Abstract
Ubiquitin-like with plant homeodomain and really interesting new gene finger domains 1 (UHRF1) functions as an epigenetic regulator recruiting PCNA, DNMT1, histone deacetylase 1, G9a, SuV39H, herpes virus-associated ubiquitin-specific protease, and Tat-interactive protein by multiple corresponding domains of DNA and H3 to maintain DNA methylation and histone modifications. Overexpression of UHRF1 has been found as a potential biomarker in various cancers resulting in either DNA hypermethylation or global DNA hypo-methylation, which participates in the occurrence, progression, and invasion of cancer. The role of UHRF1 in the reciprocal interaction between DNA methylation and histone modifications, the dynamic structural transformation of UHRF1 protein within epigenetic code replication machinery in epigenetic regulations, as well as modifications during cell cycle and chemotherapy targeting UHRF1 are evaluated in this study.
Video abstract
Point your SmartPhone at the code above. If you have a QR code reader the video abstract will appear. Or use:
Introduction
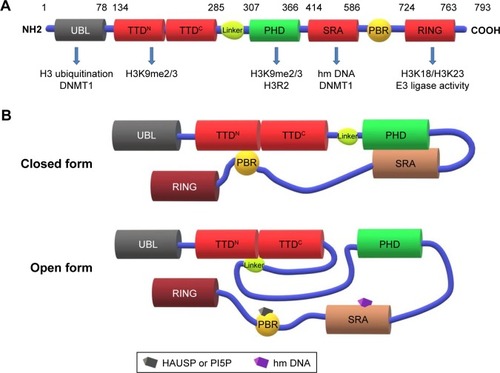
Ubiquitin-like with plant homeodomain (PHD) and really interesting new gene (RING) finger domain 1 (UHRF1, also called ICBP90 or Np95) was identified as a transcription factor which could regulate the expression of topoisomerase IIα by binding to an inverted CCAAT box located in its promoter.Citation1 The nuclear protein UHRF1 consists of multiple domains such as ubiquitin-like domain (UBL), tandem tudor domain (TTD; TTDN and TTDC), PHD, SET and RING-associated domain (SRA) as well as RING domain, which play an important role in the epigenetic regulation of gene expression and tumorigenesis.
Epigenetic modification refers to inherited changes in gene expression without variations in DNA structure or sequenceCitation2–Citation4 and includes DNA methylation, histone modification, genome imprinting, X-chromosome inactivation, and microRNA regulation,Citation4,Citation5 where DNA methylation and histone modification are most important and their aberrant changes are always involved in cancer and neurological diseases. UHRF1 serves as a key regulator that participates in both DNA methylation and histone modifications.
UHRF1 allows crosstalk between DNA methylation and histone code as an epigenetic coordinator
DNA methylation is a fundamental epigenetic process in the regulation of gene expression. The process occurs at the carbon-5 position of cytosines mainly within CpG sites mediated by DNA methyltransferases (DNMTs). In detail, DNMT3A and DNMT3B are mainly responsible for de novo methylation during gametogenesis and early embryonic development, whereas DNMT1 preferentially methylates hemi-methylated CpG sites for maintenance of DNA methylation by a self-inhibitory mechanism during DNA replication and cell division.Citation6 Aberrant DNA methylation can lead to abnormal embryogenesis,Citation7 neurological diseases,Citation8 and cancers,Citation9,Citation10 including global DNA hypomethylation and specific CpG island hypermethylation in which the former can lead to chromosomal instability,Citation11 activation of certain transcription factors,Citation12 and loss of genetic imprints;Citation13 the latter often results in the inhibition of tumor suppressor genes.
Eukaryotic DNMT1 is composed of an N-terminal nuclear localization sequence, a replication foci targeting sequence (RFTS) that localizes DNMT1 to the DNA replication fork, a zinc finger CXXC domain that specifically recognizes unmethylated CpG dinucleotides,Citation14,Citation15 a pair of bromo-adjacent homology (BAH) domains, and a C-terminal methyltransferase domain including the catalytic core and the target recognition domain (TRD). Although the occlusion of the CXXC-BAH1 linker at the catalytic site and restraint by BAH2-TRD loop preventing TRD from binding to unmethylated CpG sites facilitate DNMT1-mediated maintenance of DNA methylation,Citation16,Citation17 the auto-inhibitory role of RFTS domain and DNMT1-interacting proteins which affect its activity, such as UHRF1, have been revealed.Citation18,Citation19 UHRF1 recognizes and binds to hemi-methylated DNA (hmDNA) through its SRA domain by the thumb (444–499 residues) and NKR finger (483–496 residues) sub-domains targeting minor and major grooves, respectively,Citation20 and recruits DNMT1 by targeting the RFTS domain which leaves the catalytic pocket of DNMT1 in the S phase of the cell cycle to replication fociCitation21,Citation22 and relieves the auto-inhibitory activity of DNMT1 to maintain the cytosine in newly synthesized DNA methylation with high fidelity. In addition, the UHRF1 C-terminal RING finger functions as an ubiquitin E3 ligase to establish histone H3 ubiquitination at Lys23 and Lys18 recognized by the RFTS domain of DNMT1Citation23,Citation24 to promote its localization onto replication foci, which also has a prerequisite role in the maintenance of DNA methylation. Recently, the N-terminal UBL domain of UHRF1 was found to bind directly to DNMT1 enhancing DNMT1 enzymatic activity toward newly replicated chromatin by controlling targeted H3 ubiquitylation through a hydrophobic patch.Citation25,Citation26 Furthermore, the UBL domain can bind the E2 Ube2D and form a stable E2/E3/chromatin complex that is equally required for the DNMT1-mediated maintenance of DNA methylation.Citation26,Citation27
Posttranslational modification of histone participates in DNA replication, DNA damage response, chromosome translocation, transcription activation and suppression, X chromosome inactivation, and heterochromatin replication.Citation28 To date, the most studied histone modifications include the methylation of arginine (R) and lysine (K), acetylation of lysine, phosphorylation of serine (S) and threonine (T), and lysine ubiquitination. The histone lysine methylation is divided into monomethylation, dimethylation, and trimethylation, which greatly increases the complexity of histone modification and gene expression. Lysine acetylation of histone tails leads to transcriptional activation by neutralization of a positive charge at a lysine side chain, triggering a detachment of the side chain from the negatively charged DNA strands.
