Abstract
Background
Tumor-associated macrophages (TAMs) serve as crucial modulators of the complicated interaction between cancer cells and immune microenvironment. Sirtuin 1 (SIRT1) has an impact on immune reactions in cancer progression. Current knowledge of the role of SIRT1 in the regulation of M1-like macrophages as well as in hepatocellular carcinoma (HCC) is insufficient.
Methods
SIRT1 expression in HCC tissues was detected using quantitative reverse transcriptase PCR (qRT-PCR) and Western blot. M1 markers were detected by qRT-PCR and flow cytometry assay. Moreover, the influence of SIRT1 on HCC cell apoptosis, migration, and invasion was studied using transwell assay, flow cytometry assay, and TUNEL assays, respectively.
Results
In this study, it was revealed that SIRT1 was upregulated in patients suffering from HCC; these patients were also shown to have elevated levels of M1-like TAM infiltration. SIRT1 was able to reinforce M1-like macrophage infiltration and inhibit HCC metastasis. Furthermore, SIRT1 enhanced NF-κB stimulation, promoting phosphorylation of p65, IκB, and IκB kinase. It was further demonstrated in our study that SIRT1 had an impact on polarization of M1 through the NF-κB pathway. NF-κB repression downregulated M1 markers in macrophages, which excessively expressed SIRT1 and counteracted the influence of SIRT1 on migration of HCC cells.
Conclusion
Taken together, these results offer proof that SIRT1 is an essential regulator of the immune reaction that counteracts malignant HCC cell migration as well as growth, indicating that macrophage SIRT1 could serve as an innovative target to treat HCC.
Introduction
Hepatocellular carcinoma (HCC) is characterized by its high mortality rate, accompanied by chronic inflammatory reactions in liver.Citation1 The step-by-step progression of HCC is characterized by alterations in the hepatic microenvironment.Citation2 To date, no drug has been approved for first-line treatment of terminal HCC, which comprises 40%–70% of all HCC cases.Citation3 Consequently, it is important to discover and target essential pathways that strengthen treatment efficacy via stimulation of antitumor immune reactions for improved prognosis and survival in the future.
Cancers are often characterized by excessive infiltration of macrophages.Citation4,Citation5 The M1 subtype of macrophages represses tumors, while the M2 subtype promotes them.Citation6 Specific tumor microenvironment signals affect the function as well as the polarization of macrophages.Citation7 Tumor-associated macrophages (TAMs) serve as crucial modulators of the complicated interactions between tumors and the immune system.Citation8 Treatment approaches that affect the bioactivity and/or existence of macrophages have been proved to be promising preclinically and are currently being assessed clinically.Citation9,Citation10 Stimulated macrophages are able to phagocytose malignant cells by hindering the interaction between CD47 and signal regulatory protein alpha (SIRPa), which has been con-temporarily applied to cancer treatment in various clinical trials.Citation11–Citation13 Furthermore, it should be taken into account that macrophages contribute to antitumor reactions.Citation14 It has been shown preclinically that macrophage proliferation in tumors is repressed, and TAM half-lives are shortened in comparison to resident macrophages of corresponding normal tissues.Citation15,Citation16 As a result, aggregation of TAMs in the tumor microenvironment, particularly M1 TAMs with phagocytic capability, is indispensable to TAM quantity protection and antitumor immunity in human cancers.Citation17,Citation18 Agents participating in these reactions are innovative targets to treat the disease.
As a part of sirtuin family (SIRT1–SIRT7) in mammals, SIRT1 is a conserved deacetylase relying on nicotinamide adenine dinucleotide (NAD)+, which contributes to the regulation of transcriptional silencing and cell viability.Citation19–Citation22 The expression of SIRT1 was significantly increased in the HCC cell line. The tumorigenic influence of SIRT1 has been demonstrated in HCC in recent years, except for its influence on different biological reactions, such as inhibition of cell death and enhancement of proliferation, tumor stimulation, progression, and metastasis.Citation23,Citation24 It was verified that SIRT1 had an impact on tumorigenesis, metastasis, and clinical outcomes as well as resistance to chemotherapy in HCC, caused by deacetylation of tumor suppressor or oncogenic factors.Citation25,Citation26 Nevertheless, the role of SIRT1 in the regulation of M1-like macrophages and in HCC is unclear.
