Abstract
Background
Prostate cancer is one of the most commonly diagnosed diseases in males.
Methods
RT-qPCR was used to detect miR-129-5p expression in tumor tissues and adjacent normal tissues from patients with prostate cancer. The cell proliferation assay and colony forming assay were used to study the role of miR-129-5p in mediating prostate cancer cell growth. Bioinformatic analysis and dual luciferase assay were performed to predict and confirm ETV1 as a target gene of miR-129-5p.
Results
We found that miR-129-5p levels were decreased significantly in human prostate cancer tissues compared with matched normal tissues from patients with prostate cancer. Overexpression of miR-129-5p suppressed prostate cancer cell growth while antagonist of miR-129-5p promoted cell proliferation in immortal prostate cell line RWPE-1. In addition, elevation of miR-129-5p decreased ETV1 expression in prostate cancer cells while downregulation of miR-129-5p increased ETV1 expression in RWPE-1. Mechanistically, ETV1 is confirmed a direct target of miR-129-5p in prostate cancer cells. Through repression of ETV1 expression, miR-129-5p could inactivate YAP signaling in prostate cancer cells. In addition, overexpression of ETV1 attenuated miR-129-5p induced cell proliferation in prostate cancer cells. Correlation analysis further revealed that there was a negative correlation between miR-129-5p levels and ETV1 mRNA levels in tumor tissues from patients with prostate cancer.
Conclusion
Our results identified miR-129-5p as a tumor suppressor in prostate cancer via repression of ETV1.
Keywords:
Introduction
Prostate cancer is the most commonly diagnosed malignant cancer types in males and ranks third among cancer-related deaths in men worldwide.Citation1 Globally, there were estimated 903,500 new prostate cancer cases and 258,400 estimated deaths in 2012.Citation2 Currently, prostate-specific antigen is used for early prediction of prostate cancer.Citation3 When patients are diagnosed as early stage prostate cancer, radical prostatectomy that can greatly improve patients’ overall survival is performed.Citation4 Although many patients with prostate cancer are curable, tumor recurrence, metastasis, and development of castration resistance frequently occur and eventually lead to patient death.Citation5–Citation7 Advances in genetics have deepened our understandings on the carcinogenesis of prostate cancer,Citation8 but the precise molecular mechanism of prostate cancer progression is not fully elucidated. As a result, further investigation is still needed to provide valuable targets for prostate cancer treatment.
MicroRNAs (miRNAs) belong to a class of noncoding RNAs with the feathers of small (19–25 nucleotides), single-strand, and ubiquitously expression in human.Citation9 Mechanistically, miRNAs bind to complementary sites on 3′-untranslated region (3′UTR) of messenger RNA (mRNA) and lead to mRNA degradation or translational inhibition, resulting in repression of target gene expression.Citation10 MiRNAs are important regulators of normal biological processes including cell proliferation, cell differentiation, cell motility, and cell death. Accumulating evidences suggested that deregulation of miRNAs was associated with cancer initiation and progression.Citation11,Citation12 In prostate cancer, many miRNAs were reported as oncogenes or tumor suppressors via regulation of their numerous target genes.Citation13–Citation15 For example, miR-203 was reported as an upregulated miRNA in breast cancer and promoted breast cancer progression via targeting SOCS3.Citation16 Moreover, several miRNAs were proved to be promising biomarkers for prostate cancer diagnosis.Citation17 Profiling of miRNAs suggested that four miRNAs (miR-4289, miR-326, miR-152-3p, and miR-98-5p) were biomarkers to identify patients with prostate cancer from healthy people.Citation17 Using microarray, many cancer-associated miRNAs have been identified in prostate cancer.Citation18,Citation19 In 2016, one study involving miRNA profiling of 21 normal prostate tissues and 56 prostate tumor tissues has discovered many deregulated miRNAs in prostate cancer; among them, miR-424 was experimentally validated as an oncogene.Citation20 However, the potential function of other aberrant expressed miRNAs has not been studied yet.
ETS factors are transcription factors that can directly bind to promoters and enhancers to recruit other transcriptional machinery components.Citation21 Overexpression of ETS family members such as ETV1, ETV4, and ETV5 are associated with prostate cancer progression.Citation22 In prostate cancer cells, ETV1 cooperates with androgen receptor signaling and contributes to highly aggressive phenotype in mice and human.Citation23 ETV1 also activated transcription of TAZ to promote prostate cancer development.Citation24 Upregulated by JMJD2A, ETV1 activated transcription of YAP, a TAZ homologue, in prostate cancer.Citation25 Therefore, the regulation of ETV1 in prostate cancer needs to be investigated.
In the current study, we showed that miR-129-5p was down-regulated in prostate cancer tissues and cell lines. Overexpression of miR-129-5p inhibited cell proliferation of PC-3, while antagonizing of miR-129-5p promoted RWPE-1 cell growth. Mechanistic studies indicated that miR-129-5p negatively regulated ETV1 to inactivate YAP signaling in prostate cancer. Forced overexpression of ETV1 reversed miR-129-5p mimics inducing cell growth arrest. Reverse transcription-quantitative PCR (RT-qPCR) suggested a negative correlation between miR-129-5p and ETV1 mRNA expression in tumor tissues from patients with prostate cancer. Our findings revealed miR-129-5p as a tumor suppressor in prostate cancer via repression of ETV1.