Methylation of histone H3 plays an important role in transcription, among which H3K4 methylation often causes transcriptional activation while H3K9 methylation always coordinates gene silencing. Unmethylated or methylated lysine can be modified by some proteins to regulate gene expression. Generally, UHRF1 recognizes the dimethylated or trimethylated H3K9 (H3K9me2/3) mediated by G9aCitation29 or SuV39H through TTDN and PHD (TTD-PHD) as well as identifies unmodified arginine 2 (R2) and unmodified lysine 4 (K4) through the PHD domain.Citation30–Citation36 The association with histone marks are all required for DNA methylation.Citation34,Citation37 Moreover, UHRF1 reportedly adopts a closed conformation in the absence of chromatin in which a polybasic region (PBR) in the C-terminus binds to the TTD and inhibits its recognition of H3K9me3, whereas the SRA domain binds to the PHD domain and inhibits recognition of unmethylated histone H3 at residue R2 (H3R2); upon binding to hmDNA by the SRA domain, UHRF1 impairs the intramolecular interactions and transfers to an open state,Citation30,Citation34,Citation38 which allows TTD-PHD to recognize H3K9me3 and facilitates SRA-PBR to either recognize hmDNA or recruit DNMT1 for an accurate methylation heredityCitation36 (). In addition, phosphatidylinositol 5-phosphate (PI5P) in the nucleus can interact with the PBR through phosphorylation of S651 to participate in the regulation of heterochromatin localization of UHRF1 and crosstalk between H3K9 methylation and DNA methylation.Citation39,Citation40 Phosphorylation of S298 in the linker residue abrogates the UHRF1-H3 interaction by altering the relative position of the two reader modules, indicating the linker region may act as a functional switch of UHRF1 involved in multiple regulatory pathways such as maintenance of DNA methylation, transcriptional repression, and cell cycle progression.Citation41 In contrast, Zhao et alCitation42 found only a 10% reduction of DNA methylation in various tissues after abolishment of the association between H3K9me2/3 and UHRF1 in mammals, indicating that H3K9 methylation binding of UHRF1 is not the only process to ensure DNA methylation. A positive correlation exists between UHRF1 and EZH2 in cancer cells,Citation43,Citation44 and EZH2 participates in H3K27 methylation which mediates gene silencing.Citation45,Citation46 Ferry et alCitation47 found nonhistone mammalian DNA ligase 1 (LIG1), which contains a conserved H3K9-like mimic can also be methylated by G9a/GLP at K126 and subsequently recruit UHRF1 to replication foci, which is similar to binding H3K9me2/3 to maintain DNA methylation.
Figure 1 Schematic representation of UHRF1 with structure–function domains in the maintenance of DNA methylation.
Notes: (A) Domain architecture of UHRF1 with their corresponding epigenetic function including DNA methylation and histone modification. (B) Conformational changes of UHRF1 from close to open state mediated by hmDNA.
Abbreviations: DNMT, DNA methyltransferases; H3K9me2/3, dimethylated or trimethylated H3K9; H3R2, unmethylated histone H3 at residue R2; hmDNA, hemi-methylated DNA; HAUSP, herpes virus-associated ubiquitin-specific protease; PBR, polybasic region; PHD, plant homeodomain; PI5P, phosphatidylinositol 5-phosphate; RING, really interesting new gene; SRA, SET and RING-associated domain; TTD, tandem tudor domain; UBL, ubiquitin-like domain.
UHRF1 can also lead to global DNA hypomethylation in cancers.Citation48–Citation50 Long interspersed nucleotide element-1 (LINE1) is considered a surrogate marker of global DNA methylation.Citation51 Nakamura et alCitation48 found that overexpression of UHRF1 could drive global DNA hypomethylation in esophageal squamous cell carcinoma (ESCC). Paradoxically, Ye et alCitation52 reported that knockdown of UHRF1 results in LINE1 hypomethylation in ESCC. Because the states of promoter methylation in tumor suppressor genes were not mentioned in these two studies, Hoshimoto et alCitation53 hypothesized that ESCC was associated with global DNA hypomethylation and specific tumor-related gene hypermethylation.
Although DNMT1 has a 30- to 40-fold preference for hmDNA sites,Citation54 it can still bind to unmethylated DNA, and exert de novo methylation activity.Citation55,Citation56 UHRF1 may lead to aberrant DNA methylation state in cancer cells for two reasons: 1) UHRF1 recruits DNMT1 to hm-CpG islands or CpG islands, which have not been previously methylated because DNMT1 also has the property of de novo methylationCitation19,Citation56 which undergoes hypermethylation, over time, finally leading to the malignancy of cancer; 2) when the UHRF1 protein levels become high, ubiquitination may occur, and DNMT1 is ubiquitylated to a proper level to maintain the malignancy state of cancer, including hypermethylation of specific tumor suppressor gene promoters as well as global DNA hypomethylation.
Self-regulation of UHRF1 macromolecular complex during the cell cycle
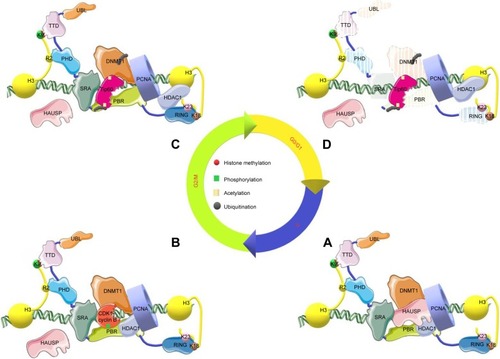
UHRF1, DNMT1, HAUSP, HDAC1, Tip60, Hsp90, Suv39H1, PCNA, and pRb form a macromolecular complex termed epigenetic code replication machinery (ECREM)Citation57 to undergo temporal and spatial control during the cell cycle. During DNA replication, DNMT1 is partially recruited into the replication forks by PCNA.Citation58–Citation60 Tip60 interacts with SRA and RING domain of UHRF1Citation61 through its enzymatic MYST domain, and overexpression of Tip60 leads to the downregulation of UHRF1.Citation62 In addition, UHRF1 can repress the activity and expression of Tip60.Citation63 Furthermore, overexpression of Tip60 leads to acetylation and subsequent ubiquitylation-dependent proteasomal degradation of DNMT1 mediated by UHRF1.Citation64 Although increased DNMT1 abundance in multiple cancers is largely due to the reduced degradation at the protein level rather than higher mRNA level,Citation65,Citation66 the consequence appears diverse based on cell cycle status or cell types,Citation63,Citation67–Citation69 and DNMT1 stability might be regulated by other upstream factors such as pRb and ATM.Citation70 However, the mechanism remains to be elucidated.