Materials and methods
Clinical specimens
Tissue specimens were acquired from the First Affiliated Hospital of Xinxiang Medical College. Fresh specimens such as tumor tissues and corresponding noncancerous liver tissues were acquired from patients suffering from HCC after neoplasm excision. Fifteen HCC specimens were acquired from 2010 to 2014. Median age of the patients was 50 years (range: 17–73 years). HCC was diagnosed based on histopathology in all patients. None of the patients underwent anticancer treatment or displayed distant metastases before operation. Written informed consent was obtained from all participants prior to the study. For any participant under 18 years of age, a parent or guardian provided written informed consent. This research was approved by the School of Basic Medical Sciences, Xinxiang Medical University First Affiliated Hospital of Xinxiang Medical College, and conducted in accordance with the Declaration of Helsinki.
Cell cultivation and treatment
The hepatic cancer cell lines, HepG2 and RAW 264.7, HL-60 were bought from Bioresource Collection and Research Center (Shanghai, China) in 2014. Cell cultivation was performed in DMEM with 10% FBS at 37°C under conditions of 5% CO2.
Lipopolysaccharide (LPS; 100 ng/mL) and interferon-gamma (IFN-γ; 20 ng/mL) were applied for 24 hours to activate RAW 264.7 macrophages in order to harvest M1 macrophages. Conditioned medium arising from RAW 264.7 or HL-60 macrophages was used for HepG2 activation for 24 hours in order to detect cell migration, death, and invasion.
Cell transfection
Plasmids of SIRT1-pcDNA3.1 (HMGB1) and pcDNA3.1 (vector) were used for transfection. Briefly, six-well plates were seeded with RAW 264.7 or HL-60 macrophages with 4×105 cells/well. The cultivating medium was spiked with 4 µg of plasmid DNA and 3 µL of Turbofect reagent and then incubated for 6 hours. The transfection admixture was removed by centrifugation after 6 hours, and cells underwent another 24-hour incubation in normal medium.
Immunohistochemistry (IHC)
HCC cells were fixed with formalin, embedded, sliced, and stained using primary antibodies targeting CD16/32 and CCR7 via standard IHC methods. Two-minute counterstaining of nuclei was carried out using DAPI (provided by Sigma-Aldrich). Confocal microscopy (LSM510, METALaser Scanning Microscope; Carl Zeiss) was used for observation as well as image acquisition.
RNA isolation and qRT-PCR
TRIzol® reagent (Thermo Fisher Scientific) was utilized in order to isolate total RNA as per the manufacturer’s instructions. Spectrophotometry (Thermo Fisher Scientific) was applied to quantify the isolated RNA using 260/280 nanometer absorbance. RNA specimens were preserved at -80°C. An ABI 7500 quantitative PCR System (Thermo Fisher Scientific) was applied to perform and analyze quantitative reverse transcriptase PCR (qRT-PCR). Relative mRNA expression was assessed according to the comparative cycle threshold (CT) (2−ΔΔCT) approach. GAPDH served as the internal reference for normalization. Primers used in this study are as follows: SIRT1 forward: ATGAAGCAC CAACCGTATC, reverse: CTGAATTGACCTTGACT GATG; GADPH forward: GTCTCCTCTGACTTCAA CAGCG, reverse: ACCACCCTGTTGCTGTAGCCAA; iNOS forward: AATGGCAACATCAGGTCGGCCATCACT, reverse: GCTGTGTGTCACAGAAGTCTCGAACTC; TNF-α forward: CATCTTCTCAAAATTCGAGTGACAA, reverse: TGGGAGTAGACAAGGTACAACCC; IL-1β forward: TCATTGTGGCTGTGGAGAAG, reverse: AGGCCA CAGGTATTTTGTCG; IL-6 forward: ATCCAGTTGC CTTCTTCTTGGGACTGA, reverse: TAAGCCTCCGAC TTGTGAAGTGGT.
Cell migration assessment
Transwell assays were carried out in order to investigate migration. Briefly, DMEM without serum including 1 µg/mL mitomycin C was applied to a suspension of 50,000 cells, which were later seeded in the top well of a poly-carbonate transwell containing 24 wells (MD Millipore, Billerica, MA, USA). The bottom well contained DMEM with 10% serum. Cells on the top surface of the filters were eliminated after an incubation of 24 hours. Cells at the bottom surface were fixed, stained with 1% crystal violet, and quantified.
Cell invasion assessment
Transwell chambers with Matrigel coating were utilized in order to perform invasion assays. Cells that underwent transfection were planted in the top chamber (1.0×105 cells/chamber) and were incubated for 24 hours under conditions of 5% CO2 and 37°C. FBS (20%) at the bottom of the chambers served as a chemoattractant A cotton swab was used to eliminate cells that did not exhibit invasion at the top surface after incubation, while those showing invasion at the bottom surface were fixed using 100% methanol and stained with 1% crystal violet. Microscope images were used to quantify the number of cells showing invasion characteristics. Six random fields were selected for quantification in each assay.