Materials and methods
Patient samples and patient tumor data analysis
Prostate tumor tissues and matched normal prostate tissues were collected from 30 patients with prostate cancer receiving radical prostatectomy during 2015–2017 in China-Japan Union Hospital of Jilin University. Written informed consents were provided by all patients before experiments. All procedures were carried out under the supervision of medical ethics committee of Jilin University. The study was conducted in accordance with the Declaration of Helsinki. For bioinformatic analysis of miRNA profiling in prostate cancer, GSE60117 containing data of 21 normal tissues and 56 prostate tumor tissues were analyzed on Gene Expression Omnibus (GEO) using GEO2R to obtain most significantly deregulated miRNAs.
Cell lines
Human normal epithelial cell line RWPE-1 and human prostate cancer cell lines PC-3, DU145, and LNCaP were purchased from American Type Culture Collection (ATCC, Manassas, VA, USA). All cell lines were used within 6 months after receipt. PC-3, DU145, and LNCaP cells were cultured in DMEM (Gibco, Thermo Fisher Scientific, Waltham, MA, USA) supplemented with 10% FBS (HyClone, Thermo Fisher Scientific) and 1% Penicillin–Streptomycin solution (Invitrogen, Thermo Fisher Scientific). RWPE-1 cells were maintained in Keratinocyte Serum Free Medium (Invitrogen, Thermo Fisher Scientific) supplemented with bovine pituitary extract (Invitrogen, Thermo Fisher Scientific) and human recombinant EGF (Invitrogen, Thermo Fisher Scientific). All cell lines were maintained in a 37°C incubator with 5% CO2.
MiR-129-5p overexpression and antagonizing
MiR-NC mimics, miR-129-5p mimics, miR-NC antagonist, and miR-129-5p antagonist were bought from RiboBio (Guangzhou, P.R. China). For miR-129-5p overexpression or downregulation, miR-129-5p mimics or miR-129-5p antagonist was mixed with Lipofectamine 2000 (Invitrogen, Thermo Fisher Scientific), maintained for 15 minutes, and then added into culture medium of cells. Seventy-two hours after transfection, the cells were harvested and the RNA and/or protein was extracted for the following experiments.
Western blot
Protein lysates were prepared using RIPA lysis buffer (Sigma-Aldrich, Merck KGaA, Darmstadt, Germany) according to the manufacturer’s protocol. Antibodies for ETV1, YAP, CTGF, and CYR61 were bought from Cell Signaling Technology (CST, Danvers, MA, USA). β-actin antibody was purchased from Sigma-Aldrich. Secondary antibodies against rabbit and mouse were products of Proteintech (Chicago, IL, USA). Western blot was performed in a standard procedure. Briefly, lysates were loaded on SDS gel; proteins were separated and then transferred on a PVDF membrane. The membrane was blocked by 5% nonfat milk and incubated in indicated primary antibodies overnight. On the next day, membrane was incubated in secondary antibodies for 1 hour. The membrane was developed by ECL Western blot substrate (Pierce, Thermo Fisher Scientific).
RNA extraction and real-time RT-PCR
Total RNA from tissues and cells were extracted using TRIzol Reagent (Invitrogen, Thermo Fisher Scientific) according to the manufacturer’s protocol. For miR-129-5p expression detection, RNA was reverse transcribed using a stem-loop primer with RevertAid First Strand cDNA kit (Thermo Fisher Scientific). For gene expression, RNA was reverse transcribed into first-stranded cDNA using PrimeScript RT Master Mix (Takara, Kusatsu, Japan). Real-time RT-PCR was carried out using SYBR Premix Ex Taq kit (Takara). U6 and β-actin were served as internal controls for miRNA and mRNA, respectively. The primer sequences were listed as follows: ETV1-forward: 5′-CTGAACCCTGTAACTCCTTTCC-3′; ETV1-reverse: 5′-AGACATCTGGCGTTGGTACATA-3′; CTGF-forward: 5′-CAGCATGGACGTTCGTCTG-3′; CTGF-reverse: 5′-AACCACGGTTTGGTCCTTGG-3′; CYR61-forward: 5′-CTCGCCTTAGTCGTCACCC-3′; CYR61-reverse: 5′-CGCCGAAGTTGCATTCCAG-3′; β-actin-forward: 5′-CATGTACGTTGCTATCCAGGC-3′; β-actin-reverse: 5′-CTCCTTAATGTCACGCACGAT-3′.
ETV1 overexpression
Full length of ETV1 open reading frame was amplified from RWPE-1 cDNA and ligated into pcDNA3.1 plasmid (OriGene, Rockville, MD, USA). For ETV1 overexpression, pcDNA3.1-ETV1 plasmid was mixed with Lipofectamine 2000 (Invitrogen, Thermo Fisher Scientific) in serum-free DMEM for 15 minutes and added into culture medium.
Cell proliferation assay
For analysis of cell growth ability, cells were seeded in 96-well plate. Every 24 hours from day 0 to day 3, 10 µL CCK-8 solution (Cell Counting Kit-8, Dojindo, Kumamoto, Japan) was added into culture medium in each well for 2 hours. After that, medium containing CCK-8 solution was transferred into a new 96-well plate and the absorbance at 450 nm was detected to reflect cell proliferation ability.