Conversely, HAUSP interacts with PBR of UHRF1 through UBL1 and UBL2 domains to stabilize UHRF1 via deubiquitination and protects it from autoubiquitination as well as promote the open state of UHRF1 to facilitate the binding of UHRF1 to H3K9me3.Citation34,Citation71–Citation73 When S652 (located in PBR) is phosphorylated by M phase-specific kinase CDK1-cyclin BCitation71 during mitosis, UHRF1 is separated from HAUSP and the PBR domain is exposed to Tip60 for acetylation and ultimate degradation.Citation39 In addition, HAUSP can deubiquitinate Tip60.Citation74
HAUSP binds to the KG linker of DNMT1 at its acidic pocket near the C-terminal to deubiquitinate DNMT1 from proteasomal degradation.Citation64 In addition, HAUSP functions as a deubiquitylating enzyme toward ubiquitylated histone H3, and is likely involved in DNMT1 recruitment to DNA replication sites and the regulation of maintenance of DNA methylation without affecting the kinetics and efficiency of DNA replication.Citation75 Simultaneously, Cheng et alCitation76 found that acetylation of the KG linker lysine residues impairs DNMT1–HAUSP interaction and promotes the degradation of DNMT1. Treatment with HDAC inhibitor not only increases the level of acetylated DNMT1 but also decreases the total DNMT1 protein level. HDACs can be recruited by both DNMT1 and UHRF1 to repress gene expression, and HDAC1 can deacetylate DNMT1 to protect it from degradation.Citation64,Citation77 In addition, the stability of DNMT1 is maintained in part through the recruitment of Hsp90 mediated by the ubiquitin-proteasome pathway.Citation78 Suv39H1 and G9a are both H3K9 histone methyltransferases found in the same macromolecular complex with UHRF1Citation43,Citation67,Citation79 to prevent transcriptional activation;Citation80,Citation81 the latter can maintain the DNA methylation state required for de novo DNA methylation and the establishment in mouse embryonic stem cells.Citation82–Citation84 UHRF1 is necessary for binding Suv39H1 and H3K9me3 modification,Citation43 however, the underlying interaction mechanisms remain to be elucidated.
The macromolecular complex may function as follows (): UHRF1 and DNMT1 adopt a closed conformation and an auto-inhibitory state after protein synthesis, respectively. During DNA replication, the ECREM binds to replication foci. First, the closed conformation of UHRF1 shifts to an open state due to the following reasons: 1) HAUSP binds to PBR as well as the hmDNA; 2) SRA domain binds to hmDNA; and 3) phosphorylation of S651 by PI5P leads to TTD relief from PBR and forms a histone binding cassette with the PHD domain and linker. Second, UHRF1 in the open state binds to H3K9me2/3 or H3K9me3e mimic LIG1 by TTD or TTD-PHD as well as binds to H3R2 via the PHD domain. Third, the RING domain of UHRF1 ubiquitinates H3K18 and/or H3K23, which recruits the RFTS domain of DNMT1. The RFTS domain of DNMT1 also binds to the SRA domain or UBL of UHRF1 to relieve the self-inhibitory activity. HDAC1 and HAUSP can prevent UHRF1 and DNMT1 from acetylation and ubiquitination for degradation mediated by Tip60 during S phase when the abundance of Tip60 is relatively low. HAUSP can also deubiquitinate H3 which recruits the RFTS domain of DNMT1 to maintain a faithful inheritance of DNA methylation. At the beginning of G2 phase, when DNA replication is completed and methylation inheritance is formed, UHRF1 and DNMT1 undergo degradation. In detail, HAUSP dislocates from the UHRF1/DNMT1 complex due to phosphorylation of the S652 position by CDK1-cyclin B followed by ubiquitination of UHRF1. Degradation of UHRF1 leads to upregulation of Tip60 and acetylation of DNMT1, which is subsequently ubiquitylated by UHRF1 and other ubiquitin-related enzymes. Finally, the abundance of UHRF1 and DNMT1 decreases to ensure the normal cell cycle.
Figure 2 Working model for dynamic regulation of UHRF1 macromolecular complex during cell cycle.
Notes: (A) The spatialization of UHRF1 macromolecular complex including UHRF1 at open state and other members of ECREM recruited into replication forks during S phase. (B) The separation of HAUSP from the complex owing to phosphorylation mediated by CDK1-cyclin B at the beginning of G2 phase. (C) Acetylation and ubiquitination of UHRF1 and DNMT1 by Tip60 due to release of HAUSP and HDAC1 from ECREM during G2-M phase. (D) The ubiquitination-mediated protein degradation of UHRF1 and DNMT1 in order to enter the normal cell cycle.
Abbreviations: DNMT, DNA methyltransferases; ECREM, epigenetic code replication machinery; HAUSP, herpes virus-associated ubiquitin-specific protease; HDAC1, histone deacetylase 1; PBR, polybasic region; PHD, plant homeodomain; RING, really interesting new gene; SRA, SET and RING-associated domain; Tip60, Tat-interactive protein; TTD, tandem tudor domain; UBL, ubiquitin-like domain; UHRF1, ubiquitin-like with PHD and RING finger domains 1; PCNA, proliferating cell nuclear antigen; CDK1, cyclin-dependent kinase 1.
