Abstract
Several epidemiological studies have reported that polymorphisms in microRNA-196a2 (miR-196a2) were associated with various cancers. However, the results remained unverified and were inconsistent in different cancers. Therefore, we carried out an updated meta-analysis to elaborate the effects of rs11614913 polymorphism on cancer susceptibility. A total of 84 articles with 35,802 cases and 41,541 controls were included to evaluate the association between the miR-196a2 rs11614913 and cancer risk by pooled odds ratios (ORs) and 95% confidence intervals (CIs). The results showed that miR-196a2 rs11614913 polymorphism is associated with cancer susceptibility, especially in lung cancer (homozygote comparison, OR =0.840, 95% CI =0.734–0.961; recessive model, OR =0.858, 95% CI =0.771–0.955), hepatocellular carcinoma (allelic contrast, OR =0.894, 95% CI =0.800–0.998; homozygote comparison, OR =0.900, 95% CI =0.813–0.997; recessive model, OR =0.800, 95% CI =0.678–0.944), and head and neck cancer (allelic contrast, OR =1.076, 95% CI =1.006–1.152; homozygote comparison, OR =1.214, 95% CI =1.043–1.413). In addition, significant association was found among Asian populations (allele model, OR =0.847, 95% CI =0.899–0.997, P=0.038; homozygote model, OR =0.878, 95% CI =0.788–0.977, P=0.017; recessive model, OR =0.895, 95% CI =0.824–0.972, P=0.008) but not in Caucasians. The updated meta-analysis confirmed the previous results that miR-196a2 rs11614913 polymorphism may serve as a risk factor for patients with cancers.
Introduction
The rising morbidity and mortality of cancer has drawn extensive attention worldwide, and finding possible risk factors of tumorigenesis has been a priority task for researchers. Recently, an increasing number of studies have focused on associations between miRNA polymorphisms and cancer susceptibility, which indicated that accumulation of genetic variants may be involved in cancer development, including oral cancer,Citation1 lung cancer,Citation2,Citation3 gastric cancer,Citation4 breast cancer,Citation5 glioma,Citation6 non-small cell lung cancer,Citation7 hepatocellular carcinoma,Citation8,Citation9 gallbladder cancer,Citation10 and head and neck cancer (HNC).Citation11 As the molecular mechanism of cancer remains unclear, further exploration of more accurate cancer treatments and prognosis would be of great importance.
MiRNAs are a class of small non-coding RNAs with 18–25 nucleotides in length, which play as oncogenes or anti-oncogenes in the pathogenesis of tumor by targeting multiple genes.Citation12–Citation14 Studies have shown that almost 10%–30% of all human gene expressions have been regulated by mature miRNAs.Citation15 MiRNAs could modulate related genes implicated in cellular processes, including cell differentiation, growth, apoptosis, and immune response.Citation16–Citation18
Hsa-microRNA-196a2 (miR-196a2), initially discovered by Lagos-Quintana et al,Citation19 has been proven to play important roles in various cancers.Citation20,Citation21 Single nucleotide polymorphisms (SNPs) provide new sources of genetic variation, which contribute to potential molecular mechanisms of cancer development.Citation22 SNPs or mutations in miRNA sequence may transform miRNA expression and/or maturation, related to miRNA function by activating the transcription of the primary transcript, pri-miRNA and pre-miRNA processing, and miRNA–mRNA interactions.Citation23 MiR-196a2 rs11614913, as a definitional miRNA polymorphism,Citation24–Citation26 is crucially associated with cancer risk.Citation23,Citation27 It is located in the 3′-untranslated region of the miR-196a2 precursor.Citation28 Hoffman et alCitation5 also showed that miR-196a2 rs11614913 not only influenced the transcription level of mature miR-196a, but also had a biological effect on target gene production. This updated meta-analysis was performed to explore the association between the hsa-miR-196a2 polymorphism and cancer risk and to further estimate the overall cancer risk by pooling all available data.
Materials and methods
Publication search
Two investigators (LYH, HAB) carried out a systematic review on PubMed, Cochrane Library, and Web of Science, by using (“microRNA-196a2” or “miR-196a2”, or “miR-196-a-2” or “miR-196-2” or “miR-196-a” or “rs11614913”), and (“cancer” or “tumor” or “carcinoma” or “neoplasm” or “malignancy”), and (“polymorphism” or “variation” or “susceptibility”) as the search terms in order to identify potentially eligible studies. We based our dates for literature retrieval from January 2008 to September 2017.
Inclusion and exclusion criteria
Relevant studies had to meet the following inclusion criteria: 1) full-text article; 2) evaluation of a link between miRNA polymorphisms and cancer risks; 3) sufficient data for estimating the odds ratio (OR) with 95% CI and a P-value. Studies containing two or more case-control groups were considered as two or more independent studies. Studies that were, 1) review, letters, and comment articles; 2) not for cancer risk; and 3) duplicate samples or publications, were excluded.
Assessment of study quality
The quality of the study was determined by the Newcastle–Ottawa Scale for cohort studies.
Data extraction
Data extraction from the eligible studies were performed independently by two authors (LYH, HAB), based on the inclusion and exclusion criteria. For each publication, the following data were recorded: first author, date of publication, country of origin, ethnicity, type of tumor, source of control groups, total numbers of cases and controls, and genotyping method.
Statistical analysis
The departure of frequencies of miR-196a2 rs11614913 polymorphisms was assessed under the Hardy–Weinberg equilibrium (HWE) for each publication by adopting the goodness-of-fit test (chi-square or Fisher exact test). The association between the miR-196a2 rs11614913 polymorphisms and the risk of cancer was evaluated by calculating pooled OR together with corresponding 95% CI based on the method published by Woolf.Citation29 Also, a P-value<0.05 was considered statistically significant. In addition, we used stratified meta-regression analyses to explore major causes of heterogeneity among the articles. We respectively examined the association between genetic mutants and cancer risk in allelic contrast (T vs C), homozygote comparisons (TT vs CC), heterozygote comparisons (TC vs CC), recessive model (TT vs TC+CC), and dominant model (TT+TC vs CC). Subgroup analyses were performed by ethnicity (Asian and Caucasian), tumor types (if one tumor type contained less than three individual studies, it was combined into “other cancer” subgroups), and source of control (hospital based and population based).
Q testsCitation30 and I2 testsCitation31 were carried out to test the heterogeneity. I2 values describe the percentage of total variation across studies that are due to heterogeneity rather than chance. I2=0% prompts no heterogeneity observed, with 25% identified as low, 50% as moderate, and 75% as high. If I2 was ≥50% or if the P-value of heterogeneity was <0.05, indicating significant heterogeneity among these articles, a random-effect model was used;Citation32 otherwise, a fixed-effect mode was used.Citation33 Sensitivity analyses were conducted to estimate the stability of the meta-analysis result. We adopted Egger’s test to assess potential publication bias by visual inspection of the Funnel plot. A P-value <0.05 was regarded as an indication of potential publication bias.Citation34 All statistical analyses were performed with the Stata software package version 12.0 (Stata Corporation, College Station, TX, USA).
Results
Study identification
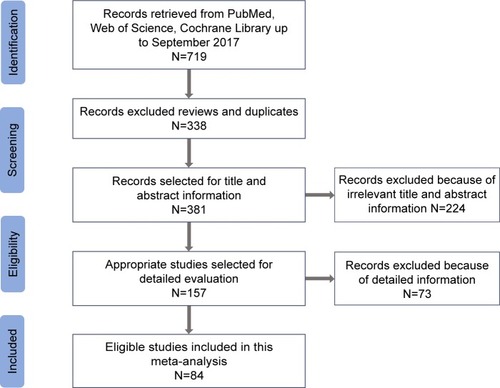
Overall, 84 articles,Citation1–Citation11,Citation26,Citation27,Citation35–Citation100 which were relevant to the search terms, were selected based on the inclusion criteria from PubMed, Cochrane, and Web of Science (). These studies with a total of 35,802 cases and 41,541 controls were subjected to further checking. In the present meta-analysis, we excluded 73 articles (36 articles were meta-analysis, 22 articles did not express concern about cancer risk, 11 articles lacked detailed allele frequency data or OR calculation, and four articles were incomplete text). The included study characteristics are provided in .
