Abstract
Background and aims
Recently, the abnormal expression of FoxM1 has been found in many malignant tumors. However, the clinicopathological and prognostic value of FoxM1 expression in hepatocellular carcinoma (HCC) patients remains controversial. We conducted a meta-analysis to establish the relationship between FoxM1 expression and the clinicopathological features and prognostic value in patients with HCC.
Methods
An electronic search for relevant articles was conducted according to a set of criteria in the PubMed, Cochrane Library, Web of Science, EMBASE, Chinese CNKI and Chinese WanFang databases. The correlation data between FoxM1 expression and clinicopathological features and survival outcomes were analyzed. Pooled odds ratios (ORs) and hazard ratios (HRs) with 95% CIs were calculated using STATA14.2.
Results
A total of 14 studies comprising of 2,036 patients were enrolled in this meta-analysis. The results showed that FoxM1 expression was related to the incidence, tumor size (>5 cm), vascular invasion, differentiation and TNM stage. Moreover, overexpression of FoxM1 indicated a poor 3- and 5-year overall survival rate (OS) and recurrence-free survival rate (disease-free survival rate).
Conclusion
Our meta-analysis indicated that FoxM1 expression was associated with incidence, tumor size (>5 cm), vascular invasion, differentiation and TNM stage. Accordingly, FoxM1 may be a reliable prognostic biomarker for patients with HCC. However, additional high-quality studies are still needed to further support these findings.
Introduction
Hepatocellular carcinoma (HCC) is one of the most common malignancies, which ranks fifth in incidence and second in mortality rate among all cancers worldwide, with about 690,000 patients’ deaths each year.Citation1 At present, radical resection and liver transplantation are the main treatment options for HCC. However, the clinical symptoms of liver cancer in its early stage are atypical and many patients who have undergone treatment progress to middle-late stage. Moreover, HCC is not sensitive to radiotherapy and chemotherapy, and it easily recurs and metastasizes, which all contribute to its poor prognosis.Citation2–Citation4 Therefore, it is very important to study the molecular mechanism of recurrence, invasion and metastasis of liver cancer and find a more suitable prognostic target for HCC.
FoxM1 is a member of the Fox transcription factor family, and the human FoxM1 gene is located on chromosome 12p13.3 and contains 10 exons.Citation5 FoxM1 plays an important role in the cell cycle phases G1/S and G2/M as a proliferation-specific transcription factor.Citation6,Citation7 Recently, studies have found that FoxM1 was highly expressed in a variety of malignant tumors, such as prostate cancer,Citation8 cervical cancer,Citation9 pancreatic cancer,Citation10 lung cancer.Citation11 Moreover, the expression of FoxM1 was related to clinical features and prognosis. Several studies also found that FoxM1 is abnormally expressed in HCC. However, the relationship between the expression of FoxM1 and the clinicopathological characteristics and prognostic value in patients with HCC remains controversial. Lin et alCitation12 found that FoxM1 expression was associated with differentiation and TNM stage, but was not related to vascular invasion, tumor size and liver cirrhosis. Egawa et alCitation13 found that FoxM1 expression was associated with alpha fetoprotein (AFP) level, tumor size differentiation, TNM stage and poor prognosis. Sun et alCitation14 found that positive FoxM1 expression was correlated with tumor size, tumor number, TNM stage, but was not associated with AFP level, liver cirrhosis, vascular invasion and differentiation. Shi et alCitation15 found that FoxM1 expression was related to differentiation, but was not associated with tumor size, tumor number, differentiation and TNM stage. Additionally, they found no significant correlation between FoxM1 expression and overall survival rate by multivariate analysis. In contrast, Meng et alCitation16 found that overexpression of FoxM1 was related to tumor size, tumor number, vascular invasion and TNM stage, but not to differentiation. In view of the above discrepancies, it is necessary to conduct a meta-analysis to assess the role of FoxM1 in HCC.
