh-Con and HT29/sh-GACAT3 cells were subcutaneously injected into BALB/c-nu/nu mice. The effects of GACAT3 on tumor growth in nude mice were reflected by growth curves. The growth curves showed that GACAT3 promote the growth of tumor (P<0.05, ). Also, the weight of tumors in the HT29/sh-Con group was obviously greater than that in the HT29/sh-GACAT3 group (P<0.05, ).
Knockdown of GACAT3 inhibited cell migration and invasion in vitro
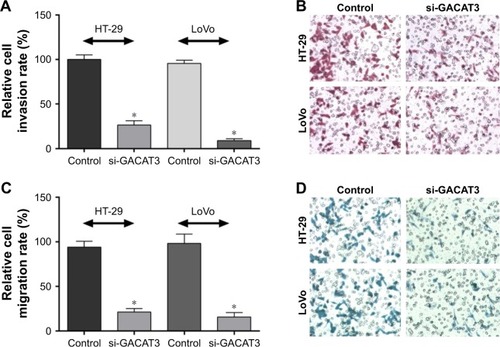
To investigate whether GACAT3 has a direct functional role in regulating the invasion and migration of CRC cells, we performed the invasion and migration assay. The results demonstrated that downregulation of GACAT3 significantly decreased the abilities of HT29 and LoVo cells to invade (). Similarly, migration of HT29 and LoVo cells was decreased following knockdown of GACAT3 (). These results indicate that GACAT3 could promote the migration and invasion of CRC cells.
Figure 3 Knockdown of GACAT3 inhibited cell migration and invasion in vitro.
Abbreviation: GACAT3, gastric cancer-associated transcript 3.
GACAT3 is a target of miR-149
Recent studies have suggested that lncRNA may function as a ceRNA or a molecular sponge in modulating the functions of miRNA. In order to screen out the miRNA associated with lncRNA, we performed the bioinformatics analysis, dual-luciferase reporter assay, and pull-down assay.
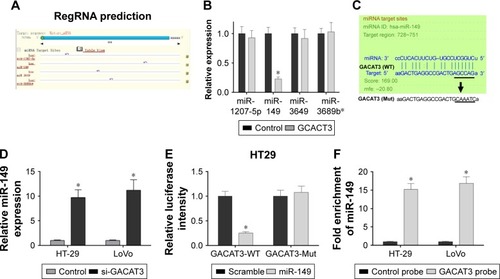
First, the results of bioinformatics analysis (miRanda) of miRNA recognition sequences revealed that four miRNA binding sites were presented in GACAT3 cDNA, and they are miR-1207-5p, miR-149, miR-3649, and miR-3689b (). Meanwhile, we detected the expression of four potentially targeted miRNAs following the overexpression of GACAT3, and the results demonstrated that the expression level of miR-149 was significantly reduced and associated with the overexpression of GACAT3 (). Second, we found si-GACAT3 significantly increased the expression of miR-149 in HT29 and LoVo cells (, P<0.05). To investigate whether GACAT3 was a functional target of miR-149, we performed the dual-luciferase reporter assay. The results showed that the luciferase intensity of miR-149 was decreased in the GACAT3-Mut group (P<0.05; ). Furthermore, we performed inverse pull-down assay to determine whether GACAT3 could target miR-149. As indicated in , miR-149 was precipitated by GACAT3 probe. Taken together, our results demonstrated that miR-149 could directly bind to GACAT3 at the miRNA recognition site.
Figure 4 GACAT3 is identified as the functional downstream target of miR-149.
Abbreviations: 3′-UTR, 3′-untranslated region; GACAT3, gastric cancer-associated transcript 3; qRT-PCR, quantitative real-time PCR.
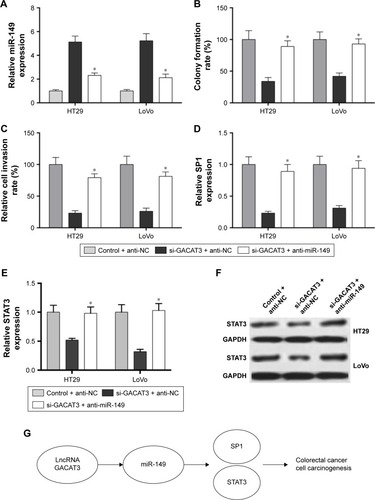
Knockdown of miR-149 significantly reversed the inhibitory effects of si-GACAT3 on colorectal cells
Although we have confirmed that GACAT3 is a target of miR-149, the role of miR-149 in GACAT3-induced promotion on colorectal cells remains unknown. As shown in , anti-miR-149, markedly downregulated miR-149 expression in HT29 and LoVo cells transfected with si-GACAT3 (P<0.01). showed that knockdown of miR-149 in HT29 and LoVo cells, which stably transfected with si-GACAT3, significantly reversed si-GACAT3-induced inhibitory effects on cell clone formation and invasion. Furthermore, anti-miR-149 significantly increased the expression of SP1 inhibited by si-GACAT3 (). Meanwhile, showed that anti-miR-149 significantly increased the expression of STAT3 inhibited by si-GACAT3. Altogether, our study demonstrated that GACAT3 promotes cell carcinogenesis of CRC through miR-149-mediated SP1 and STAT3 signaling ().