Table 1 Characteristics of studies included in the meta-analysis
In total, there were studies on hepatocellular carcinoma (n=14), breast cancer (n=14), colorectal cancer (n=10), gastric cancer (n=10), lung cancer (n=9), esophageal squamous cell carcinoma (ESCC; n=6), HNC (n=5), bladder cancer (n=2), prostate cancer (n=2), oral squamous cell carcinoma (n=2), epithelial ovarian cancer (n=2), renal cell cancer (n=1), glioma (n=1), pancreatic cancer (n=1), cervical cancer (n=1), nasopharyngeal carcinoma (n=1), gallbladder cancer (n=1), acute lymphoblastic leukemia (n=1), and non-Hodgkin lymphoma (n=1). There were 64 studies of Asians and 18 studies of Caucasians.
Among the genotyping methods used in these studies, 57 studies used polymerase chain reaction (including polymerase chain reaction restriction fragment length polymorphism and polymerase chain reaction-ligation detection reaction), 16 studies used Taqman SNP genotyping assay, and others used MassARRAY and DNA sequencing. The controls of 42 studies mainly came from a hospital-based healthy population matched for gender and age, and 42 studies had population-based controls (PB). The distribution of genotypes in the controls of all of the studies was in agreement with HWE (P>0.05).
Quantitative synthesis
In this meta-analysis, we analyzed the hsa-miR-196a2 rs11614913 polymorphism in 84 comparisons with 35,802 cases and 41,541 controls. All the studies were pooled into the meta-analysis, and the results showed that the hsa-miR-196a2 rs11614913 polymorphism was significantly associated with the risk of cancer in the following genetic models: TT vs CC: OR =0.900, 95% CI =0.813–0.987, P=0.043; TT vs TC+CC: OR =0.918, 95% CI =0.851–0.989, P=0.025.
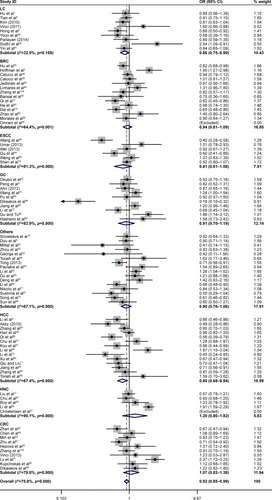
Then, we performed the subgroup analysis of different specific cancer types, genotypes, control sources, and ethnicities (). In the different cancer types, close association between rs11614913 and cancer risk was found for lung cancer (homozygote comparison, OR =0.840, 95% CI =0.734–0.961, P=0.011; recessive model, OR =0.858, 95% CI =0.771–0.955, P=0.005), hepatocellular carcinoma (allelic contrast, OR =0.894, 95% CI =0.800–0.998, P=0.047; homozygote comparison, OR =0.900, 95% CI =0.813–0.997, P=0.039; recessive model, OR =0.800, 95% CI =0.678–0.944, P=0.008), and HNC (allelic contrast, OR =1.076, 95% CI =1.006–1.152, P=0.033; homozygote comparison, OR =1.214, 95% CI =1.043–1.413, P=0.012; and ). However, the association between rs11614913 and breast cancer, ESCC, gastric cancer (GC), or colorectal cancer (CRC) is not statistically significant.
Table 2 Meta-analysis of miR-192a rs11614913 polymorphism with cancer risk
Figure 2 Forest plots of the association between miR-196a2 rs11614913 polymorphism and cancer risk in different cancer types for homozygote comparison (TT vs CC).
Abbreviations: BRC, breast cancer; CRC, colorectal cancer; ESCC, esophageal squamous cell carcinoma; GC, gastric cancer; HCC, hepatocellular carcinoma; HNC, head and neck cancer; LC, lung cancer; miR-196a2, microRNA-196a2; OR, odds ratio.
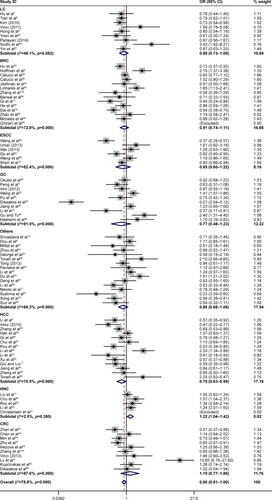
Figure 3 Forest plots of the association between miR-196a2 rs11614913 polymorphism and cancer risk in different cancer types for recessive model (TT vs TC+CC).
Abbreviations: BRC, breast cancer; CRC, colorectal cancer; ESCC, esophageal squamous cell carcinoma; GC, gastric cancer; HCC, hepatocellular carcinoma; HNC, head and neck cancer; LC, lung cancer; miR-196a2, microRNA-196a2; OR, odds ratio.
In ethnic subgroup analysis, a strong association was found between rs11614913 and cancer risk in the allelic contrast (T vs C: OR =0.847, 95% CI =0.899–0.997, P=0.038), the homozygote comparison (TT vs CC: OR =0.878, 95% CI =0.788–0.977, P=0.017), and the recessive model (OR =0.895, 95% CI =0.824–0.972, P=0.008) among Asians, whereas negative results were obtained for Caucasians in all genetic models. Additionally, decreased risk was observed in the polymerase chain reaction (PCR) method for the homozygote comparison (TT vs CC: OR =0.849, 95% CI =0.732–0.986, P=0.032) and the recessive model (TT vs TC+CC: OR =0.880, 95% CI =0.800–0.969, P=0.009), and no significant association of cancer risk was found in Taqman and other methods.
Test of heterogeneity
Among the studies of rs11614913, we found heterogeneity in overall comparisons and subgroup analysis. Moreover, the heterogeneity we evaluated for all genetic models by ethnicity, cancer type, source of controls, as well HWE status was significant. However, we found that heterogeneity could not be explained by the variable ethnicity, cancer type, source of controls, and HWE status (data not shown).
Sensitivity analysis
Sensitivity analysis was conducted to assess the effect by excluding a single study in turn. Sensitivity analysis of the rs11614913 polymorphism in an allelic comparison is presented in . Overall, we found that no individual study had an influence on the pooled OR. The results demonstrated that the pooled ORs were not materially altered, suggesting the stability of our meta-analysis.
Publication bias
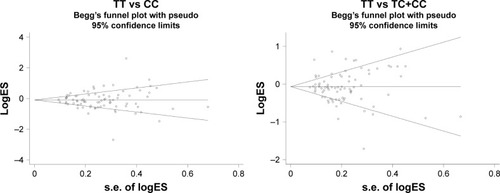
The publication bias of the present meta-analysis was assessed by Begg’s funnel plot and Egger’s test. The funnel plot for the rs11614913 polymorphism in the allelic comparison is presented in . No evidence of publication bias was noted in Begg’s funnel plot (T vs C [P-value for Begg’s test =0.660], TT vs CC [P-value for Begg’s test =0.971, ], TC vs CC [P-value for Begg’s test =0.951], TT vs TC+CC [P-value for Begg’s test =0.908, ], TC+TT vs CC [P-value for Begg’s test =0.592]) and Egger’s test (allele contrast [P=0.923], homozygous model [P=0.822], heterozygous model [P=0.761], recessive model [P=0.899], and dominant model [P=0.401]). The quality of included studies is presented in .
Table 3 Methodological quality of the included studies according to the Newcastle–Ottawa scale
Figure 4 Begg’s funnel plot for publication bias of miR-196a2 rs11614913 polymorphism and cancer risk by homozygote comparison and recessive model.
Abbreviations: miR-196a2, microRNA-196a2; OR, odds ratio.