Methods
Search strategy
A comprehensive literature search was conducted in electronic databases including the PubMed, Cochrane Library, Web of Science, EMBASE, Chinese CNKI and Chinese WanFang databases. The search ended on October 1, 2017. The following key words were used: (“FoxM1” or “FoxM1a” or “FoxM1b” or “FoxM1c”) and (“hepatocellular carcinoma” or “hepatic tumor” or “liver tumor” or “hepatic cancer” or “liver cancer” or “HCC”). No language limitation was imposed. We also searched the references cited in the identified articles to find out other applicable studies.
Inclusion criteria and exclusion criteria
The following inclusion criteria were used for all articles identified in the literature search: 1) The patients were clearly diagnosed with HCC and the FoxM1 expression status was directly examined; 2) The expression of FoxM1 was mainly tested by immunohistochemistry (IHC) analysis or RT-PCR analysis; 3) Results that included clinicopathological characteristics, disease-free (recurrence-free) survival and overall survival and Hazard ratios (HRs) were reported or could be calculated from the published data.
Studies were excluded if: 1) they were non-eligible trials including ecological studies, case reports, reviews, editorials, letters, conference abstracts and animal trials; 2) they were repeat studies based on the same database or patients.
Data extraction and quality assessment
Two authors (Chaojie Liang, Jingyang Zhao) inspected the search findings. The titles and abstracts were inspected for potentially eligible studies. Discrepancies were resolved by discussion, re-extraction, or third-party adjudication. Extracted data included first author, publication year, experimental methods, antibody, antibody concentration, cut-off value, control, clinicopathological features and HRs with 95% CI for overall survival, recurrence-free survival and disease-free survival. The Newcastle-Ottawa Scale (NOS) was used to evaluate the quality of the studies, when the NOS≥6, we considered the study to be high quality, otherwise it was deemed as low quality. All of the studies included in this meta-analysis were considered to be high quality.
Statistical analysis
In this meta-analysis, the STATA 14.2 software (StataCorp LLC., College Station, TX, USA) was used to calculate the pooled odds ratios (ORs) and hazard ratios (HRs) with 95% confidence intervals (CIs). Engauge Digitizer 10.0 software (GitHub, Inc., San Francisco, CA, USA) was used to extract the survival data from Kaplan–Meier curves in the articles. The relationship between FoxM1 expression and clinicopathological features, including gender, hepatitis B surface antigen (HBsAg), AFP, liver cirrhosis, tumor size (5 cm), vascular invasion, tumor number, differentiation and TNM stage was evaluated by fixed- or random-effects models when ICitation2 was >50% or <50%. The HR or OR was considered to be statistically significant if the 95% CI did not overlap 1. The potential publication bias was examined by the funnel plots and Begg’s test.
Results
Characteristics of studies
The details of the selection process are shown in . Fourteen studies,Citation12–Citation25 comprising a total of 2,036 patients, were included in this meta-analysis. The included studies were published from 2010 to 2017, the sample size ranged from 52 to 406 patients. Thirteen of these studies were from China, and one from Japan. Nine of these studies were published in English, and 5 in Chinese. All these studies scored ≥6 in the methodological assessment, which implied that they were of high quality.