Colony forming assay
For colony forming assay, 500 treated cells were seeded in each well in 6-well plates. After growing for 16 days, the culture medium was discarded, and each well was slightly washed with PBS. Then, cell colonies were fixed with 4% paraformaldehyde (Sigma-Aldrich) followed by staining with 0.5% crystal violet (Sigma-Aldrich). Images of colony were captured, and colony numbers were counted using ImageJ software.
Dual luciferase reporter assay
ETV1 3′UTR was cloned from cDNA of RWPE-1 and annealed into pGL3-basic (Promega, Fitchburg, WI, USA). The primer sequences were as follows: ETV1 3′UTR-forward: 5′-GCTCTAGATTCTGTAAATGTGAT-3′; ETV1 3′UTR-reverse: 5′-GCTCTAGATGAAATTGGAG TACTT-3′. Two site mutations were introduced into pGL3-ETV1 3′UTR-WT (wild type) to construct pGL3-ETV1 3′UTR-Mut (mutant) using GeneArt™ Site-Directed Mutagenesis PLUS System (Thermo Fisher Scientific). For dual luciferase reporter assay, cells were cotransfected with pGL3-ETV1 3′UTR-WT or pGL3-ETV1 3′UTR-Mut in combination with miR-NC mimics or miR-129-5p mimics and pRL plasmid. After 48 hours, relative luciferase activity was detected using Dual Luciferase Reporter Assay System (Promega) in accordance with the manufacturer’s protocol.
Statistical analysis
All data were analyzed using GraphPad Prism 7 and expressed as mean ± SD. Student’s t-test was used to compare differences between two groups. One-way ANOVA was carried out to compare differences among three groups, followed by Newman–Keuls test. Correlation analysis between miR-129-5p and ETV1 expression was conducted using Pearson correlation analysis. P-values <0.05 were considered to be statistically significant.
Results
Expression of miR-129-5p was downregulated in prostate cancer tissues and cell lines
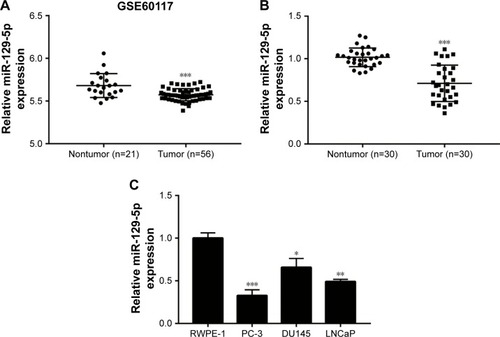
To explore deregulated miRNAs in prostate cancer, we analyzed miRNA profiling of 21 normal prostate tissues and 56 prostate cancer tissues from data set GSE60117 in GEO database. As shown in , miR-129-5p was significantly downregulated in prostate cancer tissues compared with normal prostate tissues (P<0.001). To confirm this result, RT-qPCR was performed to detect miR-129-5p expression in 30 pairs of prostate cancer tissues and matched normal tissues collected. Similarly, a decrease of miR-129-5p expression was observed in prostate cancer tissues compared with normal tissues (, P<0.001). Next, association between miR-129-5p expression and clinicopathological factors in 30 patients with prostate cancer was analyzed. No significant association was observed between miR-129-5p expression with age, Gleason score, or distant metastasis, but high expression of miR-129-5p was associated with early pathologic stage (P<0.01) (). Additionally, compared with normal epithelial prostate cell RWPE-1, expression of miR-129-5p was decreased in prostate cancer cell lines (PC-3, DU145, and LNCaP) (, P<0.001, P<0.05, and P<0.01, respectively).
Table 1 Association of miR-129-5p with clinicopathological features of 30 prostate cancer patients
Figure 1 Downregulation of miR-129-5p in prostate cancer tissues and cell lines.
Notes: (A) Through analysis of miRNA profiling in GSE60117, miR-129-5p was one of the most significantly downregulated miRNAs in 56 prostate tumor tissues compared with 21 normal prostate tissues. (B) Reverse transcription quantitative PCR indicated that miR-129-5p was decreased in 30 prostate cancer tissues in comparison with 30 matched normal prostate tissues. (C) In comparison with normal epithelial prostate cell line RWPE-1, expression of miR-129-5p was reduced in prostate cancer cell lines (PC-3, DU145, and LNCaP). *P<0.05, **P<0.01, ***P<0.001.
MiR-129-5p regulated prostate cancer cell proliferation
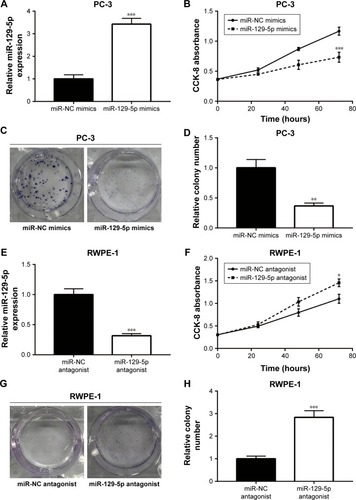
To investigate the potential role of miR-129-5p in prostate cancer, miR-129-5p mimics was transfected into PC-3 cells. Overexpression of miR-129-5p by transfection of miR-129-5p mimics significantly inhibited cell growth of PC-3 cells (, P<0.001). Additionally, in the colony forming assay, miR-129-5p mimics reduced colony number formed by PC-3 cells (, P<0.01). We next downregulated miR-129-5p expression in RWPE-1 cells by transfection of miR-129-5p antagonist (). In contrast, miR-129-5p downregulation promoted cell proliferation of RWPE-1 cells (, P<0.05). The colony forming assay showed that miR-129-5p antagonist increased colony number formed by RWPE-1 cells (, P<0.001).