UHRF1 regulation and target therapy
Due to overexpression in various types of cancers such as gastric cancer,Citation85 colorectal cancer,Citation86 hepatocellular carcinoma,Citation87 ESCC,Citation48,Citation88 prostate cancer,Citation89,Citation90 bladder cancer,Citation91 breast cancer,Citation92 lung cancer,Citation93,Citation94 melanoma,Citation95 medulloblastoma,Citation96 lymphoblastic leukemia,Citation97 and osteosarcoma.Citation98 UHRF1 appears a potential biomarker and is promising in the target therapy of cancers. High expression of UHRF1 inhibits a variety of tumor suppressor genes () such as p16INK4,Citation99–Citation101 BRCA1,Citation102 KISS1,Citation103 RASSF1,Citation94 MEG3,Citation104 CDH4,Citation85,Citation105 Keap1,Citation106 KLF17,Citation107 RIP3,Citation108 SHP1,Citation109 RUNX3, FOXO4, CDX2,Citation85 PPARG,Citation110 Slit3,Citation105 CDH1,Citation43,Citation98,Citation111 GPX3,Citation43 IGFBP3,Citation43,Citation112 RGS2,Citation113 UBE2L6,Citation114 and miR-145.Citation115 Knowledge regarding the regulation of UHRF1 is limited; overexpression of UHRF1 may be involved in aberrantly high expression of transcription factors. Moreover, cell cycle regulators such as E2F8, E2F1,Citation116,Citation117 FOXM1,Citation118 leptin, SP1,Citation119,Citation120 Hsp90,Citation121 P53/P21,Citation122 and WDR79Citation123 can also protect UHRF1 from ubiquitination. Conversely, microRNAs such as miR-124,Citation124 microRNA-9,Citation86 miR-101,Citation125 miR-378, miR-193a-3p,Citation126 miR-1455 p/miR-145-3 p,Citation127 miR-146a/b,Citation105 and let-7a-3Citation128 play an important role in the downregulation of UHRF1 (). Most of the related microRNAs have been found suppressed in multiple cancers and can regulate the expression of UHRF1 by base pairing with 3′ UTR at the mRNA level to inhibit protein synthesis. The expression and activity of UHRF1 are also regulated by posttranslational modification. Chen et alCitation129 found that casein kinase 1 delta could catalyze the phosphorylation of UHRF1 at S108 to undergo ubiquitination and degradation mediated by SCFβ-TrCP in response to DNA damage; kinase Pim1 destabilizes UHRF1 by phosphorylation at S311;Citation130 methyltransferase PRMT6 can induce H3R2me2a and inhibit the association of the PHD of UHRF1 with H3R2 leading to DNA hypomethylation,Citation131 which provides new insights for cancer therapy targeting UHRF1. Graf et alCitation132 found the LRR domain of Pramel7 in embryos interacts with the SRA and RING domain of UHRF1 which subsequently undergoes degradation and hypomethylation. Wang et alCitation120 found that globular adiponectin inhibited leptin-stimulated cell proliferation in esophageal adenocarcinoma via the inhibition of UHRF1 mediated by adiponectin receptor 2.
Table 1 Inhibition of TSGs by overexpression of UHRF1 in various types of cancers
Table 2 Regulation of UHRF1 by microRNA in various types of cancers
Anticancer drugs related to UHRF1 are as follows: uracil derivative NSC232003Citation133 functions as the only direct inhibitor by targeting fit within the SRA of UHRF1. ShikoninCitation134 induces downregulation of both UHRF1 and DNMT1 in MDF-7 and hela cell lines. In addition, hinokitiol,Citation68 dihydroartemisinin,Citation89 epigallocatechin-3-gallate,Citation135 emodin,Citation136 mTOR inhibitor torin-2,Citation137 luteolin,Citation69 ERK1/2 pathway inhibitor PD98059, LY294002, AG490,Citation138 Hsp90 inhibitor 17-AAG or 17-dimethylamino-ethylamino-17-demethoxygeldanamycin,Citation121 anisomycin,Citation139 and curcuminCitation95,Citation140 have been reported to be used in various types of cancers.
Other functions of UHRF1 apart from epigenetic modification include its participation in the oxidative stress and DNA damage responseCitation141,Citation142 to inhibit caspase-dependent apoptosis to promote the proliferation, invasion, and metastasis,Citation143 promoting the development of embryogenesis and preimplantation of embryos,Citation73,Citation144 and regulating the function and development of T lymphocytesCitation145,Citation146 as well as the plasticity of smooth muscle cells.Citation147
Prospects
Great progress has been made in the functional and modulation mechanisms of the UHRF1 protein complex, and it will likely become a universal biomarker for cancer and specific targets for cancer therapy. Emerging evidence indicates that UHRF1 modules do not act independently of each other but establish complex modes of interaction with patterns of chromatin modifications. Due to the variant abundance of ECREM and the distinct level of components in diverse cell lines, the UHRF1 protein complex possibly undergoes temporal and spatial control during the cell cycle, although the mechanisms remain to be elucidated. Furthermore, the phosphorylation sites of amino acids among the five domains are involved in the regulation of UHRF1 activity, protein stability, DNA methylation, and histone posttranscriptional modification. As mentioned above, the mechanisms regarding degradation of UHRF1 by acetylation and ubiquitination and specific drugs targeting UHRF1 need to be further clarified. Because many drugs can downregulate UHRF1 as well as DNMT1, and lower expression of DNMT1 is more sensitive to 5-aza-CdR treatment,Citation148 chemotherapy in cancer cells by using a DNMT1 inhibitor accompanied by UHRF1 inhibitor for the treatment of cancer is yet to be investigated. In summary, DNA methylation within CpG varies in different organs and timesCitation149 when cells are supposed to differentiate by epigenetic regulation, without DNA mutations or chromosomal translocation, indicating the important dynamics of UHRF1 in epigenetics and the various roles it plays over the life span.
Acknowledgments
This study was supported by National Natural Science Foundation of China (No 81272946) and Natural Science Foundation of Liaoning Province (No 2015020503 and 2015020473).
Disclosure
The authors report no conflicts of interest in this work.