The relationship between FoxM1 expression and clinicopathological features in patients with HCC
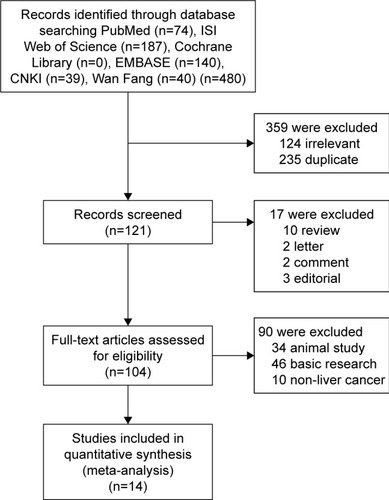
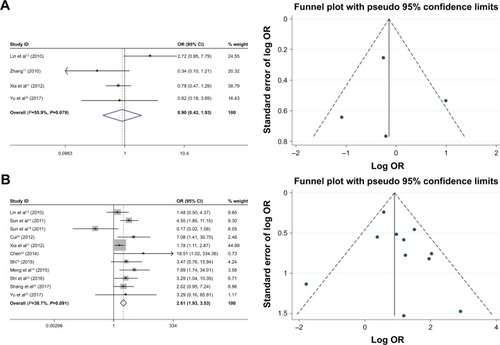
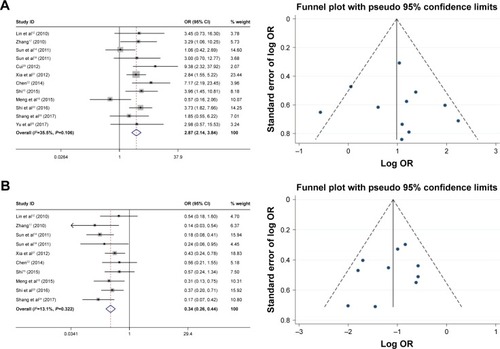
As shown in , the relationship between the FoxM1 expression and the clinicopathological features of HCC was established in this meta-analysis. Seven studies comprising 1,659 patients were used to evaluate the difference in FoxM1 expression between HCC tissue and normal tissue. The expression of FoxM1 in HCC was significantly higher than that in normal tissue (OR=6.54, 95% CI=3.84–11.14, P<0.001, ). However, the expression of FoxM1 was not significantly associated with gender (OR=0.89, 95% CI=0.67–1.17, P=0.391, ), HBsAg (OR=1.33, 95% CI=0.93–1.91, P=0.871, ), liver cirrhosis (OR=1.05, 95% CI=0.81–1.37, P=0.693, ), AFP (OR=1.26, 95% CI=1.00–1.59, P=0.054, ) and tumor number (OR=0.90, 95% CI=0.42–1.93, P=0.793, ). Six studies comprising 783 patients exhibited a significant relationship between FoxM1 expression and tumor size (≤5 cm: >5 cm) (OR=0.47, 95% CI=0.35–0.64, P<0.001, ). The ORs for vascular invasion were calculated for 11 studies, including 1,491 patients. The results indicated a significant association between FoxM1 expression and vascular invasion (OR=2.61, 95% CI=1.93–3.53, P<0.001, ). Additionally, FoxM1 expression was related to differentiation (OR=2.87, 95% CI=2.14–3.84, P=0.000, ) and TNM stage (OR=0.34, 95% CI=0.26–0.44, P<0.001, ).
Table 1 FoxM1 clinicopathological features for HCC
Figure 2 Forest plot and funnel plots of the studies evaluating the relationship between FoxM1 expression and clinicopathological features.
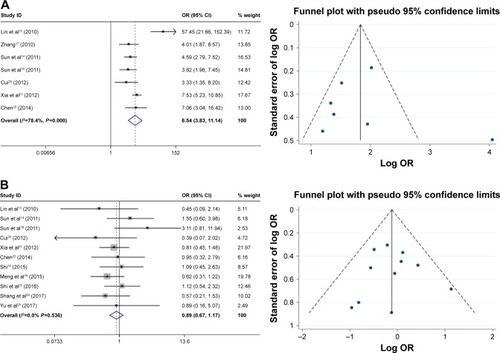
Figure 3 Forest plot and funnel plots of studies evaluating the relationship between FoxM1 expression and clinicopathological features.
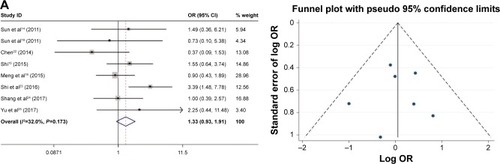
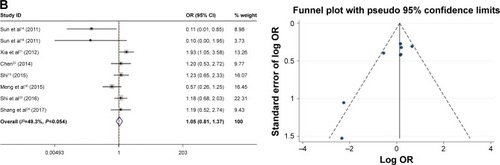
Figure 4 Forest plot and funnel plots of studies evaluating the relationship between FoxM1 expression and clinicopathological features.