Discussion
MiRNAs are reported as critical posttranscriptional regulators in gene expression and are involved in various diseases. The associations between miR-196a2 rs11614913 polymorphism and susceptibility to different cancers are widely explored. Guo et alCitation101 found that the C allele had the effect of increasing cancer risk in gastric cancer, and Ma et alCitation102 found that TT could decrease the risk of colorectal cancer. Moreover, Wang et alCitation103 and Zhang et alCitation104 showed that the rs11614913 polymorphism has no association with the risk of hepatocellular carcinoma. However, the regulatory effects of miRNA in carcinogenesis remain unclear. Therefore, we performed this updated meta-analysis to explore the molecular mechanisms of the genetic associations between miRNA and SNPs with cancer risk.
MiR-196a2 is composed of two distinct mature miRNAs (miR-196a-3P and miR-196a-5P), which are processed from the same stem loop;Citation105 thus, the potential targets of miR-196a could be influenced by its altered expression patterns. SNPs in miRNAs could potentially affect the processing or target selection of miRNAs,Citation106,Citation107 which is identified as a key factor in oncogenesis, and contributes to regulate the translation or degradation of messenger RNA (mRNA).Citation23 Hoffman et alCitation5 found that the expression of mature miR-196a2 was increased 9.3-fold in cells transfected with pre-miR-196a2-C but upregulated only by 4.4-fold with pre-miR-196a2-T, and that the C allele of rs11614913 increased mature miR-196a2 levels in lung cancerCitation7 and CRCCitation42 tissues. Xu et alCitation108 have shown that miR-196a2 rs11614913 CC is associated with significantly increased expression of mature miR-196a (lower cycle threshold corresponding to a higher expression) in cardiac tissue specimens of congenital heart disease, and the increased miR-196a expression could further decrease mRNA target of HOXB8. These results indicated that the rs11614913 polymorphism may affect the processing of the pre-miRNA to its mature form.
Several meta-analyses have been performed to analyse the SNP of this miRNA that is associated with the cancer risk.Citation104,Citation109 In our present work, we screened out all the studies published to date and included more papers and cancer types than the previously published meta-analyses. For example, Kang et alCitation109 conducted a meta-analysis encompassing the rs11614913 polymorphism in miR-196a2 and cancer risks, which suggested that the rs11614913 polymorphism may contribute to decreased susceptibility to liver cancer (allele model, homozygous model, dominant model, and heterozygous model) and lung cancer (allele model, homozygous model, and recessive model); however, this was not duplicated in our meta-analysis. In this study, we concluded that the rs11614913 polymorphism conferred a decreased susceptibility to lung cancer (homozygote comparison, recessive model) and hepatocellular carcinoma (allelic contrast, homozygote comparison, recessive model) or an increased susceptibility to HNC (allelic contrast, homozygote comparison). Our study had a larger sample size than the previous ones, which might influence the results. In addition, the previous meta-analyses did not evaluate the quality of the included studies.
According to the procedure of seeking for the source of heterogeneity, we performed subgroup studies according to cancer type, ethnicity, and source of control. A strong association was found between rs11614913 and cancer risk in lung cancers, hepatocellular carcinoma, and HNC, but not in breast cancer, gastric cancer, ESCC, or CRC, which was not similar to the findings of previous studies.Citation101–Citation103,Citation109 The present meta-analysis showed that homozygote TT had the effect of decreasing the risk of lung cancer or hepatocellular carcinoma compared with that of CC homozygote or C allele carriers. We conducted another subgroup analysis by population to determine the association between these miRNA polymorphisms and tumorigenesis. The results suggested that individuals with alterative T allele could decrease cancer susceptibility in Asians but not in Caucasians, indicating that the difference of ethnic background and the living environment may also be a risk factor.
To determine the hsa-miR-196a2 rs11614913 polymorphism, PCR, Taqman, and other methods have been adopted. We found that the hsa-miR-196a2 rs11614913 polymorphism significantly decreased cancer risk in homozygous models and the recessive model when using the PCR method, but this result was not shown when selecting Taqman and other methods. Therefore, more effort may be necessary for further progress in SNP analysis. We found sources of heterogeneity in the studies from cancer type and ethnicity suggesting cancer and population playing important roles. When detecting the source of control, we observed significant associations in population-based and hospital-based controls. This may be due to the included studies matching age, gender, and residential area to control selection bias.
Nevertheless, several defects of this meta-analysis should be emphasized. Firstly, although we strictly screened articles and precisely extracted the data, the differences in the selection of subjects could not be eliminated. Secondly, in our meta-analysis, only Asian and Caucasian ethnicities were included, and the impact of the differences in racial descent should not be ignored. Thirdly, potential language bias could not be avoided due to limitation of studies published in English or Chinese. Therefore, it is not possible to avoid potential publication bias in this meta-analysis.
In summary, miR-196a2 rs11614913 polymorphism may contribute to the development of cancer, especially in lung cancer, hepatocellular carcinoma, and HNC. It might be useful as a candidate marker for the diagnosis of these cancers, and could also be a potential protective factor for cancer risks in Asians. Furthermore, more significant studies and investigations with larger populations focusing on cancer types or ethnicities should be performed to confirm the results.
Acknowledgments
This review was supported by Health Care 3F Project of Shenzhen (Peking University First Hospital-The Second People’s Hospital of Shenzhen, Academician Yinglu Guo’s Team), the Shenzhen Key Medical Discipline Fund, Special Support Funds of Shenzhen for Introduced High-Level Medical Team, Shenzhen Foundation of Science and Technology (JCYJ20150330102720182), and Shenzhen Health and Family Planning Commission Scientific Research Project (201506026, 201601025, and 201606019).
Supplementary materials
Table S1 Details of the sensitivity analyses of the association between rs11614913 polymorphism and cancer risk homozygous model (TT vs CC) and recessive model (TT vs TC+CC).
Table S2 P-values of Begg’s and Egger’s test for the polymorphism rs11614913
References
- LiuCJTsaiMMTuHFLuiMTChengHWLinSCmiR-196a overexpression and miR-196a2 gene polymorphism are prognostic predictors of oral carcinomasAnn Surg Oncol201320Suppl 3S406S41423138850
- HongYSKangHJKwakJYAssociation between microR-NA196a2 rs11614913 genotypes and the risk of non-small cell lung cancer in Korean populationJ Prev Med Public Health201144312513021617338
- TianTShuYChenJA functional genetic variant in microRNA-196a2 is associated with increased susceptibility of lung cancer in ChineseCancer Epidemiology, Biomarkers & Prev200918411831187