Figure 2 MiR-129-5p regulated cell proliferation in PC-3 and RWPE-1 cells.
Notes: (A) Transfection of miR-129-5p mimics elevated miR-129-5p expression by more than three-fold in PC-3 cells. (B) MiR-129-5p mimics inhibited cell proliferation of PC-3 cells in the CCK-8 assay. (C) In the colony forming assay, overexpression of miR-129-5p decreased colony number formed by PC-3 cells. (D) Quantitative analysis of colony number in (C). (E) Transfection of miR-129-5p antagonist decreased miR-129-5p expression in RWPE-1 cells. (F) Cell growth ability was elevated toward miR-129-5p downregulation in RWPE-1 cells. (G) MiR-129-5p antagonist increased colony number formed by RWPE-1 cells. (H) Quantitative analysis of colony number in (G). *P<0.05, **P<0.01, ***P<0.001.
Abbreviation: CCK-8, Cell Counting Kit-8.
Thus, these data suggested that miR-129-5p could suppress prostate cancer cell proliferation.
MiR-129-5p repressed ETV1 expression by directly binding to its 3′UTR
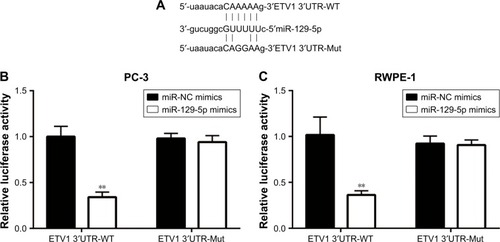
MiRNA exerts its function via binding to 3′UTR on its target gene mRNA.Citation10 ETV1 functions as an oncogene and is over-expressed in prostate cancer.Citation23 Using RT-qPCR, we found that ETV1 was overexpressed in all prostate cancer cell lines examined including PC-3 (P<0.01), DU145 (P<0.05), and LNCaP (P<0.01) compared with normal epithelial prostate cell RWPE-1 (). In PC-3, overexpression of miR-129-5p decreased ETV1 mRNA levels (, P<0.001). Additionally, Western blot showed that miR-129-5p mimics induced reduction of ETV1 protein expression () and quantification of protein levels suggested that ETV1 protein expression was significantly downregulated toward miR-129-5p overexpression (, P<0.01). Moreover, in RWPE-1 cells, downregulation of miR-129-5p elevated ETV1 mRNA levels (). Western blot showed that miR-129-5p antagonist induced elevation of ETV1 protein expression () and quantification of protein levels suggested that ETV1 was significantly upregulated toward miR-129-5p overexpression (, P<0.01). Using miRanda, our bioinformatic analysis showed that there was a complementary site between miR-129-5p sequence and ETV1 mRNA 3′UTR sequence (). To validate ETV1 as a target gene of miR-129-5p, we carried out the Dual Luciferase Reporter Assay in PC-3 cells. As expected, miR-129-5p mimics reduced relative luciferase activity in PC-3 cells transfected with ETV1 3′UTR-WT not ETV1 3′UTR-Mut (). Similarly, in RWPE-1 cells, overexpression of miR-129-5p repressed luciferase activity of ETV1 3′UTR-WT (, P<0.01). These data suggested that miR-129-5p might inhibit prostate cancer progression via regulation of ETV1.
Figure 3 ETV1 was negatively regulated by miR-129-5p.
Notes: (A) In comparison with normal epithelial prostate cell line RWPE-1, expression of ETV1 mRNA was reduced in prostate cancer cell lines (PC-3, DU145, and LNCaP). (B) MiR-129-5p mimics decreased ETV1 mRNA expression in PC-3 cells. (C) MiR-129-5p mimics decreased ETV1 protein expression in PC-3 cells. (D) Quantitative analysis of ETV1 protein expression in (C). (E) MiR-129-5p antagonist increased ETV1 mRNA expression in RWPE-1 cells. (F) MiR-129-5p antagonist increased ETV1 protein expression in RWPE-1 cells. (G) Quantitative analysis of ETV1 protein expression in (F). *P<0.05, **P<0.01, ***P<0.001.
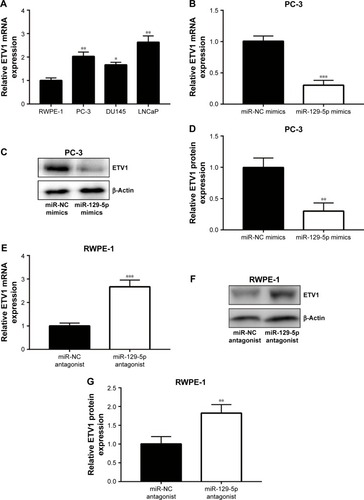
Figure 4 ETV1 was a direct target of miR-129-5p.