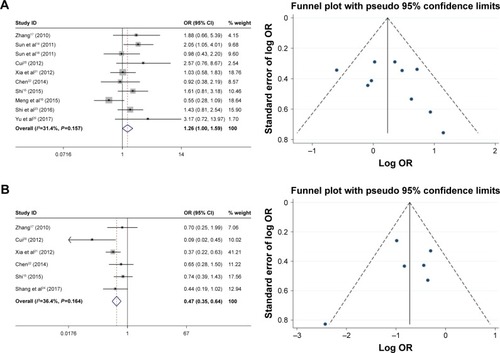
The association between FoxM1 expression and overall survival in patients with HCC
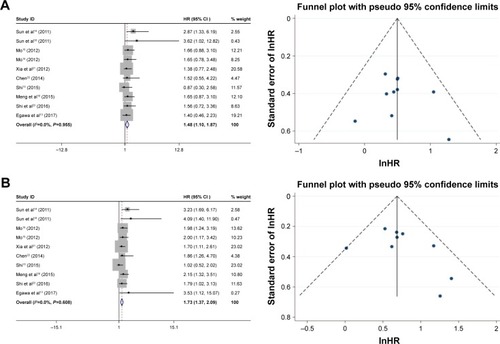
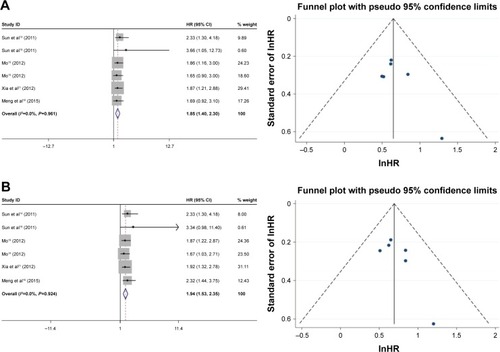
The HRs for the 3-year and 5-year overall survival rate were calculated for 9 studies (Mo Ping had two data sets), which comprised 981 cases with positive FoxM1 expression and 705 cases with negative FoxM1. The results indicated that the expression of FoxM1 was related to poor 3-year overall survival (positive: negative, HR=1.48, 95% CI=1.10–1.87, P=0.000, ) and 5-year overall survival (positive: negative, HR=1.73, 95% CI=1.37–2.09, P=0.000, ).
The association between FoxM1 expression and recurrence-free (disease-free) survival in patients with HCC
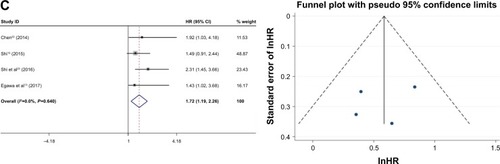
The HRs for the 3-year and 5-year recurrence-free survival rate were assessed for five studies (Mo Ping had two data sets), which included 676 cases with positive FoxM1 expression and 457 cases with negative FoxM1. The results indicated that expression of FoxM1 was related to poor 3-year recurrence-free survival (positive: negative, HR=1.85, 95% CI=1.40–2.30, P=0.000, ) and 5-year recurrence-free survival (positive: negative, HR=1.94, 95% CI=1.53–2.35, P=0.000, ). The HRs for disease-free survival rate were assessed for four studies, which included 305 cases with FoxM1 expression and 248 cases with FoxM1 suppression, the results showed that expression of FoxM1 was associated with poor disease-free survival (positive: negative, HR=1.72, 95% CI=1.19–2.35, P=0.000, ).
Publication bias and sensitivity analysis
According to the results of the Begg’s test, there was no publication bias for FoxM1 expression (P=0.734), gender (P=1), HBsAg (P=1), liver cirrhosis (P=0.063), AFP (P=0.296), tumor size (P=1), vascular invasion (P=0.755), differentiation (P=0.945), TNM stage (P=0.592), 3-year overall survival rate (P=0.592), 5-year overall survival rate (P=0.474), 3-year recurrence-free survival rate (P=1), 5-year recurrence-free survival rate (P=0.452) and disease-free survival rate (P=0.452).