Figure 5 Regulation of CRC cell clone formation, invasion, and the expression of SP1 and STAT3 by GACAT3 requires miR-149.
Abbreviations: CRC, colorectal cancer; GACAT3, gastric cancer-associated transcript 3; qRT-PCR, quantitative real-time PCR; SP1, specificity protein 1.
Discussion
Recently, many large-scale gene expression studies of tumor samples have revealed that many lncRNAs are misregulated in multiple cancers,Citation27–Citation29 including CRC.Citation30 For example, lncUBC1 is associated with poor prognosis and facilitates cell proliferation, migration, and invasion in CRC.Citation31 Meanwhile, lncRNA NEAT1 impacts cell proliferation and apoptosis of CRC via regulation of Akt signaling.Citation32 Moreover, the novel lncRNA TUSC7 could inhibit proliferation by sponging miR-211 in CRC.Citation33 In this study, we found that lncRNA GACAT3 is significantly overexpressed in CRC tissues and correlated with the proliferation and invasion of CRC cells, indicating that GACAT3 plays an important role in CRC implicating the potential application of GACAT3 in the treatment of CRC.
Previous researches have convinced that high expression of GACAT3 was first found in gastric cancer,Citation22 and its expression level was significantly associated with poor prognosis. Overexpressed GACAT3 showed extensive potentials in cell proliferation, migration, and invasion through different mechanisms. For example, upregulated expression of GACAT3 enhanced cell proliferation in gastric cancer by inhibiting the relative expression of miR-129-5P. Besides, GACAT3 promotes gastric cancer cell proliferation through the IL-6/STAT3 signaling pathway.Citation34 Similarly, in our study, highly expressed GACAT3 was found in CRC tissues and cell lines, as compared with adjacent histologically normal tissues and cell lines.
To verify the role of GACAT3 in CRC cell lines, HT29 and LoVo cells were transfected with GACAT3 siRNA and GACAT3 scramble, respectively. Downregulated GACAT3 was identified to suppress cell proliferation in HT29 and LoVo cells through MTT assay and the colony formation assay. The above results suggest that GACAT3 is higher expressed in CRC tissues and cell lines, and overexpressed GACAT3 promotes cell proliferation. Having identified that GACAT3 promoted the proliferation of CRC cells in vitro, we further explored the effect of GACAT3 in vivo. Down-regulated expression of GACAT3 was found to significantly suppress tumor growth in si-GACAT3 model mice. The above results indicated that GACAT3 siRNA inhibits CRC growth in vivo. Meanwhile, Transwell assays were conducted to verify whether GACAT3 played an important role in migration and invasion of CRC. The result suggested that both the migration and invasion abilities of CRC cells were largely suppressed by GACAT3 siRNA.
GACAT3 has been identified to possess oncogenic roles in tumor growth and metastasis, and it acts as a potential biomarker and therapeutic target in many types of human cancers. In accordance, overexpressed GACAT3 was detected in human CRC tissues and cell lines in this study. However, the underlying mechanism of GACAT3 in CRC is less well characterized. Thus, it is of a great significance to get a better understanding of the mechanism of GACAT3 in CRC. In this study, we revealed that GACAT3 regulates the growth and metastasis of CRC by targeting miR-149 for the first time.
Emerging evidence confirmed that lncRNA can act as endogenous miRNA sponges and competitively bind to miRNAs to regulate their expression.Citation35,Citation36 To confirm whether GACAT3 serves as an miRNA sponge, we performed the bioinformatics analysis. The results showed that there was a binding site for miR-149 on GACAT3, and the expression of miR-149 was negatively correlated with GACAT3. Meanwhile, the level of miR-149 was upregulated by GACAT3 siRNA in HT29 and LoVo cells. Furthermore, luciferase activity assay showed that the intensity of fluorescence signal was largely weakened by miR-149 mimic in lncRNA GACAT3 WT group. Furthermore, the reverse pull-down assay verified that miR-149 can directly bind to GACAT3. Therefore, we are the first to prove that GACAT3 can directly be combined with miR-149.