References
- HopfnerRMousliMJeltschJMICBP90, a novel human CCAAT binding protein, involved in the regulation of topoisomerase IIalpha expressionCancer Res200060112112810646863
- LevensonJMSweattJDEpigenetic mechanisms in memory formationNat Rev Neurosci20056210811815654323
- MartinCZhangYMechanisms of epigenetic inheritanceCurr Opin Cell Biol200719326627217466502
- ProbstAVDunleavyEAlmouzniGEpigenetic inheritance during the cell cycleNat Rev Mol Cell Biol200910319220619234478
- la RosaAHManoharanMGoolamASCurrent concepts of epigenetics in testicular cancerIndian J Surg Oncol20178216917428546713
- GaoLTanXFZhangSAn intramolecular interaction of UHRF1 reveals dual control for its histone associationStructure201826230431129395786
- JähnerDStuhlmannHStewartCLDe novo methylation and expression of retroviral genomes during mouse embryogenesisNature198229858756236286285203
- KleinCJBotuyanM-VWuYMutations in DNMT1 cause hereditary sensory neuropathy with dementia and hearing lossNature Genetics201143659560021532572
- YangLRodriguezBMayleADNMT3A loss drives enhancer hypomethylation in FLT3-ITD-associated leukemiasCancer Cell201629692293427300438
- Al-KharashiLAAl-MohannaFHTulbahAAboussekhraAThe DNA methyl-transferase protein DNMT1 enhances tumor-promoting properties of breast stromal fibroblastsOncotarget2018922329234329416775
- YangASEstecioMRGarcia-ManeroGKantarjianHMIssaJPComment on “chromosomal instability and tumors promoted by DNA hypomethylation” and “induction of tumors in mice by genomic hypomethylation”Science20033025648115314615517
- OgishimaTShiinaHBreaultJEPromoter CpG hypomethylation and transcription factor EGR1 hyperactivate heparanase expression in bladder cancerOncogene200524456765677216007175
- TianFTangZSongGLoss of imprinting of IGF2 correlates with hypomethylation of the H19 differentially methylated region in the tumor tissue of colorectal cancer patientsMol Med Rep2012561536154022427002
- LeeJHVooKSSkalnikDGIdentification and characterization of the DNA binding domain of CpG-binding proteinJ Biol Chem200127648446694467611572867
- PradhanMEstèvePOChinHGSamaranaykeMKimGDPradhanSCXXC domain of human DNMT1 is essential for enzymatic activityBiochemistry20084738100001000918754681
- SongJRechkoblitOBestorTHPatelDJStructure of DNMT1-DNA complex reveals a role for autoinhibition in maintenance DNA methylationScience201133160201036104021163962
- SongJTeplovaMIshibe-MurakamiSPatelDJStructure-based mechanistic insights into DNMT1-mediated maintenance DNA methylationScience2012335606970971222323818
- TakeshitaKSuetakeIYamashitaEStructural insight into maintenance methylation by mouse DNA methyltransferase 1 (Dnmt1)Proc Natl Acad Sci U S A2011108229055905921518897
- BashtrykovPJankeviciusGJurkowskaRZRagozinSJeltschAThe UHRF1 protein stimulates the activity and specificity of the maintenance DNA methyltransferase DNMT1 by an allosteric mechanismJ Biol Chem201428974106411524368767
- AvvakumovGVWalkerJRXueSStructural basis for recognition of hemi-methylated DNA by the SRA domain of human UHRF1Nature2008455721482282518772889
- BostickMKimJKEstèvePOClarkAPradhanSJacobsenSEUHRF1 plays a role in maintaining DNA methylation in mammalian cellsScience200731758451760176417673620
- BerkyurekACSuetakeIAritaKThe DNA methyltransferase Dnmt1 directly interacts with the SET and RING finger-associated (SRA) domain of the multifunctional protein Uhrf1 to facilitate accession of the catalytic center to hemi-methylated DNAJ Biol Chem2014289137938624253042
- NishiyamaAYamaguchiLSharifJUhrf1-dependent H3K23 ubiquitylation couples maintenance DNA methylation and replicationNature2013502747024925324013172
- QinWWolfPLiuNDNA methylation requires a DNMT1 ubiquitin interacting motif (UIM) and histone ubiquitinationCell Res201525891192926065575
- LiTWangLDuYStructural and mechanistic insights into UHRF1-mediated DNMT1 activation in the maintenance DNA methylationNucleic Acids Res20184663218323129471350
- FosterBMStolzPMulhollandCBCritical role of the UBL domain in stimulating the E3 ubiquitin ligase activity of UHRF1 toward chromatinMol Cell201872473975230392929
- DarosaPAHarrisonJSZelterAA bifunctional role for the UHRF1 UBL domain in the control of hemi-methylated DNA-dependent histone ubiquitylationMol Cell201872475376530392931
- MurataniMTanseyWPHow the ubiquitin-proteasome system controls transcriptionNat Rev Mol Cell Biol20034319220112612638
- WozniakRJKlimeckiWTLauSSFeinsteinYFutscherBW5-Aza-2′-deoxycytidine-mediated reductions in G9A histone methyltransferase and histone H3 K9 di-methylation levels are linked to tumor suppressor gene reactivationOncogene2007261779016799634
- ChengJYangYFangJStructural insight into coordinated recognition of trimethylated histone H3 lysine 9 (H3K9me3) by the plant homeodomain (PHD) and tandem tudor domain (TTD) of UHRF1 (ubiquitin-like, containing PHD and RING finger domains, 1) proteinJ Biol Chem201328821329133923161542
- HuLLiZWangPLinYXuYCrystal structure of PHD domain of UHRF1 and insights into recognition of unmodified histone H3 arginine residue 2Cell Res20112191374137821808300
- RajakumaraEWangZMaHPHD finger recognition of unmodified histone H3R2 links UHRF1 to regulation of euchromatic gene expressionMol Cell201143227528421777816
- WangCShenJYangZStructural basis for site-specific reading of unmodified R2 of histone H3 tail by UHRF1 PHD fingerCell Res20112191379138221808299
- RothbartSBDicksonBMOngMSMultivalent histone engagement by the linked tandem Tudor and PHD domains of UHRF1 is required for the epigenetic inheritance of DNA methylationGenes Dev201327111288129823752590
- LallousNLegrandPMcEwenAGRamón-MaiquesSSamamaJPBirckCThe PHD finger of human UHRF1 reveals a new subgroup of unmethylated histone H3 tail readersPLoS One2011611e2759922096602
- FangJChengJWangJHemi-methylated DNA opens a closed conformation of UHRF1 to facilitate its histone recognitionNat Commun2016711119727045799
- RothbartSBKrajewskiKNadyNAssociation of UHRF1 with methylated H3K9 directs the maintenance of DNA methylationNat Struct Mol Biol201219111155116023022729
- AritaKIsogaiSOdaTRecognition of modification status on a histone H3 tail by linked histone reader modules of the epi-genetic regulator UHRF1Proc Natl Acad Sci U S A201210932129501295522837395