- PengSKuangZShengCZhangYXuHChengQAssociation of microRNA-196a-2 gene polymorphism with gastric cancer risk in a Chinese populationDig Dis Sci20105582288229319834808
- HoffmanAEZhengTYiCmicroRNA miR-196a-2 and breast cancer: a genetic and epigenetic association study and functional analysisCancer research200969145970597719567675
- DouTWuQChenXA polymorphism of microRNA196a genome region was associated with decreased risk of glioma in Chinese populationJ Cancer Res Clin Oncol2010136121853185920229273
- HuZChenJTianTGenetic variants of miRNA sequences and non-small cell lung cancer survivalJ Clin Invest200811872600260818521189
- AkkizHBayramSBekarAAkgolluEUlgerYA functional polymorphism in pre-microRNA-196a-2 contributes to the susceptibility of hepatocellular carcinoma in a Turkish population: a case-control studyJ Viral Hepat2011187e399e40721692953
- LiXDLiZGSongXXLiuCFA variant in microRNA-196a2 is associated with susceptibility to hepatocellular carcinoma in Chinese patients with cirrhosisPathology201042766967321080878
- SrivastavaKSrivastavaAMittalBCommon genetic variants in pre-microRNAs and risk of gallbladder cancer in North Indian populationJ Hum Genet201055849549920520619
- LiuZLiGWeiSGenetic variants in selected pre-microRNA genes and the risk of squamous cell carcinoma of the head and neckCancer152010116204753476020549817
- ChoWCMicroRNAs: potential biomarkers for cancer diagnosis, prognosis and targets for therapyInt J Biochem Cell Biol20104281273128120026422
- Esquela-KerscherASlackFJOncomirs – microRNAs with a role in cancerNat Rev Cancer20066425926916557279
- LuJGetzGMiskaEAMicroRNA expression profiles classify human cancersNature2005435704383483815944708
- BerezikovEGuryevVvan de BeltJWienholdsEPlasterkRHCuppenEPhylogenetic shadowing and computational identification of human microRNA genesCell20051201212415652478
- Lopez-CimaMFGonzalez-ArriagaPGarcia-CastroLPolymorphisms in XPC, XPD, XRCC1, and XRCC3 DNA repair genes and lung cancer risk in a population of northern SpainBMC cancer2007716217705814
- BartelDPMicroRNAs: genomics, biogenesis, mechanism, and functionCell2004116228129714744438
- RuanKFangXOuyangGMicroRNAs: novel regulators in the hallmarks of human cancerCancer Lett2009285211612619464788
- Lagos-QuintanaMRauhutRMeyerJBorkhardtATuschlTNew microRNAs from mouse and humanRNA20039217517912554859
- Valencia-SanchezMALiuJHannonGJParkerRControl of translation and mRNA degradation by miRNAs and siRNAsGenes Dev200620551552416510870
- AmbrosVThe functions of animal microRNAsNature2004431700635035515372042
- ChenKSongFCalinGAWeiQHaoXZhangWPolymorphisms in microRNA targets: a gold mine for molecular epidemiologyCarcinogenesis20082971306131118477647
- RyanBMRoblesAIHarrisCCGenetic variation in microRNA networks: the implications for cancer researchNature Rev Cancer201010638940220495573
- MinKTKimJWJeonYJAssociation of the miR-146aC>G, 149C>T, 196a2C>T, and 499A>G polymorphisms with colorectal cancer in the Korean populationMol Carcinog201251Suppl 1E65E7322161766
- HezovaRKovarikovaABienertova-VaskuJEvaluation of SNPs in miR-196-a2, miR-27a and miR-146a as risk factors of colorectal cancerWorld J Gastroenterol14201218222827283122719192
- JiangJJiaZFCaoDHWuYHSunZWCaoXYAssociation of the miR-146a rs2910164 polymorphism with gastric cancer susceptibility and prognosisFuture Oncol201612192215222627267319
- HuYYuCYWangJLGuanJChenHYFangJYMicroRNA sequence polymorphisms and the risk of different types of cancerSci Rep20144364824413317
- JohnnidisJBHarrisMHWheelerRTRegulation of progenitor cell proliferation and granulocyte function by microRNA-223Nature200845171821125112918278031
- WoolfBOn estimating the relation between blood group and diseaseAnn Hum Genet195519425125314388528
- LiuYZhanYChenZDirecting cellular information flow via CRISPR signal conductorsNat Methods2016131193894427595406
- VangelMGRukhinALMaximum likelihood analysis for heteroscedastic one-way random effects ANOVA in interlaboratory studiesBiometrics199955112913611318146
- DerSimonianRKackerRRandom-effects model for meta-analysis of clinical trials: an updateContemp Clin Trials200728210511416807131
- MantelNHaenszelWStatistical aspects of the analysis of data from retrospective studies of diseaseJ Natl Cancer Inst195922471974813655060
- EggerMDavey SmithGSchneiderMMinderCBias in meta-analysis detected by a simple, graphical testBMJ199731571096296349310563
- HuZLiangJWangZCommon genetic variants in pre-microRNAs were associated with increased risk of breast cancer in Chinese womenHum Mutat2009301798418634034
- CatucciIYangRVerderioPEvaluation of SNPs in miR-146a, miR196a2 and miR-499 as low-penetrance alleles in German and Italian familial breast cancer casesHum Mutat2010311E1052E105719847796
- ChristensenBCAvissar-WhitingMOuelletLGMature microRNA sequence polymorphism in MIR196A2 is associated with risk and prognosis of head and neck cancerClin Cancer Res201016143713372020501619
- WangKGuoHHuHA functional variation in pre-microRNA-196a is associated with susceptibility of esophageal squamous cell carcinoma risk in Chinese HanBiomarkers201015761461820722507
- GeorgeGPGangwarRMandalRKSankhwarSNMittalRDGenetic variation in microRNA genes and prostate cancer risk in North Indian populationMol Biol Rep20113831609161520842445
- JedlinskiDJGabrovskaPNWeinsteinSRSmithRAGriffithsLRSingle nucleotide polymorphism in hsa-mir-196a-2 and breast cancer risk: a case control studyTwin Res Hum Genet201114541742121962133
- MittalRDGangwarRGeorgeGPMittalTKapoorRInvestigative role of pre-microRNAs in bladder cancer patients: a case-control study in North IndiaDNA Cell Biol201130640140621345130
- ZhanJFChenLHChenZXA functional variant in microRNA-196a2 is associated with susceptibility of colorectal cancer in a Chinese populationArchiv Med Res2011422144148
- ZhouBWangKWangYCommon genetic polymorphisms in pre-microRNAs and risk of cervical squamous cell carcinomaMol Carcinog201150749950521319225
- ChenHSunLYChenLLZhengHQZhangQFA variant in micro RNA-196a2 is not associated with susceptibility to and progression of colorectal cancer in ChineseInt Med J2012426e115e119
- LinharesJJAzevedoMJrSiufiAAEvaluation of single nucleotide polymorphisms in microRNAs (hsa-miR-196a2 rs11614913 C/T) from Brazilian women with breast cancerBMC Med Genet20121311923228090
- YoonKAYoonHParkSThe prognostic impact of microRNA sequence polymorphisms on the recurrence of patients with completely resected non-small cell lung cancerJ Thorac Cardiovasc Surg2012144479480722818121
- ZhuLChuHGuDA functional polymorphism in miRNA-196a2 is associated with colorectal cancer risk in a Chinese populationDNA Cell Biol201231335035421815818
- AhnDHRahHChoiY-KAssociation of the miR-146aC>G, miR-149T>C, miR-196a2T>C, and miR-499A>G polymorphisms with gastric cancer risk and survival in the Korean populationMol Carcinog201352S1395122006587
- HanYPuRHanXAssociations of pri-miR-34b/c and pre-miR-196a2 polymorphisms and their multiplicative interactions with hepatitis B virus mutations with hepatocellular carcinoma riskPLoS One201383e5856423516510
- HongMJChoiYYJangJAAssociation between genetic variants in pre-microRNAs and survival of early-stage NSCLCJ Thorac Oncol20138670371023470291
- LvMDongWLiLAssociation between genetic variants in pre-miRNA and colorectal cancer risk in a Chinese populationJ Cancer Res Clin Oncol201313981405141023728616