Evaluation of cell apoptosis
Flow cytometry (FC) was used for cell-death assessment. Cells were washed twice in cold PBS, spun down for 5 minutes at 100 rpm, and resuspended in binding buffer. FITC-Annexin V and propidium iodide (PI) were added before incubating for 10 minutes at room temperature. A FACScan flow cytometer (Becton, Dickinson and Company, Franklin Lakes, NJ, USA) was used to detect fluorescence signals.
TUNEL assay
Cells on the slide were fixed with the help of 4% paraformaldehyde. An in situ cell death detection kit (provided by Roche) was used for TUNEL labeling of dead cells according to the manufacturer’s instructions.
FC
Cells were incubated with anti-mouse CD16/32 antibody conjugated to PE (R&D Systems, Inc., Minneapolis, MN, USA) and were washed before FC was carried out as instructed (Guava Easy Cyte™8; EMD Millipore).
Western blotting analysis
Lysis buffer (Beyotime, Jiangsu, China) was used for homogenization of cell lysates. Bradford assay (Bio-Rad, Hercules, CA, USA) was used for protein quantification. Proteins were checked for quality with a standard SDS–PAGE. Enhanced chemiluminescence and a detection reagent (Pierce, Rockford, IL, USA) was utilized in order to examine immunoreactive bands, which were analyzed using an Omega 16ic chemiluminescence imaging system (Ultra-Lum, Claremont, CA, USA). Primary antibodies used were as follows: rabbit anti-PhosphoIKKα/β (2697; Cell Signaling Technology, Beverly, MA, USA), rabbit anti-IKKβ (8943; Cell Signaling Technology), rabbit anti-Phospho-NF-κB p65 (3033; Cell Signaling Technology), and rabbit anti-NF-κB p65 (8242; Cell Signaling Technology).
Statistical analysis
Results are displayed as mean ± SEM. Evaluation of differences was performed using a two-tailed, unequal-variance Student’s t-test or ANOVA prior to Tukey’s post hoc analysis. Values with P<0.05 were considered to be statistically significant.
Results
Expression of SIRT1 is increased in HCC tissues
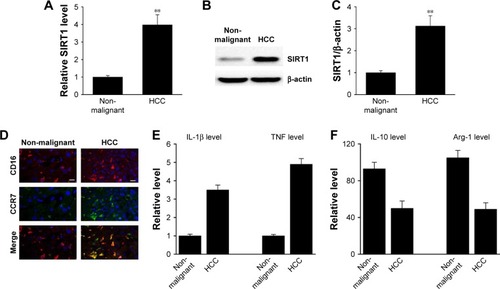
In order to measure the SIRT1 protein expression levels in clinical HCC tissues, RT-PCR and Western blotting were carried out in a cohort of 15 HCC tissues and paired non-malignant tissues. As shown in , not only mRNA but also protein SIRT1 levels were elevated in HCC tissues compared to non-malignant tissues. In addition, it has been shown via IHC that the HCC tissues contained more M1 macrophages (). M1 markers were increased in the HCC tissues together with a decrease in M1 markers (). The above-mentioned findings indicate that SIRT1 expression and the density of M1-like macrophages were increased in HCC.
Figure 1 SIRT1 upregulation was correlated with M1-like macrophage density in HCC tissues.
Notes: (A) The expression of SIRT1 in paired non-malignant tissues and primary HCC tumor tissue was determined by qRT-PCR. (B, C) Representative immunoblots (B) and quantitative assessment of SIRT1 (C) in normal hepatic tissue (Con) as well as primary HCC tissue. (D) Immunofluorescence staining of M1 macrophage marker CD16 (red) and CCR7 (green) in normal liver tissue (Con) and primary HCC tissue. DAPI (blue) was used for nuclei staining. Scale bar, 10 µm. (E, F) M1 markers and M2 markers were determined by qRT-PCR. Results are displayed as mean ± SEM, n=15, **P<0.01 vs normal group.
Abbreviations: HCC, hepatocellular carcinoma; qRT-PCR, quantitative reverse transcriptase PCR; Con, control; SEM, standard error of mean; TNF, tumor necrosis factor.