Recently, an increasing number of researches revealed that miRNAs acted as tumor suppressors or oncogenes in many human cancers.Citation37–Citation39 Previous studies proved that miR-149 was downregulated in human cancers,Citation40 and miR-149 inhibits non-small cell lung cancer cells EMT by targeting FOXM1.Citation41 Other research showed that miR-149 inhibits proliferation and cell cycle progression through the targeting of ZBTB2 in human gastric cancer.Citation42 Subsequently, miR-149 was identified to offset the role of GACAT3 on the viability and motility of CRC cells. Furthermore, we have detected the expression of specificity protein 1 (SP1, Sp1) as a target of miR-149.Citation43 Meanwhile, we demonstrated that the expression of STAT3 inhibited by si-GACAT3 was reversed by anti-miR149. These above results indicate that GACAT3 regulates the proliferation and migration of CRC by targeting miR-149.
Conclusion
We found that GACAT3 was overexpressed in CRC tissues and cell lines. Downregulated GACAT3 was verified to suppress cell proliferation and migration in CRC cell lines. Further researches revealed that miR-149 was a direct target of GACAT3. Moreover, GACAT3 siRNA was verified to suppress CRC growth in vivo. Our research is the first to reveal the possible link between miR-149 and GACAT3 in CRC. The GACAT3–miR-149 pathway provides a new sight for CRC treatment.
Acknowledgments
This study was funded by the key project of Cangzhou Science and Technology Research and Development Program of China (No 162302105).
Supplementary material
Table S1 The sequences of primer used for qRT-PCR
Disclosure
The authors report no conflicts of interest in this work.
References
- KarsaLVLigniniTAPatnickJLambertRSauvagetCThe dimensions of the CRC problemBest Pract Res Clin Gastroenterol201024438139620833343
- EliasDFaronMIugaBSPrognostic similarities and differences in optimally resected liver metastases and peritoneal metastases from colorectal cancersAnn Surg2015261115716324509197
- WitalisonEECuiXCauseyCPThompsonPRHofsethLJMolecular targeting of protein arginine deiminases to suppress colitis and prevent colon cancerOncotarget2015634360533606226440311
- DickinsonBTKisielJAhlquistDAGradyWMMolecular markers for colorectal cancer screeningGut20156491485149425994221
- MercerTRDingerMEMattickJSLong non-coding RNAs: insights into functionsNat Rev Genet200910315515919188922
- PontingCPOliverPLReikWEvolution and functions of long non-coding RNAsCell2009136462964119239885
- NissanAStojadinovicAMitrani-RosenbaumSColon cancer associated transcript-1: a novel RNA expressed in malignant and pre-malignant human tissuesInt J Cancer201213071598160621547902
- FaticaABozzoniILong non-coding RNAs: new players in cell differentiation and developmentNat Rev Genet201415172124296535
- St LaurentGWahlestedtCKapranovPThe landscape of long noncoding RNA classificationTrends Genet201531523925125869999
- ShenFCaiWSFengZLong non-coding RNA SPRY4-IT1 promotes colorectal cancer metastasis by regulate epithelial-mesenchymal transitionOncotarget201789144791448627391336
- ChenXLiuBYangRIntegrated analysis of long non-coding RNAs in human colorectal cancerOncotarget2016717238972390827004403
- ZhuMLiuJFXiaoJLnc-mg is a long non-coding RNA that promotes myogenesisNat Commun201781471828281528
- RaoAKDMRajkumarTManiSPerspectives of long non-coding RNAs in cancerMol Biol Rep201744220321828391434
- MercerTRMattickJSStructure and function of long noncoding RNAs in epigenetic regulationNat Struct Mol Biol201320330030723463315
- YoonJHAbdelmohsenKGorospeMPosttranscriptional gene regulation by long noncoding RNAJ Mol Biol2013425193723373023178169
- WangKLongBZhouLYCARL lncRNA inhibits anoxia-induced mitochondrial fission and apoptosis in cardiomyocytes by impairing miR-539-dependent PHB2 downregulationNat Commun20145359624710105
- TayYRinnJPandolfiPPThe multilayered complexity of ceRNA crosstalk and competitionNature2014505748334435224429633