- SongXSturgisEMLiuJMicroRNA variants increase the risk of HPV-associated squamous cell carcinoma of the oropharynx in never smokersPLoS One201382e5662223457596
- WangSTaoGWuDA functional polymorphism in MIR196A2 is associated with risk and prognosis of gastric cancerMol Carcinog201352Suppl 1E87E9523423813
- YuanZZengXYangDWangWLiuZEffects of common polymorphism rs11614913 in Hsa-miR-196a2 on lung cancer riskPLoS One201384e6104723593385
- ZhangJWangRMaY-YAssociation Between Single Nucleotide Polymorphisms in miRNA196a-2 and miRNA146a and Susceptibility to Hepatocellular Carcinoma in a Chinese PopulationAsian Pac J Cancer Prev201314116427643124377545
- BansalCSharmaKLMisraSSrivastavaANMittalBSinghUSCommon genetic variants in pre-microRNAs and risk of breast cancer in the North Indian populationEcancermedicalscience2014847325374621
- ChuYHHsiehMJChiouHLMicroRNA gene polymorphisms and environmental factors increase patient susceptibility to hepatocellular carcinomaPLoS One201492e8993024587132
- DikeakosPTheodoropoulosGRizosSTzanakisNZografosGGazouliMAssociation of the miR-146aC>G, miR-149T>C, and miR-196a2T>C polymorphisms with gastric cancer risk and survival in the Greek populationMol Biol Rep20144121075108024379078
- DuMLuDWangQGenetic variations in microRNAs and the risk and survival of renal cell cancerCarcinogenesis20143571629163524681820
- DuWMaX-LZhaoCAssociations of Single Nucleotide Polymorphisms in miR-146a, miR-196a, miR-149 and miR-499 with Colorectal Cancer SusceptibilityAsian Pac J Cancer Prev20141521047105524568449
- FanXWuZEffects of four single nucleotide polymorphisms in microRNA-coding genes on lung cancer riskTumour Biol20143511108151082425077922
- KupcinskasJBruzaiteIJuzenasSLack of association between miR-27a, miR-146a, miR-196a-2, miR-492 and miR-608 gene polymorphisms and colorectal cancerSci Rep20144599325103961
- LiPYanHZhangHA functional polymorphism in MIR196A2 is associated with risk and progression of nasopharyngeal carcinoma in the Chinese population. Genet Test Mol BiomarkersMar2014183149155
- QuYQuHLuoMMicroRNAs related polymorphisms and genetic susceptibility to esophageal squamous cell carcinomaMol Genet Genom2014289611231130
- TongNXuBShiDHsa-miR-196a2 polymorphism increases the risk of acute lymphoblastic leukemia in Chinese childrenMutat Res2014759162124291415
- WangNLiYZhouRMHsa-miR-196a2 functional SNP is associated with the risk of ESCC in individuals under 60 years oldBiomarkers2014191434824320161
- DengSWangWLiXZhangPCommon genetic polymorphisms in pre-microRNAs and risk of bladder cancerWorld J Surg Oncol20151329726458899
- DikaiakosPGazouliMRizosSZografosGTheodoropoulosGEEvaluation of genetic variants in miRNAs in patients with colorectal cancerCancer Biom2015152157162
- LiXLiKWuZAssociation of four common SNPs in microRNA polymorphisms with the risk of hepatocellular carcinomaInt J Clin Exp Pathol2015889560956626464719
- NiQJiAYinJWangXLiuXEffects of Two Common Polymorphisms rs2910164 in miR-146a and rs11614913 in miR-196a2 on Gastric Cancer SusceptibilityGastroenterol Res Pract2015201576416325983750
- NikolicZSavic PavicevicDVucicNAssessment of association between genetic variants in microRNA genes hsa-miR-499, hsa-miR-196a2 and hsa-miR-27a and prostate cancer risk in Serbian populationExp Mol Pathol201599114515026112096
- QiPWangLZhouBAssociations of miRNA polymorphisms and expression levels with breast cancer risk in the Chinese populationGenet Mol Res20151426289629626125831
- WuYHaoXFengZLiuYGenetic polymorphisms in miRNAs and susceptibility to colorectal cancerCell Biochem Biophysics2015711271278
- DaiZMKangHFZhangWGThe Associations of single nucleotide polymorphisms in miR196a2, miR-499, and miR-608 with breast cancer susceptibility: A STROBE-Compliant Observational StudyMedicine2016957e282626886638
- LiJChengGWangSA single-nucleotide polymorphism of miR-196a2T>C rs11614913 Is associated with hepatocellular carcinoma in the Chinese populationGenet Test Mol Biomarkers201620421321526866381
- LiMLiRJBaiHAssociation between the pre-miR-196a2 rs11614913 polymorphism and gastric cancer susceptibility in a Chinese populationGenet Mol Res2016152
- QiuGPLiuJMicroRNA gene polymorphisms in evaluating therapeutic efficacy after transcatheter arterial chemoembolization for primary hepatocellular carcinomaGenet Test Mol Biomarkers2016
- ShenFChenJGuoSGenetic variants in miR-196a2 and miR-499 are associated with susceptibility to esophageal squamous cell carcinoma in Chinese Han populationTumour Biol20163744777478426518769
- SongZSWuYZhaoHGAssociation between the rs11614913 variant of miRNA-196a-2 and the risk of epithelial ovarian cancerOncol Lett201611119420026870188
- XuXLingQWangJDonor miR-196a-2 polymorphism is associated with hepatocellular carcinoma recurrence after liver transplantation in a Han Chinese populationInt J Cancer2016138362062926365437
- YinZCuiZRenYAssociation between polymorphisms in pre-miRNA genes and risk of lung cancer in a Chinese non-smoking female populationLung Cancer201694152126973201
- ZhaoHXuJZhaoDSomatic mutation of the SNP rs11614913 and its association with increased MIR 196A2 expression in breast cancerDNA Cell Biol2016352818726710106
- OkuboMTaharaTShibataTAssociation between common genetic variants in pre-microRNAs and gastric cancer risk in Japanese populationHelicobacter201015652453121073609
- PuJYDongWZhangLLiangWBYangYLvMLNo association between single nucleotide polymorphisms in pre-mirnas and the risk of gastric cancer in Chinese populationIran J Basic Med Sci201417212813324711897
- OmraniMHashemiMEskandari-NasabEhsa-mir-499 rs3746444 gene polymorphism is associated with susceptibility to breast cancer in an Iranian populationBiomark Med20148225926724521023
- QiJHWangJChenJHigh-resolution melting analysis reveals genetic polymorphisms in microRNAs confer hepatocellular carcinoma risk in Chinese patientsBMC Cancer20141464325176041
- ChuYHTzengSLLinCWChienMHChenMKYangSFImpacts of microRNA gene polymorphisms on the susceptibility of environmental factors leading to carcinogenesis in oral cancerPLoS One201276e3977722761899
- GuJYTuLInvestigating the role of polymorphisms in miR-146a, -149, and -196a2 in the development of gastric cancerGenet Mol Res2016152 gmr.15027839
- HashemiMMoradiNZiaeeSAAssociation between single nucleotide polymorphism in miR-499, miR-196a2, miR-146a and miR-149 and prostate cancer risk in a sample of Iranian populationJ Adv Res20167349149827222754
- HeBPanYXuYAssociations of polymorphisms in microRNAs with female breast cancer risk in Chinese populationTumour Biol20153664575458225613069
- KouJTFanHHanDAssociation between four common microRNA polymorphisms and the risk of hepatocellular carcinoma and HBV infectionOncol Lett2014831255126025120701
- MoralesSGulppiFGonzalez-HormazabalPAssociation of single nucleotide polymorphisms in Pre-miR-27a, Pre-miR-196a2, Pre-miR-423, miR-608 and Pre-miR-618 with breast cancer susceptibility in a South American populationBMC Genet15201617110927421647
- PavlakisEPapaconstantinouIGazouliMMicroRNA gene polymorphisms in pancreatic cancerPancreatology201313327327823719600
- RoyRDe SarkarNGhoseSGenetic variations at microRNA and processing genes and risk of oral cancerTumour Bio20143543409341424297336
- SodhiKKBahlCSinghNBeheraDSharmaSFunctional genetic variants in pre-miR-146a and 196a2 genes are associated with risk of lung cancer in North IndiansFuture Oncol201511152159217326235181
- SunXCZhangACTongLLmiR-146a and miR-196a2 polymorphisms in ovarian cancer riskGenet Mol Res2016153 gmr.15038468