Notes: (A) Sequence alignment of ETV1 mRNA 3′UTR-WT (wild type), 3′UTR-Mut (mutant), and miR-129-5p sequence. (B) Dual Luciferase Reporter Assay showed that miR-129-5p mimics reduced luciferase activity of PC-3 cells transfected with pGL3-ETV1 3′UTR-WT not mutant ETV1 3′UTR. (C) Dual Luciferase Reporter Assay showed that miR-129-5p mimics reduced luciferase activity of RWPE-1 cells transfected with pGL3-ETV1 3′UTR-WT not mutant ETV1 3′UTR. **P<0.01.
MiR-129-5p repressed YAP and its target gene expression
ETV1 is a transcription factor and accelerates prostate cancer cell growth via transcriptional activation of YAP.Citation25 Consistent with ETV1 downregulation, miR-129-5p mimics also triggered the decrease of YAP mRNA expression (). Also, the protein levels of YAP were decreased upon miR-129-5p overexpression (, P<0.001). As a transcription coactivator, YAP stimulated transcription of CTGF and CYR61.Citation26 RT-qPCR results indicated that CTGF and CYR61 expression was decreased () and Western blot showed that CTGF and CYR61 protein expression was decreased (, P<0.01 and P<0.001, respectively) toward miR-129-5p overexpression, suggesting that miR-129-5p suppressed YAP target gene expression. In contrast, downregulation of miR-129-5p significantly elevated YAP mRNA and protein (P<0.01) expression in RWPE-1 cells (). Additionally, miR-129-5p antagonist also stimulated transcription of CTGF and CYR61 (, P<0.001), leading to enhanced protein expression of CTGF and CYR61 in RWPE-1 cells (, P<0.05 and P<0.01, respectively). The results demonstrated that miR-129-5p could repress YAP signaling via targeting ETV1 in prostate cancer.
Figure 5 MiR-129-5p mimics repressed YAP and its target gene expression in PC-3 cells.
Notes: (A) MiR-129-5p overexpression decreased YAP mRNA level in PC-3 cells. (B) Western blot showed that YAP protein level was reduced toward miR-129-5p overexpression. (C) Quantitative analysis of YAP protein level in (B). (D) MiR-129-5p overexpression decreased CTGF and CYR61 mRNA levels in PC-3 cells. (E) Western blot showed that CTGF and CYR61 protein levels were reduced toward miR-129-5p overexpression. (F) Quantitative analysis of CTGF and CYR61 protein levels in (E). **P<0.01, ***P<0.001.
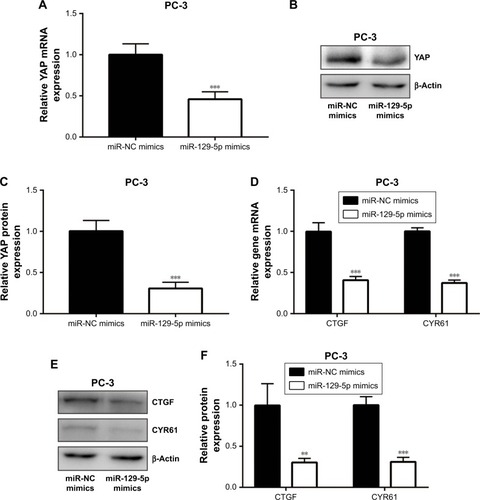
Figure 6 MiR-129-5p downregulation elevated YAP and its target gene expression in RWPE-1 cells.
Notes: (A) MiR-129-5p downregulation increased YAP mRNA level in RWPE-1 cells. (B) Western blot showed that YAP protein level was elevated toward miR-129-5p downregulation. (C) Quantitative analysis of YAP protein level in (B). (D) MiR-129-5p downregulation increased CTGF and CYR61 mRNA levels in RWPE-1 cells. (E) Western blot showed that CTGF and CYR61 protein levels were elevated toward miR-129-5p downregulation. (F) Quantitative analysis of CTGF and CYR61 protein levels in (E). *P<0.05, **P<0.01, ***P<0.001.
ETV1 overexpression reversed miR-129-5p mimics-induced cell proliferation inhibition in PC-3 cells
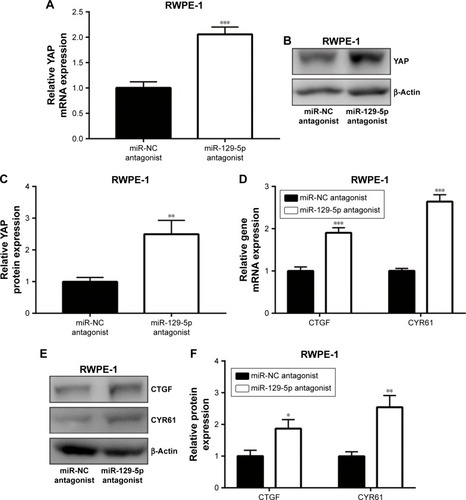
To examine whether ETV1 was pivotal for miR-129-5p function in prostate cancer cells, we constructed plasmid containing recombinant ETV1. Western blot showed that miR-129-5p mimics reduced ETV1 protein (P<0.001) expression, which could be reversed (P<0.01) by pcDNA3.1-ETV1 transfection in PC-3 cells (, P<0.001 and P<0.01, respectively). In the cell proliferation assay, forced expression of ETV1 reversed cell growth arrest induced by miR-129-5p mimics in PC-3 (, P<0.001 and P<0.05, respectively). Moreover, the colony forming assay showed that reduced colony number toward miR-129-5p mimics was recovered after ETV1 overexpression (, P<0.001 and P<0.01, respectively). Collectively, these results manifested that miR-129-5p relied on regulation of ETV1 to inhibit prostate cancer cell proliferation.