- LanevePPoAFaviaAThe long noncoding RNA linc-NeD125 controls the expression of medulloblastoma driver genes by microRNA sponge activityOncotarget2017819310033101528415684
- CaiHYaoJAnYLncRNA HOTAIR acts a competing endogenous RNA to control the expression of notch3 via sponging miR-613 in pancreatic cancerOncotarget2017820329053291728415631
- LiCMiaoRLiuSDown-regulation of miR-146b-5p by long noncoding RNA MALAT1 in hepatocellular carcinoma promotes cancer growth and metastasisOncotarget2017817286832869528404923
- ChenSCLiPFXiaoBXGuoJMLong noncoding RNA HMlin-cRNA717 and AC1,30,710 have been officially named as gastric cancer associated transcript 2 (GACAT2) and GACAT3, respectivelyTumor Biol201435183518352
- XuCShaoYXiaTlncRNA-AC130710 targeting by miR-129-5p is upregulated in gastric cancer and associates with poor prognosisTumor Biol2014351097019706
- YuXSongHXiaTGrowth inhibitory effects of three miR-129 family members on gastric cancerGene20135321879324055727
- WangPLiuYHYaoYLLong non-coding RNA CASC2 suppresses malignancy in human gliomas by miR-21Cell Signal201527227528225446261
- HanPLiJWZhangBMThe lncRNA CRNDE promotes colorectal cancer cell proliferation and chemoresistance via miR-181a-5p-mediated regulation of Wnt/beta-catenin signalingMol Cancer2017161928086904
- YinDDLiuZJZhangEKongRZhangZHGuoRHDecreased expression of long noncoding RNA MEG3 affects cell proliferation and predicts a poor prognosis in patients with colorectal cancerTumour Biol20153664851485925636452
- WangXSunWShenWFLong non-coding RNA DILC regulates liver cancer stem cells via IL-6/STAT3 axisJ Hepatol20166411283129426812074
- LiuFYangFWuXLong non-coding RNA RBMY2FP promotes proliferation of male hepatocellular carcinoma by directing DNA methylation and activating RBMY1A1 via DNMT1Oncotarget2016
- JiangBSunRFangSLnc-CC3 increases metastasis in cervical cancer by increasing Slug expressionOncotarget2016727416504166127223436
- SuJZhangEHanLLong noncoding RNA BLACAT1 indicates a poor prognosis of colorectal cancer and affects cell proliferation by epigenetically silencing of p15Cell Death Dis201783e266528277544
- GaoXWenJGaoPZhangGZhangGOverexpression of the long non-coding RNA, linc-UBC1, is associated with poor prognosis and facilitates cell proliferation, migration, and invasion in colorectal cancerOnco Targets Ther2017101017102628260919
- PengWWangZFanHLncRNA NEAT1 impacts cell proliferation and apoptosis of colorectal cancer via regulation of Akt signalingPathol Oncol Res201723365165628013491
- XuJZhangRZhaoJThe novel long noncoding RNA TUSC7 inhibits proliferation by sponging MiR-211 in colorectal cancerCell Physiol Biochem201741263564428214867
- ShenWYuanYZhaoMNovel long non-coding RNA GACAT3 promotes gastric cancer cell proliferation through the IL-6/STAT3 signaling pathwayTumor Biol201637111489514902
- HuangGWuXLiSXuXZhuHChenXThe long noncoding RNA CASC2 functions as a competing endogenous RNA by sponging miR-18a in colorectal cancerSci Rep201662652427198161
- LiuFYuanJHHuangJFLong noncoding RNA FTX inhibits hepatocellular carcinoma proliferation and metastasis by binding MCM2 and miR-374aOncogene201635415422543427065331
- WangLBuPAiYA long non-coding RNA targets microRNA miR-34a to regulate colon cancer stem cell asymmetric divisionElife20165e1462027077950
- LinSGregoryRIMicroRNA biogenesis pathways in cancerNat Rev Cancer201515632133325998712
- GarofaloMCroceCMRole of microRNAs in maintaining cancer stem cellsAdv Drug Deliv Rev201581536125446141
- BischoffAHuckBKellerBmiR149 functions as a tumor suppressor by controlling breast epithelial cell migration and invasionCancer Res201474185256526525035394
- KeYZhaoWXiongJCaoRmiR-149 inhibits non-small-cell lung cancer cells EMT by targeting FOXM1Biochem Res Int2013201350673123762558
- WangYZhengXZhangZMicroRNA-149 inhibits proliferation and cell cycle progression through the targeting of ZBTB2 in human gastric cancerPLoS One2012710e4169323144691
- WangFMaYLZhangPSP1 mediates the link between methylation of the tumour suppressor miR-149 and outcome in colorectal cancerJ Pathol20132291122422821729