- SushmaPSJamilKKumarPUSatyanarayanaURamakrishnaMTriveniBGenetic variation in MicroRNAs and risk of oral squamous cell carcinoma in South Indian populationAsian Pac J Cancer Prev201516177589759426625766
- ToraihEAFawzMSElgazzazMGHusseinMHShehataRHDaoudHGCombined genotype analyses of precursor miRNA196a2 and 499a variants with hepatic and renal cancer susceptibility a Preliminary StudyAsian Pac J Cancer Prev20161773369337527509977
- ZhangLHHaoBBZhangCYDaiXZZhangFContributions of polymorphisms in miR146a, miR196a, and miR499 to the development of hepatocellular carcinomaGenet Mol Res2016153
- ZhangMJinMYuYAssociations of miRNA polymorphisms and female physiological characteristics with breast cancer risk in Chinese populationEur J Cancer Care (Engl)201221227428022074121
- KimMJYooSSChoiYYParkJYA functional polymorphism in the pre-microRNA-196a2 and the risk of lung cancer in a Korean populationLung Cancer201069112712920466450
- VinciSGelminiSPratesiNGenetic variants in miR-146a, miR-149, miR-196a2, miR-499 and their influence on relative expression in lung cancersClin Chem Lab Med201149122073208021902575
- AhnDHRahHChoiYKAssociation of the miR-146aC>G, miR-149T>C, miR-196a2T>C, and miR-499A>G polymorphisms with gastric cancer risk and survival in the Korean populationMol Carcinog201352Suppl 1E39E5123001871
- UmarMUpadhyayRPrakashGKumarSGhoshalUCMittalBEvaluation of common genetic variants in pre-microRNA in susceptibility and prognosis of esophageal cancerMol Carcinog201352Suppl 1E10E1822692992
- VinciSGelminiSManciniIGenetic and epigenetic factors in regulation of microRNA in colorectal cancersMethods201359113814622989523
- WeiJZhengLLiuSMiR-196a2 rs11614913 T > C polymorphism and risk of esophageal cancer in a Chinese populationHuman Immunology20137491199120523792053
- ParlayanCIkedaSSatoNSawabeMMuramatsuMAraiTAssociation analysis of single nucleotide polymorphisms in miR-146a and miR-196a2 on the prevalence of cancer in elderly Japanese: a case-control studyAsian Pacific journal of cancer prevention: APJCP20141552101210724716941
- GuoJJinMZhangMChenKA genetic variant in miR-196a2 increased digestive system cancer risks: a meta-analysis of 15 case-control studiesPLoS One201271e3058522291993
- MaXPZhangTPengBYuLJiang deKAssociation between microRNA polymorphisms and cancer risk based on the findings of 66 case-control studiesPLoS One2013811e7958424278149
- WangJWangQLiuHThe association of miR-146a rs2910164 and miR-196a2 rs11614913 polymorphisms with cancer risk: a meta-analysis of 32 studiesMutagenesis201227677978822952151
- ZhangHSuYLYuHQianBYMeta-Analysis of the association between Mir-196a-2 polymorphism and cancer susceptibilityCancer2012916372
- ChenCZhangYZhangLWeakleySMYaoQMicroRNA-196: critical roles and clinical applications in development and cancerJ Cell Mol Med2011151142321091634
- GarzonRMarcucciGCroceCMTargeting microRNAs in cancer: rationale, strategies and challengesNature Rev Drug Discov201091077578920885409
- VoliniaSCalinGALiuCGA microRNA expression signature of human solid tumors defines cancer gene targetsProc Natl Acad Sci U S A200610372257226116461460
- XuJHuZXuZFunctional variant in microRNA-196a2 contributes to the susceptibility of congenital heart disease in a Chinese populationHum Mut20093081231123619514064
- KangZLiYHeXQuantitative assessment of the association between miR-196a2 rs11614913 polymorphism and cancer risk: evidence based on 45,816 subjectsTumour Biol20143576271628224633889
Disclosure
The authors report no conflicts of interest in this work.
References
- LiuCJTsaiMMTuHFLuiMTChengHWLinSCmiR-196a overexpression and miR-196a2 gene polymorphism are prognostic predictors of oral carcinomasAnn Surg Oncol201320Suppl 3S406S41423138850
- HongYSKangHJKwakJYAssociation between microRNA196a2 rs11614913 genotypes and the risk of non-small cell lung cancer in Korean populationJ Prev Med Public Health201144312513021617338
- TianTShuYChenJA functional genetic variant in microRNA-196a2 is associated with increased susceptibility of lung cancer in ChineseCancer Epidemiol, Biomarkers & Prev200918411831187
- PengSKuangZShengCZhangYXuHChengQAssociation of microRNA-196a-2 gene polymorphism with gastric cancer risk in a Chinese populationDig Dis Sci20105582288229319834808
- HoffmanAEZhengTYiCmicroRNA miR-196a-2 and breast cancer: a genetic and epigenetic association study and functional analysisCancer Res200969145970597719567675
- DouTWuQChenXA polymorphism of microRNA196a genome region was associated with decreased risk of glioma in Chinese populationJ Cancer Res Clin Oncol2010136121853185920229273
- HuZChenJTianTGenetic variants of miRNA sequences and non-small cell lung cancer survivalJ Clin Invest200811872600260818521189
- AkkizHBayramSBekarAAkgolluEUlgerYA functional polymorphism in pre-microRNA-196a-2 contributes to the susceptibility of hepatocellular carcinoma in a Turkish population: a case-control studyJ Viral Hepat2011187e399e40721692953
- LiXDLiZGSongXXLiuCFA variant in microRNA-196a2 is associated with susceptibility to hepatocellular carcinoma in Chinese patients with cirrhosisPathology201042766967321080878
- SrivastavaKSrivastavaAMittalBCommon genetic variants in pre-microRNAs and risk of gallbladder cancer in North Indian populationJ Hum Genet201055849549920520619
- LiuZLiGWeiSGenetic variants in selected pre-microRNA genes and the risk of squamous cell carcinoma of the head and neckCancer2010116204753476020549817
- ChoWCMicroRNAs: potential biomarkers for cancer diagnosis, prognosis and targets for therapyInt J Biochem Cell Biol20104281273128120026422
- Esquela-KerscherASlackFJOncomirs – microRNAs with a role in cancerNat Rev Cancer20066425926916557279
- LuJGetzGMiskaEAMicroRNA expression profiles classify human cancersNature2005435704383483815944708
- BerezikovEGuryevVvan de BeltJWienholdsEPlasterkRHCuppenEPhylogenetic shadowing and computational identification of human microRNA genesCell20051201212415652478
- Lopez-CimaMFGonzalez-ArriagaPGarcia-CastroLPolymorphisms in XPC, XPD, XRCC1, and XRCC3 DNA repair genes and lung cancer risk in a population of northern SpainBMC Cancer2007716217705814
- BartelDPMicroRNAs: genomics, biogenesis, mechanism, and functionCell2004116228129714744438
- RuanKFangXOuyangGMicroRNAs: novel regulators in the hallmarks of human cancerCancer Lett2009285211612619464788
- Lagos-QuintanaMRauhutRMeyerJBorkhardtATuschlTNew microRNAs from mouse and humanRNA20039217517912554859
- Valencia-SanchezMALiuJHannonGJParkerRControl of translation and mRNA degradation by miRNAs and siRNAsGenes Dev200620551552416510870
- AmbrosVThe functions of animal microRNAsNature2004431700635035515372042
- ChenKSongFCalinGAWeiQHaoXZhangWPolymorphisms in microRNA targets: a gold mine for molecular epidemiologyCarcinogenesis20082971306131118477647
- RyanBMRoblesAIHarrisCCGenetic variation in microRNA networks: the implications for cancer researchNature Rev Cancer201010638940220495573
- MinKTKimJWJeonYJAssociation of the miR-146aC>G, 149C>T, 196a2C>T, and 499A>G polymorphisms with colorectal cancer in the Korean populationMol Carcinog201251Suppl 1E65E7322161766
- HezovaRKovarikovaABienertova-VaskuJEvaluation of SNPs in miR-196-a2, miR-27a and miR-146a as risk factors of colorectal cancerWorld J Gastroenterol201218222827283122719192
- JiangJJiaZFCaoDHWuYHSunZWCaoXYAssociation of the miR-146a rs2910164 polymorphism with gastric cancer susceptibility and prognosisFuture Oncol201612192215222627267319