Figure 7 Forced elevation of ETV1 expression rescued cell growth arrest and inhibition of colony forming ability induced by miR-129-5p mimics in PC-3 cells.
Notes: (A) MiR-129-5p mimics decreased ETV1 protein level, which could be reversed by transfection of pcDNA3.1-ETV1. (B) Quantitative analysis of ETV1 protein level in (A). (C) Overexpression of ETV1 attenuated the suppression effect of miR-129-5p mimics on cell growth. (D) In a colony forming assay, ETV1 overexpression attenuated the decrease of colony number induced by miR-129-5p mimics. (E) Quantitative analysis of colony number in (D). *P<0.05, **P<0.01, ***P<0.001.
Abbreviation: CCK-8, Cell Counting Kit-8.
Expression of miR-129-5p was negatively associated with ETV1 mRNA levels in prostate cancer tissues
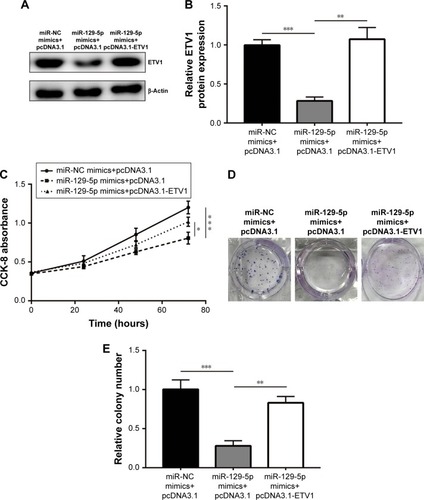
Next, we sought to investigate the relationship between miR-129-5p expression and ETV1 expression levels in prostate cancer tissues. Consistent with previous study, ETV1 protein (P<0.001) and mRNA levels were elevated in prostate cancer tissues compared with matched normal prostate tissues (, P<0.001 and P<0.05, respectively). Furthermore, Pearson correlation assay showed that miR-129-5p expression was inversely correlated with ETV1 mRNA levels in prostate cancer tissues (, P=0.0018). Our data suggested an miR-129-5p/ETV1/Hippo-YAP axis in regulating proliferation of prostate cancer cells ().
Figure 8 Expression of miR-129-5p was inversely associated with ETV1 mRNA levels in prostate cancer tissues.
Notes: (A) Representative Western blot of ETV1 expression in three pairs of prostate tumor tissues and matched normal tissues. (B) Quantification of ETV1 protein levels in prostate tumor tissues and matched normal tissues in (A). (C) Reverse transcription-quantitative PCR showed that ETV1 mRNA levels were significantly elevated in 30 prostate tumor tissues compared with adjacent normal prostate tissues. (D) Analysis of miR-129-5p expression and ETV mRNA levels showed a negative correlation between them. (E) Diagram of an miR-129-5p/ETV1/Hippo-YAP axis in prostate cancer cells. *P<0.05, ***P<0.001.
Discussion
It is well characterized that aberrant expression of miRNAs is one of hallmarks of carcinogenesis.Citation27 Recent years, many researches carried out miRNA profiling of prostate cancer tissues and discovered several miRNAs with oncogene or tumor suppressor potential.Citation20,Citation28,Citation29 Although studies have identified numerous differentially expressed miRNAs in prostate cancer tissues compared with normal tissues, little was known about the function and underlying molecular mechanism of specific miRNA in prostate cancer. In the current study, we found that miR-129-5p was downregulated in prostate cancer tissues and functioned as a tumor suppressor via regulation of ETV1.
MiR-129-5p has been reported as a tumor suppressor in many cancer types including glioblastoma, papillary thyroid cancer, colorectal cancer, and breast cancer.Citation30–Citation33 Several studies provided evidences that miR-129-5p inhibited cancer progression via repression of HMGB1.Citation34,Citation35 In prostate cancer, miR-129-5p was found as one of the most significantly downregulated miRNA in prostate cancer cell and was involved in the regulation of metabolism and cell proliferation via repression of key proteins in carnitine cycle.Citation36 Most recently, miR-129 was discovered to inhibit cell proliferation, migration, and invasiveness in prostate cancer cells.Citation37 Here, through screen of differentially expressed miRNAs in GSE60117, we also observed the decreased expression of miR-129-5p in prostate tumor tissues and further validated its downregulation in 30 pairs of prostate tumor and normal tissues we collected. Our cell functional assays validated tumor suppressor role of miR-129-5p in PC-3 and RWPE-1 cells; these results were consistent with previous reports.