- GelatoKATauberMOngMSAccessibility of different histone H3-binding domains of UHRF1 is allosterically regulated by phosphatidylinositol 5-phosphateMol Cell201454690591924813945
- RigboltKTProkhorovaTAAkimovVSystem-wide temporal characterization of the proteome and phosphoproteome of human embryonic stem cell differentiationSci Signal20114164rs321406692
- UnokiMBrunetJMousliMDrug discovery targeting epigenetic codes: the great potential of UHRF1, which links DNA methylation and histone modifications, as a drug target in cancers and toxoplasmosisBiochem Pharmacol200978101279128819501055
- ZhaoQZhangJChenRDissecting the precise role of H3K9 methylation in crosstalk with DNA maintenance methylation in mammalsNat Commun201671246427554592
- BabbioFPistoreCCurtiLThe SRA protein UHRF1 promotes epigenetic crosstalks and is involved in prostate cancer progressionOncogene201231464878488722330138
- SpodickDHECG diagnosis of MI in LBBBAm Heart J198911761409
- McCabeMTOttHMGanjiGEZH2 inhibition as a therapeutic strategy for lymphoma with EZH2-activating mutationsNature2012492742710811223051747
- SneeringerCJScottMPKuntzKWCoordinated activities of wild-type plus mutant EZH2 drive tumor-associated hypertrimethylation of lysine 27 on histone H3 (H3K27) in human B-cell lymphomasProc Natl Acad Sci U S A201010749209802098521078963
- FerryLFournierATsusakaTMethylation of DNA ligase 1 by G9a/GLP recruits UHRF1 to replicating DNA and regulates DNA methylationMol Cell201767455056528803780
- NakamuraKBabaYKosumiKUHRF1 regulates global DNA hypomethylation and is associated with poor prognosis in esophageal squamous cell carcinomaOncotarget2016736578215783127507047
- JiaYLiPFangLNegative regulation of DNMT3A de novo DNA methylation by frequently overexpressed UHRF family proteins as a mechanism for widespread DNA hypomethylation in cancerCell Discov201621600727462454
- HongJHJinEHKimSSongKSSungJKLINE-1 hypomethylation is inversely correlated with UHRF1 overexpression in gastric cancerOncol Lett20181556666667029616129
- CordauxRBatzerMAThe impact of retrotransposons on human genome evolutionNat Rev Genet2009101069170319763152
- YeJZhangYLiangWHuangJWangLZhongXUHRF1 is an independent prognostic factor and a potential therapeutic target of esophageal squamous cell carcinomaJ Cancer20178194027403929187878
- HoshimotoSTakeuchiHOnoSGenome-wide hypomethylation and specific tumor-related gene hypermethylation are associated with esophageal squamous cell carcinoma outcomeJ Thorac Oncol201510350951725514805
- JeltschAOn the enzymatic properties of Dnmt1: specificity, processivity, mechanism of linear diffusion and allosteric regulation of the enzymeEpigenetics200612636617965604
- ArandJSpielerDKariusTIn vivo control of CpG and non-CpG DNA methylation by DNA methyltransferasesPLoS Genetics201286e100275022761581
- LiYZhangZChenJStella safeguards the oocyte methylome by preventing de novo methylation mediated by DNMT1Nature2018564773413614030487604
- BronnerCChataigneauTSchini-KerthVBLandryYThe “Epigenetic Code Replication Machinery”, ECREM: a promising drugable target of the epigenetic cell memoryCurr Med Chem200714252629264117979715
- ChuangLSIanHIKohTWNgHHXuGLiBFHuman DNA-(cytosine-5) methyltransferase-PCNA complex as a target for p21WAF1Science19972775334199620009302295
- SchermellehLHaemmerASpadaFDynamics of Dnmt1 interaction with the replication machinery and its role in postreplicative maintenance of DNA methylationNucleic Acids Res200735134301431217576694
- CartronPFBlanquartCHervouetEGregoireMValletteFMHDAC1-mSin3a-NCOR1, Dnmt3b-HDAC1-Egr1 and Dnmt1-PCNA-UHRF1-G9a regulate the NY-ESO1 gene expressionMol Oncol20137345246323312906
- DaiCShiDGuWNegative regulation of the acetyltransferase TIP60-p53 interplay by UHRF1 (ubiquitin-like with PHD and RING finger domains 1)J Biol Chem201328827195811959223677994
- AshrafWBronnerCZaayterLInteraction of the epigenetic integrator UHRF1 with the MYST domain of TIP60 inside the cellJ Exp Clin Cancer Res201736118829268763
- AchourMFuhrmannGAlhosinMUHRF1 recruits the histone acetyltransferase Tip60 and controls its expression and activityBiochem Biophys Res Commun2009390352352819800870
- duZSongJWangYDNMT1 stability is regulated by proteins coordinating deubiquitination and acetylation-driven ubiquitinationSci Signal20103146ra8021045206
- de MarzoAMMarchiVLYangESVeeraswamyRLinXNelsonWGAbnormal regulation of DNA methyltransferase expression during colorectal carcinogenesisCancer Res199959163855386010463569
- AgostonATArganiPYegnasubramanianSIncreased protein stability causes DNA methyltransferase 1 dysregulation in breast cancerJ Biol Chem200528018183021831015755728
- JungHJByunHOJeeBAThe ubiquitin-like with PHD and ring finger domains 1 (UHRF1)/DNA methyltransferase 1 (DNMT1) axis is a primary regulator of cell senescenceJ Biol Chem201729293729373928100769
- SeoJSChoiYHMoonJWKimHSParkSHHinokitiol induces DNA demethylation via DNMT1 and UHRF1 inhibition in colon cancer cellsBMC Cell Biol20171811428241740
- KrifaMLeloupLGhediraKMousliMChekir-GhediraLLuteolin induces apoptosis in BE colorectal cancer cells by downregulating calpain, UHRF1, and DNMT1 expressionsNutr Cancer20146671220122725207720
- ShammaASuzukiMHayashiNATM mediates pRB function to control DNMT1 protein stability and DNA methylationMol Cell Biol201333163113312423754744
- MaHChenHGuoXM phase phosphorylation of the epigenetic regulator UHRF1 regulates its physical association with the deubiquitylase USP7 and stabilityProc Natl Acad Sci U S A2012109134828483322411829
- ZhangZMRothbartSBAllisonDFAn allosteric interaction links USP7 to deubiquitination and chromatin targeting of UHRF1Cell Rep20151291400140626299963
- ChuJLoughlinEAGaurNAUHRF1 phosphorylation by cyclin A2/cyclin-dependent kinase 2 is required for zebrafish embryogenesisMol Biol Cell2012231597022072796
- DarAShibataEDuttaADeubiquitination of Tip60 by USP7 determines the activity of the p53-dependent apoptotic pathwayMol Cell Biol201333163309332023775119
- YamaguchiLNishiyamaAMisakiTUsp7-dependent histone H3 deubiquitylation regulates maintenance of DNA methylationSci Rep2017715528246399