Discussion
FoxM1 is a member of the fork head transcription factor family, which is located at chromosome 12p13. The FoxM1 gene encodes three isoforms: FoxM1A is a transcriptional repressor, whereas FoxM1B and FoxM1C are both transcriptional activators.Citation26–Citation28 Previous studies have shown that FoxM1 plays a key role in cell cycle progression, through the regulation of the transcription of Cyclin B1, Cyclin D1, CDC25A and CDC25B to promote cell cycles from the G1 to S phase and the G2 to M phase.Citation26,Citation29 Downregulation of FoxM1 leads to cell cycle progression arrest and abnormal chromatin accumulation.Citation30,Citation31 In addition, as a typical transcription factor, FoxM1 plays an important role in embryonic development and tissue regeneration as it is involved in DNA repair and protein synthesis through the transcriptional regulation of many cell proliferation-related genes.Citation32
Recently, FoxM1 was found to be highly expressed in a variety of malignant tumor tissues and its expression is closely related to the occurrence and development of tumor, and indicated a poor differentiation, high degree of malignancy, distant metastasis, and poor prognosis.Citation33,Citation34 However, the relationship between FoxM1 expression and clinicopathological and prognostic value in patients with HCC remains controversial. The 14 articles included in this study, comprising 2,036 patients, indicated that FoxM1 was highly expressed in HCC, and FoxM1 expression was related to tumor size, vascular invasion, differentiation and TNM stage. Five of 11, six of 12 and three of 10 studies suggested that FoxM1 expression was not correlated to vascular invasion, differentiation and TNM stage, but the overall results indicated the opposite. This suggested that the FoxM1 expression was associated with tumor progression.
Two previous meta-analysesCitation35,Citation36 investigated the relationship between FoxM1 expression and the prognosis of solid tumors. Their results suggested that the FoxM1 expression was associated with poor prognosis, and the same result was obtained in the subgroup of HCC. However, the number of studies and patients included in these meta-analyses were limited, and there was no report about the relationship between FoxM1 expression and recurrence-free survival. Our meta-analysis included nine studies and 1,686 patients, suggesting that overexpression of FoxM1 predicted a poor 3-year, 5-year overall survival. Moreover, the results also indicated a poor 3-year, 5-year recurrence-free survival and disease-free survival. FoxM1 may be one of the reliable prognostic indicators of liver cancer.
With the advances in the study of FoxM1, researchers have found that FoxM1 is involved in a variety of malignant biological behaviors of tumor, which suggest that FoxM1 has potential as a target for future cancer therapies.Citation33,Citation37 FoxM1 is involved in the development of tumors mainly through its role in promoting tumor cell proliferation and cell cycle progression. Yu et alCitation38 found that upregulation of FoxM1 expression in HCC cell lines upregulated the level of cyclin B1 and cyclin D1 in order to enhance cell proliferation. Chen et alCitation39 reported that downregulation of FoxM1 can significantly inhibit the tumorigenic ability of HCC cells. Also, Yu et alCitation25 found that FoxM1 enhanced the invasion and metastatic ability of hepatoma cells by upregulating SNAI1 expression and promoting the epithelial–mesenchymal transition (EMT) process in hepatocellular carcinoma. In addition, Meng et alCitation16 found that FoxM1 expression was negatively correlated with E-cadherin expression in HCC tissues, and the expression level of E-cadherin was decreased after overexpression of FoxM1, the invasion ability was enhanced and the EMT process was promoted by activating the SNAI1. Shang et alCitation24 found that FoxM1 regulated glycolysis in hepatoma cells by downregulating GLUT1 expression.
Limitations
This meta-analysis, however, had some limitations: 1) there are some unpublished studies with different results, which may cause publication bias; 2) most of the articles included in this meta-analysis were from China, and the results may only be applicable for Chinese or Asian populations, and more articles from other regional areas should be included to support this meta-analysis; 3) the experimental methods in most of the articles included immunohistochemistry analysis, which is a semi-quantitative assay, and the results were likely to be affected by the antibody used, concentration of antibody and cut-off value. As shown in , the antibodies and concentrations were different, while the cut-off values were not the same, and these would have an impact on the results and may be the reason for the heterogeneity; 4) the number of articles and patients included in this meta-analysis are insufficient, and more high-quality studies are still needed to support the results; 5) the HRs were obtained using the Engauge Digitizer 10.0 software from the K-M curve, which may cause some errors.