Several oncogenes, such as PAK5 and RET, have been validated as target genes of miR-129-5p in cancer cells.Citation38,Citation39 Study in mice and human showed that ETV1 cooperated with androgen receptor signaling to promote highly aggressive nature of advanced prostate cancer.Citation23 ETV1 was proved to be regulated by several miRNAs such as miR-17-5p and miR-34b.Citation40,Citation41 However, whether and how ETV1 was regulated by miR-129-5p was not known. In PC-3, overexpression of miR-129-5p decreased ETV1 at both mRNA and protein levels. In contrast, downregulation of miR-129-5p increased ETV1 mRNA and protein expression in RWPE-1 cells. Bioinformatic analysis of miR-129-5p and ETV1 3′UTR sequences indicated a complementary site between them. The Dual Luciferase Reporter Assay confirmed ETV1 as a target gene of miR-129-5p in PC-3 and RWPE-1 cells. Similar to ETV1, YAP was overexpressed in prostate cancer and mediated prostate cancer metastasis, stemness, cell growth, and development of castration resistance.Citation42,Citation43 A recent study declared that ETV1 stimulated prostate cancer development through activation of YAP and its target gene expression.Citation25 In PC-3 and RWPE-1 cells, miR-129-5p suppressed YAP and its target gene CTGF and CYR61 expression at both mRNA and protein levels, suggesting that miR-129-5p might suppress YAP expression via repression of ETV1. RT-qPCR further showed a strong negative correlation between miR-129-5p expression and ETV1 mRNA levels in prostate cancer tissues, implying a regulatory relationship between ETV1 and miR-129-5p in prostate cancer. In a recent study, overexpression of miR-129 was shown to inactivate PI3K/AKT signaling in the PC-3.Citation37 Since the cross talk between PI3K/AKT and YAP signaling existed in cancer cells,Citation44 miR-129-5p might play a central role in prostate cancer via regulating a complicated signaling network.
Conclusion
Our study provided evidences confirming that miR-129-5p was a tumor suppressor in prostate cancer and was downregulated in tissues of prostate cancer. Mechanistic study discovered an miR-129-5p/ETV1/YAP axis in prostate cancer cells. Thus, miR-129-5p might be a promising target for the treatment of patients with prostate cancer.
Data sharing statement
Data are available on request.
Disclosure
The authors report no conflicts of interest in this work.
References
- LitwinMSTanHJThe diagnosis and treatment of prostate cancerJAMA2017317242532254228655021
- JemalABrayFCenterMMFerlayJWardEFormanDGlobal cancer statisticsCA Cancer J Clin2011612699021296855
- AntenorJARoehlKAEggenerSEKunduSDHanMCatalonaWJPreoperative PSA and progression-free survival after radical prostatectomy for stage T1c diseaseUrology200566115616015992903
- CozziGMusiGBianchiRMeta-analysis of studies comparing oncologic outcomes of radical prostatectomy and brachytherapy for localized prostate cancerTher Adv Urol201791124125029662542
- HuangEYChangYJHuangSPA common regulatory variant in SLC35B4 influences the recurrence and survival of prostate cancerJ Cell Mol Med20182273661367029682886
- ZhengQSChenSHWuYPIncreased paxillin expression in prostate cancer is associated with advanced pathological features, lymph node metastases and biochemical recurrenceJ Cancer20189695996729581775
- MillerETChamieKKwanLLewisMSKnudsenBSGarrawayIPImpact of treatment on progression to castration-resistance, metastases, and death in men with localized high-grade prostate cancerCancer Med20176116317227997745
- BoströmPJBjartellASCattoJWGenomic predictors of outcome in prostate cancerEur Urol20156861033104425913390
- FilipowiczWBhattacharyyaSNSonenbergNMechanisms of post-transcriptional regulation by microRNAs: are the answers in sight?Nat Rev Genet20089210211418197166
- BartelDPMicroRNAs: genomics, biogenesis, mechanism, and functionCell2004116228129714744438
- AndersenGBKnudsenAHagerHHansenLLTostJmiRNA profiling identifies deregulated miRNAs associated with osteosarcoma development and time to metastasis in two large cohortsMol Oncol201812111413129120535
- HuXWangYLiangHmiR-23a/b promote tumor growth and suppress apoptosis by targeting PDCD4 in gastric cancerCell Death Dis2017810e305928981115
- OkatoAAraiTYamadaYRegulation of NCAPG by miR-99a-3p (passenger strand) inhibits cancer cell aggressiveness and is involved in CRPCCancer Med2018751988200229608247
- HaoPKangBYaoGHaoWMaFMicroRNA-211 suppresses prostate cancer proliferation by targeting SPARCOncol Lett20181544323432929541199
- ShaoNMaGZhangJZhuWmiR-221-5p enhances cell proliferation and metastasis through post-transcriptional regulation of SOCS1 in human prostate cancerBMC Urol20181811429506516