- ChengJYangHFangJMolecular mechanism for USP7-mediated DNMT1 stabilization by acetylationNat Commun20156702325960197
- CedarHBergmanYLinking DNA methylation and histone modification: patterns and paradigmsNat Rev Genet200910529530419308066
- ZhouQAgostonATAtadjaPNelsonWGDavidsonNEInhibition of histone deacetylases promotes ubiquitin-dependent proteasomal degradation of DNA methyltransferase 1 in human breast cancer cellsMol Cancer Res20086587388318505931
- KimJKEstèvePOJacobsenSEPradhanSUHRF1 binds G9a and participates in p21 transcriptional regulation in mammalian cellsNucleic Acids Res200937249350519056828
- LachnerMO’CarrollDReaSMechtlerKJenuweinTMethylation of histone H3 lysine 9 creates a binding site for HP1 proteinsNature2001410682411612011242053
- TachibanaMSugimotoKFukushimaTShinkaiYSet domain-containing protein, G9a, is a novel lysine-preferring mammalian histone methyltransferase with hyperactivity and specific selectivity to lysines 9 and 27 of histone H3J Biol Chem200127627253092531711316813
- IkegamiKIwataniMSuzukiMGenome-wide and locus-specific DNA hypomethylation in G9a deficient mouse embryonic stem cellsGenes Cells200712111117212651
- DongKBMaksakovaIAMohnFDNA methylation in ES cells requires the lysine methyltransferase G9a but not its catalytic activityEMBO J200827202691270118818693
- LeungDCDongKBMaksakovaIALysine methyltransferase G9a is required for de novo DNA methylation and the establishment, but not the maintenance, of proviral silencingProc Natl Acad Sci U S A2011108145718572321427230
- ZhouLShangYJinZUHRF1 promotes proliferation of gastric cancer via mediating tumor suppressor gene hypermethylationCancer Biol Ther20151681241125126147747
- ZhuMXuYGeMGuiZYanFRegulation of UHRF1 by microRNA-9 modulates colorectal cancer cell proliferation and apoptosisCancer Sci2015106783383925940709
- MudbharyRHoshidaYChernyavskayaYUHRF1 overexpression drives DNA hypomethylation and hepatocellular carcinomaCancer Cell201425219620924486181
- YangCWangYZhangFInhibiting UHRF1 expression enhances radiosensitivity in human esophageal squamous cell carcinomaMol Biol Rep20134095225523523943380
- DuSXuGZouWXiangTLuoZEffect of dihydroartemisinin on UHRF1 gene expression in human prostate cancer PC-3 cellsAnticancer Drugs201728438439128059831
- JazirehiARArleDWennPBUHRF1: a master regulator in prostate cancerEpigenomics20124325125222690660
- SaidiSPopovZJanevskaVPanovSOverexpression of UHRF1 gene correlates with the major clinicopathological parameters in urinary bladder cancerInt Braz J Urol201743222422928128913
- FangLShanquLPingGGene therapy with RNAi targeting UHRF1 driven by tumor-specific promoter inhibits tumor growth and enhances the sensitivity of chemotherapeutic drug in breast cancer in vitro and in vivoCancer Chemother Pharmacol20126941079108722205202
- UnokiMDaigoYKoinumaJTsuchiyaEHamamotoRNakamuraYUHRF1 is a novel diagnostic marker of lung cancerBr J Cancer2010103221722220517312
- DaskalosAOleksiewiczUFiliaAUHRF1-mediated tumor suppressor gene inactivation in nonsmall cell lung cancerCancer201111751027103721351083
- AbusninaAKeravisTYougbaréIBronnerCLugnierCAnti-proliferative effect of curcumin on melanoma cells is mediated by PDE1A inhibition that regulates the epigenetic integrator UHRF1Mol Nutr Food Res201155111677168922045655
- ZhangZYZhuBZhaoXWRegulation of UHRF1 by microRNA-378 modulates medulloblastoma cell proliferation and apoptosisOncol Rep20173853078308428901497
- AlhosinMAbusninaAAchourMInduction of apoptosis by thymoquinone in lymphoblastic leukemia Jurkat cells is mediated by a p73-dependent pathway which targets the epigenetic integrator UHRF1Biochem Pharmacol20107991251126020026309
- LiuWQiaoRHWangDMHuangXWLiBWangDUHRF1 promotes human osteosarcoma cell invasion by downregulating the expression of E-cadherin in an Rb1-dependent mannerMol Med Rep201613131532026548607
- BoukhariAAlhosinMBronnerCCD47 activation-induced UHRF1 over-expression is associated with silencing of tumor suppressor gene p16INK4A in glioblastoma cellsAnticancer Res201535114915725550546
- WangFYangYZShiCZUHRF1 promotes cell growth and metastasis through repression of p16(ink4a) in colorectal cancerAnn Surg Oncol20121982753276222219067
- KrifaMAlhosinMMullerCDLimoniastrum guyonianum aqueous gall extract induces apoptosis in human cervical cancer cells involving p16INK4A re-expression related to UHRF1 and DNMT1 down-regulationJ Exp Clin Cancer Res20133213023688286
- JinWChenLChenYUHRF1 is associated with epigenetic silencing of BRCA1 in sporadic breast cancerBreast Cancer Res Treat2010123235937319943104
- ZhangYHuangZZhuZUpregulated UHRF1 promotes bladder cancer cell invasion by epigenetic silencing of KiSS1PLoS One2014910e10425225272010
- ZhuoHTangJLinZThe aberrant expression of MEG3 regulated by UHRF1 predicts the prognosis of hepatocellular carcinomaMol Carcinog201655220921925641194
- ZhouLZhaoXHanYRegulation of UHRF1 by miR-146a/b modulates gastric cancer invasion and metastasisFASEB J201327124929493923982143
- Abu-AlaininWGanaTLiloglouTUHRF1 regulation of the Keap1-Nrf2 pathway in pancreatic cancer contributes to oncogenesisJ Pathol2016238342343326497117
- GaoSPSunHFLiLDFuWYJinWUHRF1 promotes breast cancer progression by suppressing KLF17 expression by hypermethylating its promoterAm J Cancer Res2017771554156528744404
- YangCLiJYuLRegulation of RIP3 by the transcription factor Sp1 and the epigenetic regulator UHRF1 modulates cancer cell necroptosisCell Death Dis2017810e308428981102
- ShengYWangHLiuDMethylation of tumor suppressor gene CDH13 and SHP1 promoters and their epigenetic regulation by the UHRF1/PRMT5 complex in endometrial carcinomaGynecol Oncol2016140114515126597461
- SabatinoLFucciAPancioneMUHRF1 coordinates peroxisome proliferator activated receptor gamma (PPARG) epigenetic silencing and mediates colorectal cancer progressionOncogene201231495061507222286757
- MagnaniEMacchiFManciniMUHRF1 regulates CDH1 via promoter associated non-coding RNAs in prostate cancer cellsBiochimica et biophysica acta2018186125827029466696