- HuYYuCYWangJLGuanJChenHYFangJYMicroRNA sequence polymorphisms and the risk of different types of cancerSci Rep20144364824413317
- JohnnidisJBHarrisMHWheelerRTRegulation of progenitor cell proliferation and granulocyte function by microRNA-223Nature200845171821125112918278031
- WoolfBOn estimating the relation between blood group and diseaseAnn Hum Genet195519425125314388528
- LiuYZhanYChenZDirecting cellular information flow via CRISPR signal conductorsNat Methods2016131193894427595406
- VangelMGRukhinALMaximum likelihood analysis for heteroscedastic one-way random effects ANOVA in interlaboratory studiesBiometrics199955112913611318146
- DerSimonianRKackerRRandom-effects model for meta-analysis of clinical trials: an updateContemp Clin Trials200728210511416807131
- MantelNHaenszelWStatistical aspects of the analysis of data from retrospective studies of diseaseJ Natl Cancer Inst195922471974813655060
- EggerMDavey SmithGSchneiderMMinderCBias in meta-analysis detected by a simple, graphical testBMJ199731571096296349310563
- HuZLiangJWangZCommon genetic variants in pre-microRNAs were associated with increased risk of breast cancer in Chinese womenHum Mutat2009301798418634034
- CatucciIYangRVerderioPEvaluation of SNPs in miR-146a, miR196a2 and miR-499 as low-penetrance alleles in German and Italian familial breast cancer casesHum Mutat2010311E1052E105719847796
- ChristensenBCAvissar-WhitingMOuelletLGMature microRNA sequence polymorphism in MIR196A2 is associated with risk and prognosis of head and neck cancerClin Cancer Res201016143713372020501619
- WangKGuoHHuHA functional variation in pre-microRNA-196a is associated with susceptibility of esophageal squamous cell carcinoma risk in Chinese HanBiomarkers201015761461820722507
- GeorgeGPGangwarRMandalRKSankhwarSNMittalRDGenetic variation in microRNA genes and prostate cancer risk in North Indian populationMol Biol Rep20113831609161520842445
- JedlinskiDJGabrovskaPNWeinsteinSRSmithRAGriffithsLRSingle nucleotide polymorphism in hsa-mir-196a-2 and breast cancer risk: a case control studyTwin Res Hum Genet201114541742121962133
- MittalRDGangwarRGeorgeGPMittalTKapoorRInvestigative role of pre-microRNAs in bladder cancer patients: a case-control study in North IndiaDNA Cell Biol201130640140621345130
- ZhanJFChenLHChenZXA functional variant in microRNA-196a2 is associated with susceptibility of colorectal cancer in a Chinese populationArchiv Med Res2011422144148
- ZhouBWangKWangYCommon genetic polymorphisms in pre-microRNAs and risk of cervical squamous cell carcinomaMol Carcinog201150749950521319225
- ChenHSunLYChenLLZhengHQZhangQFA variant in microRNA-196a2 is not associated with susceptibility to and progression of colorectal cancer in ChineseInt Med J2012426e115e119
- LinharesJJAzevedoMJrSiufiAAEvaluation of single nucleotide polymorphisms in microRNAs (hsa-miR-196a2 rs11614913 C/T) from Brazilian women with breast cancerBMC Med Genet20121311923228090
- YoonKAYoonHParkSThe prognostic impact of microRNA sequence polymorphisms on the recurrence of patients with completely resected non-small cell lung cancerJ Thorac Cardiovasc Surg2012144479480722818121
- ZhuLChuHGuDA functional polymorphism in miRNA-196a2 is associated with colorectal cancer risk in a Chinese populationDNA Cell Biol201231335035421815818
- AhnDHRahHChoiY-KAssociation of the miR-146aC>G, miR-149T>C, miR-196a2T>C, and miR-499A>G polymorphisms with gastric cancer risk and survival in the Korean populationMol Carcinog201352Suppl 1395122006587
- HanYPuRHanXAssociations of pri-miR-34b/c and pre-miR-196a2 polymorphisms and their multiplicative interactions with hepatitis B virus mutations with hepatocellular carcinoma riskPLoS One201383e5856423516510
- HongMJChoiYYJangJAAssociation between genetic variants in pre-microRNAs and survival of early-stage NSCLCJ Thorac Oncol20138670371023470291
- LvMDongWLiLAssociation between genetic variants in pre-miRNA and colorectal cancer risk in a Chinese populationJ Cancer Res Clin Oncol201313981405141023728616
- SongXSturgisEMLiuJMicroRNA variants increase the risk of HPV-associated squamous cell carcinoma of the oropharynx in never smokersPLoS One201382e5662223457596
- WangSTaoGWuDA functional polymorphism in MIR196A2 is associated with risk and prognosis of gastric cancerMol Carcinog201352Suppl 1E87E9523423813
- YuanZZengXYangDWangWLiuZEffects of common polymorphism rs11614913 in Hsa-miR-196a2 on lung cancer riskPLoS One201384e6104723593385
- ZhangJWangRMaY-YAssociation between single nucleotide polymorphisms in miRNA196a-2 and miRNA146a and susceptibility to hepatocellular carcinoma in a Chinese populationAsian Pac J Cancer Prev201314116427643124377545
- BansalCSharmaKLMisraSSrivastavaANMittalBSinghUSCommon genetic variants in pre-microRNAs and risk of breast cancer in the North Indian populationEcancermedicalscience2014847325374621
- ChuYHHsiehMJChiouHLMicroRNA gene polymorphisms and environmental factors increase patient susceptibility to hepatocellular carcinomaPLoS One201492e8993024587132
- DikeakosPTheodoropoulosGRizosSTzanakisNZografosGGazouliMAssociation of the miR-146aC>G, miR-149T>C, and miR-196a2T>C polymorphisms with gastric cancer risk and survival in the Greek populationMol Biol Rep20144121075108024379078
- DuMLuDWangQGenetic variations in microRNAs and the risk and survival of renal cell cancerCarcinogenesis20143571629163524681820
- DuWMaX-LZhaoCAssociations of single nucleotide polymorphisms in miR-146a, miR-196a, miR-149 and miR-499 with colorectal cancer susceptibilityAsian Pac J Cancer Prev20141521047105524568449
- FanXWuZEffects of four single nucleotide polymorphisms in microRNA-coding genes on lung cancer riskTumour Biol20143511108151082425077922
- KupcinskasJBruzaiteIJuzenasSLack of association between miR-27a, miR-146a, miR-196a-2, miR-492 and miR-608 gene polymorphisms and colorectal cancerSci Rep20144599325103961
- LiPYanHZhangHA functional polymorphism in MIR196A2 is associated with risk and progression of nasopharyngeal carcinoma in the Chinese populationGenet Test Mol Biomarkers201418314915524328526
- QuYQuHLuoMMicroRNAs related polymorphisms and genetic susceptibility to esophageal squamous cell carcinomaMol Genet Genom2014289611231130
- TongNXuBShiDHsa-miR-196a2 polymorphism increases the risk of acute lymphoblastic leukemia in Chinese childrenMutat Res2014759162124291415
- WangNLiYZhouRMHsa-miR-196a2 functional SNP is associated with the risk of ESCC in individuals under 60 years oldBiomarkers2014191434824320161
- DengSWangWLiXZhangPCommon genetic polymorphisms in pre-microRNAs and risk of bladder cancerWorld J Surg Oncol20151329726458899
- DikaiakosPGazouliMRizosSZografosGTheodoropoulosGEEvaluation of genetic variants in miRNAs in patients with colorectal cancerCancer Biom2015152157162
- LiXLiKWuZAssociation of four common SNPs in microRNA polymorphisms with the risk of hepatocellular carcinomaInt J Clin Exp Pathol2015889560956626464719
- NiQJiAYinJWangXLiuXEffects of two common polymorphisms rs2910164 in miR-146a and rs11614913 in miR-196a2 on gastric cancer susceptibilityGastroenterol Res Pract2015201576416325983750
- NikolicZSavic PavicevicDVucicNAssessment of association between genetic variants in microRNA genes hsa-miR-499, hsa-miR-196a2 and hsa-miR-27a and prostate cancer risk in Serbian populationExp Mol Pathol201599114515026112096
- QiPWangLZhouBAssociations of miRNA polymorphisms and expression levels with breast cancer risk in the Chinese populationGenet Mol Res20151426289629626125831