- MuhammadNBhattacharyaSSteeleRRayRBAnti-miR-203 suppresses ER-positive breast cancer growth and stemness by targeting SOCS3Oncotarget2016736585955860527517632
- MatinFJeetVMoyaLA plasma biomarker panel of four microRNAs for the diagnosis of prostate cancerSci Rep201881665329703916
- JossonSGururajanMHuPmiR-409-3p/-5p promotes tumorigenesis, epithelial-to-mesenchymal transition, and bone metastasis of human prostate cancerClin Cancer Res201420174636464624963047
- Casanova-SalasIRubio-BrionesJCalatravaAIdentification of miR-187 and miR-182 as biomarkers of early diagnosis and prognosis in patients with prostate cancer treated with radical prostatectomyJ Urol2014192125225924518785
- DallavalleCAlbinoDCivenniGMicroRNA-424 impairs ubiquitination to activate STAT3 and promote prostate tumor progressionJ Clin Invest2016126124585460227820701
- HollenhorstPCMcIntoshLPGravesBJGenomic and biochemical insights into the specificity of Ets transcription factorsAnnu Rev Biochem201180143747121548782
- CurrieSLLauDKWDoaneJJStructured and disordered regions cooperatively mediate DNA-binding autoinhibition of ETS factors ETV1, ETV4 and ETV5Nucleic Acids Res20174552223224128161714
- BaenaEShaoZLinnDEETV1 directs androgen metabolism and confers aggressive prostate cancer in targeted mice and patientsGenes Dev201327668369823512661
- LiuCYYuTHuangYCuiLHongWETS (E26 transformation-specific) up-regulation of the transcriptional co-activator TAZ promotes cell migration and metastasis in prostate cancerJ Biol Chem2017292229420943028408625
- KimTDJinFShinSHistone demethylase JMJD2A drives prostate tumorigenesis through transcription factor ETV1J Clin Invest2016126270672026731476
- ZhouYHuangTChengASYuJKangWToKFThe TEAD family and its oncogenic role in promoting tumorigenesisInt J Mol Sci2016171pii:E138
- AshrafNMImranKKastnerDWPotential involvement of mi-RNA 574-3p in progression of prostate cancer: a bioinformatic studyMol Cell Probes201736212828734841
- DoldiVPennatiMForteBGandelliniPZaffaroniNDissecting the role of microRNAs in prostate cancer metastasis: implications for the design of novel therapeutic approachesCell Mol Life Sci201673132531254226970978
- ZhangYJiangFHeHIdentification of a novel microRNA-mRNA regulatory biomodule in human prostate cancerCell Death Dis20189330129467540
- ZengAYinJLiYmiR-129-5p targets Wnt5a to block PKC/ERK/NF-κB and JNK pathways in glioblastomaCell Death Dis20189339429531296
- ZhangHCaiYZhengLZhangZLinXJiangNLong noncoding RNA NEAT1 regulate papillary thyroid cancer progression by modulating miR-129-5p/KLK7 expressionJ Cell Physiol2018233106638664829319165
- LinJShiZYuZHeZLncRNA HIF1A-AS2 positively affects the progression and EMT formation of colorectal cancer through regulating miR-129-5p and Dnmt3aBiomed Pharmacother20189843343929278853
- LuXMaJChuJMiR-129-5p sensitizes the response of HER-2 positive breast cancer to trastuzumab by reducing rpS6Cell Physiol Biochem20174462346235629258115
- LiuKHuangJNiJMALAT1 promotes osteosarcoma development by regulation of HMGB1 via miR-142-3p and miR-129-5pCell Cycle201716657858728346809
- WuQWyMJieYZhaoHLncRNA MALAT1 induces colon cancer development by regulating miR-129-5p/HMGB1 axisJ Cell Physiol201823396750675729226325
- ValentinoACalarcoADi SalleADeregulation of microR-NAs mediated control of carnitine cycle in prostate cancer: molecular basis and pathophysiological consequencesOncogene201736436030604028671672
- XuSGeJZhangZZhouWMiR-129 inhibits cell proliferation and metastasis by targeting Ets1 via PI3K/Akt/mTOR pathway in prostate cancerBiomed Pharmacother20179663464129035829
- ZhaiJQuSLiXmiR-129 suppresses tumor cell growth and invasion by targeting PAK5 in hepatocellular carcinomaBiochem Biophys Res Commun2015464116116726116538
- DuanLHaoXLiuZZhangYZhangGMiR-129-5p is down-regulated and involved in the growth, apoptosis and migration of medullary thyroid carcinoma cells through targeting RETFEBS Lett201458891644165124631532
- LiJLaiYMaJmiR-17-5p suppresses cell proliferation and invasion by targeting ETV1 in triple-negative breast cancerBMC Cancer201717174529126392
- ShiinaMHashimotoYKatoTDifferential expression of miR-34b and androgen receptor pathway regulate prostate cancer aggressiveness between African-Americans and CaucasiansOncotarget2017858356836828039468
- GuoYCuiJJiZmiR-302/367/LATS2/YAP pathway is essential for prostate tumor-propagating cells and promotes the development of castration resistanceOncogene201736456336634728745315
- LiuHDaiXCaoXPRDM4 mediates YAP-induced cell invasion by activating leukocyte-specific integrin beta2 expressionEMBO Rep2018196pii:e45180
- YuSCaiXWuCAdhesion glycoprotein CD44 functions as an upstream regulator of a network connecting ERK, Akt and Hippo-YAP pathways in cancer progressionOncotarget2015652951296525605020