- BeckATrippelFWagnerAOverexpression of UHRF1 promotes silencing of tumor suppressor genes and predicts outcome in hepatoblastomaClin Epigenetics2018102729507645
- YingLLinJQiuFEpigenetic repression of regulator of G-protein signaling 2 by ubiquitin-like with PHD and ring-finger domain 1 promotes bladder cancer progressionFEBS J2015282117418225323766
- ZhangQQiaoLWangXDingCChenJJUHRF1 epigenetically down-regulates UbcH8 to inhibit apoptosis in cervical cancer cellsCell Cycle201817330030829157076
- YeZShenNWengYLow miR-145 silenced by DNA methylation promotes NSCLC cell proliferation, migration and invasion by targeting mucin 1Cancer Biol Ther20151671071107925961369
- UnokiMNishidateTNakamuraYICBP90, an E2F-1 target, recruits HDAC1 and binds to methyl-CpG through its SRA domainOncogene200423467601761015361834
- ParkS-APlattJLeeJWLópez-GiráldezFHerbstRSKooJSE2F8 as a Novel Therapeutic Target for Lung CancerJ Natl Cancer Inst2015107djv15126089541
- SandersDAGormallyMVMarsicoGBeraldiDTannahillDBalasubramanianSFOXM1 binds directly to non-consensus sequences in the human genomeGenome Biol20151613026100407
- WuSMChengWLLiaoCJNegative modulation of the epi-genetic regulator, UHRF1, by thyroid hormone receptors suppresses liver cancer cell growthInt J Cancer20151371374925430639
- WangJChengYYinXGlobular adiponectin inhibits leptin-stimulated esophageal adenocarcinoma cell proliferation via adiponectin receptor 2-mediated suppression of UHRF1Mol Cell Biochem20174311–210311228285359
- DingGChenPZhangHRegulation of ubiquitin-like with plant homeodomain and RING finger domain 1 (UHRF1) protein stability by heat shock protein 90 chaperone machineryJ Biol Chem201629138201252013527489107
- ArimaYHirotaTBronnerCDown-regulation of nuclear protein ICBP90 by p53/p21Cip1/WAF1-dependent DNA-damage checkpoint signals contributes to cell cycle arrest at G1/S transitionGenes Cells20049213114215009091
- ChenJShengXMaHWDR79 mediates the proliferation of non-small cell lung cancer cells by regulating the stability of UHRF1J Cell Mol Med20182252856286429516630
- WangXWuQXuBmiR-124 exerts tumor suppressive functions on the cell proliferation, motility and angiogenesis of bladder cancer by fine-tuning UHRF1FEBS J2015282224376438826310391
- GotoYKurozumiANohataNThe microRNA signature of patients with sunitinib failure: regulation of UHRF1 pathways by microRNA-101 in renal cell carcinomaOncotarget2016737590705908627487138
- DengWYanMYuTQuantitative proteomic analysis of the metastasis-inhibitory mechanism of miR-193a-3p in non-small cell lung cancerCell Physiol Biochem20153551677168825833338
- AtalaARe: Regulation of UHRF1 by dual-strand tumor-suppressor microRNA-145 (miR-145-5p and miR-145-3p): inhibition of bladder cancer cell aggressivenessJ Urol20161964131427628846
- PengRLiuHPengHPromoter hypermethylation of let-7a-3 is relevant to its down-expression in diabetic nephropathy by targeting UHRF1Gene20155701576326049093
- ChenHMaHInuzukaHDNA damage regulates UHRF1 stability via the SCF(β-TrCP) E3 ligaseMol Cell Biol20133361139114823297342
- YangJLiuKYangJPIM1 induces cellular senescence through phosphorylation of UHRF1 at Ser311Oncogene201736344828484228394343
- VelandNHardikarSZhongYThe arginine methyltransferase PRMT6 regulates DNA methylation and contributes to global DNA hypomethylation in cancerCell Rep201721123390339729262320
- GrafUCasanovaEAWyckSPramel7 mediates ground-state pluripotency through proteasomal-epigenetic combined pathwaysNat Cell Biol201719776377328604677
- MyrianthopoulosVCartronPFLiutkevičiūtėZTandem virtual screening targeting the SRA domain of UHRF1 identifies a novel chemical tool modulating DNA methylationEur J Med Chem201611439039627049577
- JangSYHongDJeongSYKimJHShikonin causes apoptosis by up-regulating p73 and down-regulating ICBP90 in human cancer cellsBiochem Biophys Res Commun20154651717626235879
- AchourMMousliMAlhosinMEpigallocatechin-3-gallate up-regulates tumor suppressor gene expression via a reactive oxygen species-dependent down-regulation of UHRF1Biochem Biophys Res Commun2013430120821223201574
- LinYChenWWangZCaiPEmodin promotes the arrest of human lymphoma Raji cell proliferation through the UHRF1-DNMT3A-∆Np73 pathwaysMol Med Rep20171656544655128901428
- WangCWangXSuZFeiHLiuXPanQThe novel mTOR inhibitor Torin-2 induces autophagy and downregulates the expression of UHRF1 to suppress hepatocarcinoma cell growthOncol Rep20153441708171626239364
- FangZXingFBronnerCTengZGuoZICBP90 mediates the ERK1/2 signaling to regulate the proliferation of Jurkat T cellsCell Immunol20092571–2808719328461
- YuCXingFTangZAnisomycin suppresses Jurkat T cell growth by the cell cycle-regulating proteinsPharmacol Rep201365243544423744428
- ParasharGCapalashNPromoter methylation-independent reactivation of PAX1 by curcumin and resveratrol is mediated by UHRF1Clin Exp Med201616347147826081871
- TienALSenbanerjeeSKulkarniAUHRF1 depletion causes a G2/M arrest, activation of DNA damage response and apoptosisBiochem J2011435117518521214517
- LiXMengQRosenEMFanSUHRF1 confers radioresistance to human breast cancer cellsInt J Radiat Biol201187326327321067293
- GeTTYangMChenZLouGGuTUHRF1 gene silencing inhibits cell proliferation and promotes cell apoptosis in human cervical squamous cell carcinoma CaSki cellsJ Ovarian Res2016914227431502
- MaenoharaSUnokiMTohHRole of UHRF1 in de novo DNA methylation in oocytes and maintenance methylation in preimplantation embryosPLoS Genet20171310e100704228976982
- CuiYChenXZhangJUhrf1 controls iNKT cell survival and differentiation through the Akt-mTOR axisCell Rep201615225626327050515
- ObataYFurusawaYEndoTAThe epigenetic regulator Uhrf1 facilitates the proliferation and maturation of colonic regulatory T cellsNat Immunol201415657157924777532
- EliaLKunderfrancoPCarulloPUHRF1 epigenetically orchestrates smooth muscle cell plasticity in arterial diseaseJ Clin Invest201812862473248629558369
- PanFPZhouHKBuHQEmodin enhances the demethylation by 5-Aza-CdR of pancreatic cancer cell tumor-suppressor genes P16, RASSF1A and ppENKOncol Rep20163541941194926782786
- EhrlichMGama-SosaMAHuangLHAmount and distribution of 5-methylcytosine in human DNA from different types of tissues of cellsNucleic Acids Res1982108270927217079182