Table 2 Characteristics of studies included in the meta-analysis
Although there are some limitations in the article, the results are still meaningful, our meta-analysis found that FOXM1 expression was related to the incidence, tumor size, vascular invasion, differentiation, and TNM stage. Additionally, the results also indicated a poor 3-year, 5-year OS and recurrence-free survival (disease-free survival), and FOXM1 may be a reliable biomarker for the prognosis of HCC patients. However, more high-quality studies to support these findings are still needed.
Acknowledgments
This study was supported by the Beijing Municipal Administration of the Hospital Clinical Medicine Development of Special Funding Support (ZYLX201612), Capital Foundation of Medical Development (shoufa2016-2-2053), Beijing Tongren Hospital Funds (TRYY-KYJJ-2015-032), and the Municipal Administration of Hospital Clinical Medicine Development of Special Funding Support (trzdyxzy201705).
Disclosure
The authors report no conflicts of interest in this work.
References
- GongXLQinSKProgress in systemic therapy of advanced hepatocel-lular carcinomaWorld J Gastroenterol201622296582659427547002
- FornerALlovetJMBruixJHepatocellular carcinomaLancet20123799821245125522353262
- AvilaMABerasainCSangroBPrietoJNew therapies for hepatocellular carcinomaOncogene200625273866388416799628
- NishikawaHKimuraTKitaROsakiYTreatment for hepatocellular carcinoma in elderly patients: a literature reviewJ Cancer20134863564324155775
- MyattSSLamEWThe emerging roles of forkhead box (Fox) proteins in cancerNat Rev Cancer200771184785917943136
- Alvarez-FernándezMHalimVAApreliaMLaoukiliJMohammedSMedemaRHProtein phosphatase 2A (B55alpha) prevents premature activation of forkhead transcription factor FoxM1 by antagonizing cyclin A/cyclin-dependent kinase-mediated phosphorylationJ Biol Chem201128638330293303621813648
- BolteCZhangYWangICKalinTVMolkentinJDKalinichenkoVVExpression of FoxM1 transcription factor in cardiomyocytes is required for myocardial developmentPLoS One201167e2221721779394
- LiuYLiuYYuanBFOXM1 promotes the progression of prostate cancer by regulating PSA gene transcriptionOncotarget2017810170271703728199985
- ChenHWangJYangHChenDLiPAssociation between FOXM1 and hedgehog signaling pathway in human cervical carcinoma by tissue microarray analysisOncol Lett20161242664267327698840
- CuiJShiMXieDFOXM1 promotes the warburg effect and pancreatic cancer progression via transactivation of LDHA expressionClin Cancer Res201420102595260624634381
- KongFFQuZQYuanHHOverexpression of FOXM1 is associated with EMT and is a predictor of poor prognosis in non-small cell lung cancerOncology Rep201431626602668
- LinMGuoLMLiuHNuclear accumulation of glioma-associated oncogene 2 protein and enhanced expression of forkhead-box transcription factor M1 protein in human hepatocellular carcinomaHistol Histopathol201025101269127520712011
- EgawaMYoshidaYOguraSIncreased expression of Forkhead box M1 transcription factor is associated with clinicopathological features and confers a poor prognosis in human hepatocellular carcinomaHepatol Res201747111196120528002884
- SunHCLiMLuJLOverexpression of Forkhead box M1 protein associates with aggressive tumor features and poor prognosis of hepatocellular carcinomaOncology Rep201125615331539
- ShiCThe molecular mechanisms by which Gli2 promotes hepatocellular cancer cell proliferation through induction of FoxM1/KIF20A expressionThesis NanChang University2015
- MengFDWeiJ-CQuKFoxM1 overexpression promotes epithelial-mesenchymal transition and metastasis of hepatocellular carcinomaWorld J Gastroenterol201521119621325574092
- ZhangLThe expression and clinical significance of FoxMB1 in hepatocellular carcinoma and the relationship with MTA1 and MMP-2ThesisGuangxi Medical University (In Chinese)2010