- WuYHaoXFengZLiuYGenetic polymorphisms in miRNAs and susceptibility to colorectal cancerCell Biochem Biophys201571127127825213291
- DaiZMKangHFZhangWGThe Associations of single nucleotide polymorphisms in miR196a2, miR-499, and miR-608 with breast cancer susceptibility: A STROBE-Compliant Observational StudyMedicine2016957e282626886638
- LiJChengGWangSA single-nucleotide polymorphism of miR-196a2T>C rs11614913 is associated with hepatocellular carcinoma in the Chinese populationGenet Test Mol Biomarkers201620421321526866381
- LiMLiRJBaiHAssociation between the pre-miR-196a2 rs11614913 polymorphism and gastric cancer susceptibility in a Chinese populationGenet Mol Res2016152
- QiuGPLiuJMicroRNA gene polymorphisms in evaluating therapeutic efficacy after transcatheter arterial chemoembolization for primary hepatocellular carcinomaGenet Test Mol Biomarkers2016201057958627525669
- ShenFChenJGuoSGenetic variants in miR-196a2 and miR-499 are associated with susceptibility to esophageal squamous cell carcinoma in Chinese Han populationTumour Biol20163744777478426518769
- SongZSWuYZhaoHGAssociation between the rs11614913 variant of miRNA-196a-2 and the risk of epithelial ovarian cancerOncol Lett201611119420026870188
- XuXLingQWangJDonor miR-196a-2 polymorphism is associated with hepatocellular carcinoma recurrence after liver transplantation in a Han Chinese populationInt J Cancer2016138362062926365437
- YinZCuiZRenYAssociation between polymorphisms in pre-miRNA genes and risk of lung cancer in a Chinese non-smoking female populationLung Cancer201694152126973201
- ZhaoHXuJZhaoDSomatic mutation of the SNP rs11614913 and its association with increased MIR 196A2 expression in breast cancerDNA Cell Biol2016352818726710106
- OkuboMTaharaTShibataTAssociation between common genetic variants in pre-microRNAs and gastric cancer risk in Japanese populationHelicobacter201015652453121073609
- PuJYDongWZhangLLiangWBYangYLvMLNo association between single nucleotide polymorphisms in pre-mirnas and the risk of gastric cancer in Chinese populationIran J Basic Med Sci201417212813324711897
- OmraniMHashemiMEskandari-NasabEhsa-mir-499 rs3746444 gene polymorphism is associated with susceptibility to breast cancer in an Iranian populationBiomark Med20148225926724521023
- QiJHWangJChenJHigh-resolution melting analysis reveals genetic polymorphisms in microRNAs confer hepatocellular carcinoma risk in Chinese patientsBMC Cancer20141464325176041
- ChuYHTzengSLLinCWChienMHChenMKYangSFImpacts of microRNA gene polymorphisms on the susceptibility of environmental factors leading to carcinogenesis in oral cancerPLoS One201276e3977722761899
- GuJYTuLInvestigating the role of polymorphisms in miR-146a, -149, and -196a2 in the development of gastric cancerGenet Mol Res2016152 gmr.15027839
- HashemiMMoradiNZiaeeSAAssociation between single nucleotide polymorphism in miR-499, miR-196a2, miR-146a and miR-149 and prostate cancer risk in a sample of Iranian populationJ Adv Res20167349149827222754
- HeBPanYXuYAssociations of polymorphisms in microRNAs with female breast cancer risk in Chinese populationTumour Biol20153664575458225613069
- KouJTFanHHanDAssociation between four common microRNA polymorphisms and the risk of hepatocellular carcinoma and HBV infectionOncol Lett2014831255126025120701
- MoralesSGulppiFGonzalez-HormazabalPAssociation of single nucleotide polymorphisms in Pre-miR-27a, Pre-miR-196a2, Pre-miR-423, miR-608 and Pre-miR-618 with breast cancer susceptibility in a South American populationBMC Genet201617110927421647
- PavlakisEPapaconstantinouIGazouliMMicroRNA gene polymorphisms in pancreatic cancerPancreatology201313327327823719600
- RoyRDe SarkarNGhoseSGenetic variations at microRNA and processing genes and risk of oral cancerTumour Biol20143543409341424297336
- SodhiKKBahlCSinghNBeheraDSharmaSFunctional genetic variants in pre-miR-146a and 196a2 genes are associated with risk of lung cancer in North IndiansFuture Oncol201511152159217326235181
- SunXCZhangACTongLLmiR-146a and miR-196a2 polymorphisms in ovarian cancer riskGenet Mol Res292016153 gmr.15038468
- SushmaPSJamilKKumarPUSatyanarayanaURamakrishnaMTriveniBGenetic variation in MicroRNAs and risk of oral squamous cell carcinoma in South Indian populationAsian Pac J Cancer Prev201516177589759426625766
- ToraihEAFawzMSElgazzazMGHusseinMHShehataRHDaoudHGCombined genotype analyses of precursor miRNA196a2 and 499a variants with hepatic and renal cancer susceptibility a Preliminary StudyAsian Pac J Cancer Prev20161773369337527509977
- ZhangLHHaoBBZhangCYDaiXZZhangFContributions of polymorphisms in miR146a, miR196a, and miR499 to the development of hepatocellular carcinomaGenet Mol Res2016153
- ZhangMJinMYuYAssociations of miRNA polymorphisms and female physiological characteristics with breast cancer risk in Chinese populationEur J Cancer Care (Engl)201221227428022074121
- GuoJJinMZhangMChenKA genetic variant in miR-196a2 increased digestive system cancer risks: a meta-analysis of 15 case-control studiesPLoS One201271e3058522291993
- MaXPZhangTPengBYuLJiang deKAssociation between microRNA polymorphisms and cancer risk based on the findings of 66 case-control studiesPLoS One2013811e7958424278149
- WangJWangQLiuHThe association of miR-146a rs2910164 and miR-196a2 rs11614913 polymorphisms with cancer risk: a meta-analysis of 32 studiesMutagenesis201227677978822952151
- ZhangHSuYLYuHQianBYMeta-analysis of the association between Mir-196a-2 polymorphism and cancer susceptibilityCancer Biol Med201291637223691458
- ChenCZhangYZhangLWeakleySMYaoQMicroRNA-196: critical roles and clinical applications in development and cancerJ Cell Mol Med2011151142321091634
- GarzonRMarcucciGCroceCMTargeting microRNAs in cancer: rationale, strategies and challengesNature Rev Drug Discov201091077578920885409
- VoliniaSCalinGALiuCGA microRNA expression signature of human solid tumors defines cancer gene targetsProc Natl Acad Sci U S A200610372257226116461460
- XuJHuZXuZFunctional variant in microRNA-196a2 contributes to the susceptibility of congenital heart disease in a Chinese populationHum Mut20093081231123619514064
- KangZLiYHeXQuantitative assessment of the association between miR-196a2 rs11614913 polymorphism and cancer risk: evidence based on 45,816 subjectsTumour Biol20143576271628224633889
- KimMJYooSSChoiYYParkJYA functional polymorphism in the pre-microRNA-196a2 and the risk of lung cancer in a Korean populationLung cancer201069112712920466450
- VinciSGelminiSPratesiNGenetic variants in miR-146a, miR-149, miR-196a2, miR-499 and their influence on relative expression in lung cancersClinical chemistry and laboratory medicine201149122073208021902575
- UmarMUpadhyayRPrakashGKumarSGhoshalUCMittalBEvaluation of common genetic variants in pre-microRNA in susceptibility and prognosis of esophageal cancerMol Carcinog201352Suppl 1E101822692992
- VinciSGelminiSManciniIGenetic and epigenetic factors in regulation of microRNA in colorectal cancersMethods201359113814622989523
- WeiJZhengLLiuSMiR-196a2 rs11614913 T > C polymorphism and risk of esophageal cancer in a Chinese populationHuman immunology20137491199120523792053
- ParlayanCIkedaSSatoNSawabeMMuramatsuMAraiTAssociation analysis of single nucleotide polymorphisms in miR-146a and miR-196a2 on the prevalence of cancer in elderly Japanese: a case-control studyAsian Pacific journal of cancer prevention: APJCP20141552101210724716941