- SunHTengMLiuJFOXM1 expression predicts the prognosis in hepatocellular carcinoma patients after orthotopic liver transplantation combined with the Milan criteriaCancer Lett2011306221422221482449
- Upregulated FoxMo PM1 expression induced by TNF-α/ROS/HIF-1α pathway promotes HCC proliferation and resistance to apoptosisThesis Fourth Military Medical University2012 Available from: http://kns.cnki.net/KCMS/detail/detail.aspx?dbcode=CDFD&dbname=CDFD1214&filename=1012038774.nh&v=MTUwNzRFYlBJUjhlWDFMdXhZ-UzdEaDFUM3FUcldNMUZyQ1VSTEtmWXVkb0Z5cmhVTDNPVkYyNkhMTzdGdGJMcTU=
- CuiKThe expression and significance of FoxM1 and MMP-2 in hepatocellular carcinomaThesisHuazhong University of Science and Technology2012
- XiaLHuangWTianDUpregulated FoxM1 expression induced by hepatitis B virus X protein promotes tumor metastasis and indicates poor prognosis in hepatitis B virus-related hepatocellular carcinomaJ Hepatol201257360061222613004
- ChenDExpression of Gli2 and FoxM1 in hepatocellular carcinoma and their relationship with clinical prognosisThesis NanChang University2014
- ShiCHuangDLuNAberrantly activated Gli2-KIF20A axis is crucial for growth of hepatocellular carcinoma and predicts poor prognosisOncotarget2016718262062621927036048
- ShangRPuMLiYWangDFOXM1 regulates glycolysis in hepatocellular carcinoma by transactivating glucose transporter 1 expressionOncology Rep201737422612269
- YuCPYuSShiLFoxM1 promotes epithelial-mesenchymal transition of hepatocellular carcinoma by targeting Snai1Mol Med Rep20171645181518828849004
- YaoKMShaMLuZWongGGMolecular analysis of a novel winged helix protein, WIN. Expression pattern, DNA binding property, and alternative splicing within the DNA binding domainJ Biol Chem19972723219827198369242644
- YeHHoltermanAXYooKWFranksRRCostaRHPremature expression of the winged helix transcription factor HFH-11B in regenerating mouse liver accelerates hepatocyte entry into S phaseMol Cell Biol199919128570858010567581
- KimIMAckersonTRamakrishnaSThe Forkhead Box m1 transcription factor stimulates the proliferation of tumor cells during development of lung cancerCancer Res20066642153216116489016
- Krupczak-HollisKWangXKalinichenkoVVThe mouse Fork-head Box m1 transcription factor is essential for hepatoblast mitosis and development of intrahepatic bile ducts and vessels during liver morphogenesisDev Biol20042761748815531365
- LeungTWLinSSTsangACOver-expression of FoxM1 stimulates cyclin B1 expressionFEBS Lett20015071596611682060
- WangXKrupczak-HollisKTanYIncreased hepatic Forkhead Box M1B (FoxM1B) levels in old-aged mice stimulated liver regeneration through diminished p27Kip1 protein levels and increased Cdc25B expressionJ Biol Chem200227746443104431612221098
- WierstraIThe transcription factor FOXM1 (Forkhead box M1): proliferation-specific expression, transcription factor function, target genes, mouse models, and normal biological rolesAdv Cancer Res20131189739823768511
- GartelALFOXM1 in cancer: interactions and vulnerabilitiesCancer Res201777123135313928584182
- WangJLiWZhaoYMembers of FOX family could be drug targets of cancersPharmacol Ther201818118319628830838
- LiLWuDYuQLiLWuPPrognostic value of FOXM1 in solid tumors: a systematic review and meta-analysisOncotarget2017819322983230828427178
- DaiJYangLWangJXiaoYRuanQPrognostic value of FOXM1 in patients with malignant solid tumor: a meta-analysis and system reviewDisease Mark20152015352478
- NandiDCheemaPSJaiswalNNagAFoxM1: repurposing an oncogene as a biomarkerSemin Cancer Biol Epub9102017
- YuMTangZMengFElevated expression of FoxM1 promotes the tumor cell proliferation in hepatocellular carcinomaTumour Biol20163711289129726289845
- ChenTXiongJYangCSilencing of FOXM1 transcription factor expression by adenovirus-mediated RNA interference inhibits human hepatocellular carcinoma growthCancer Gene Ther201421313313824577129