SIRT1 promoted M1 macrophage polarization in vitro
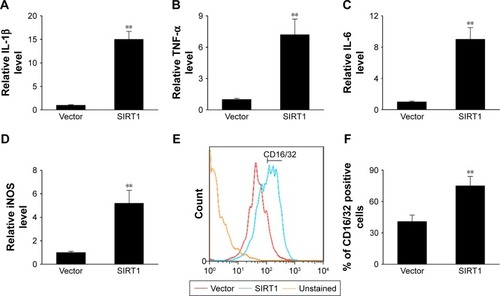
In this study, we transiently induced SIRT1 expression to evaluate its impact on the polarization of M1 macrophages. The expression of M1 markers including IL-1β, inducible nitric oxide synthase (iNOS), IL-6, and TNF-α was noticeably elevated in cells that underwent transfection with SIRT1 in comparison with those that underwent vector transfection after LPS and IFN-γ treatment (). Treatment of LPS+ IFN-γ significantly upregulated M1 markers including CD16/32 as demonstrated via FC. However, the upregulation was more noticeable in cells with SIRT1 transfection compared to those with vector transfection (). The above-mentioned findings suggested that SIRT1 promoted polarization of M1 macrophages.
Figure 2 SIRT1 promoted M1 macrophage polarization in vitro.
Notes: RAW 264.7 macrophages were transfected with SIRT1 or vector and were subsequently activated utilizing 100 ng/mL LPS and 20 ng/mL IFN-γ for 24 hours. The expression of IL-1β (A), TNF-α (B), IL-6 (C), and iNOS (D) were assessed by qPCR in RAW 264.7 macrophages. (E, F) FC was used to evaluate expression of the biomarker CD16/32 associated with M1. Results are displayed as mean ± SEM, **P<0.01 vs vector group.
Abbreviations: LPS, lipopolysaccharide; TNF, tumor necrosis factor; iNOS, inducible nitric oxide synthase; FC, flow cytometry; SEM, standard error of mean.
SIRT1 in macrophage promoted HCC cell death
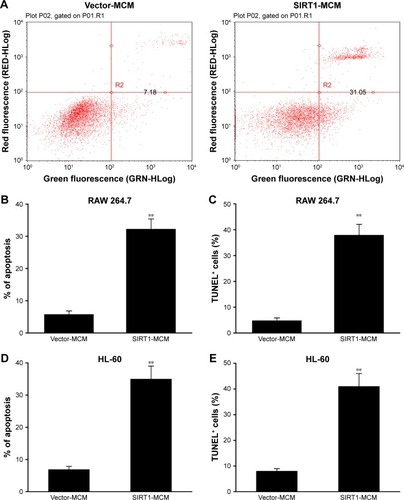
To examine the role of SIRT1 in HCC cell apoptosis, FC with Annexin V/PI staining was carried out. Cells received a supplement of conditioned medium from macrophage transfected with SIRT1 or vector. The rate of apoptosis was remarkably increased by conditioned medium generated from SIRT1-transfected macrophages when compared with that of vector-transfected macrophages (). Cell death was additionally demonstrated via a subsequent TUNEL assay. Cells with positive TUNEL staining were markedly increased in HepG2 cells by conditioned medium generated from SIRT1-transfected macrophages, when compared with that from vector-transfected macrophages (). The similar results were obtained from conditioned medium from human macrophage line HL-60 transfected with SIRT1 or vector (). These findings demonstrated that SIRT1 promotes apoptosis of HCC cells via modulation of polarization of M1 macrophages.
Figure 3 SIRT1 promoted HCC cell death.
Notes: HepG2 cells received a supplement of conditioned medium from RAW 264.7 macrophages (MCM), which were transfected with SIRT1 or vector. (A, B) SIRT1 promoted cell death as demonstrated by FC. (C) SIRT1 promoted cell death as demonstrated by TUNEL assays. HepG2 cells received a supplement of conditioned medium from HL-60 macrophages (MCM), which were transfected with SIRT1 or vector. (D, E) Cell death as demonstrated by FC (D) and TUNEL assays (E). Results are displayed as mean ± SEM. **P<0.01 vs vector-MCM group.
Abbreviations: HCC, hepatocellular carcinoma; FC, flow cytometry; MCM, macrophage-conditioned medium; SEM, standard error of mean.
SIRT1 in macrophage inhibited migration as well as invasion of HCC cells
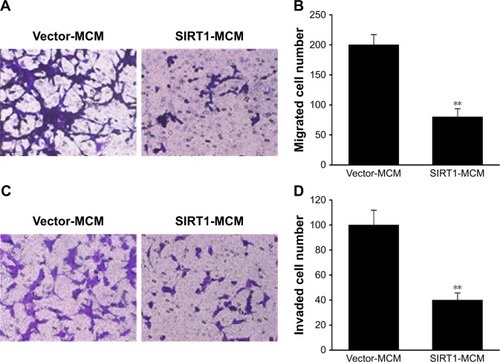
Next, we investigated the contribution of SIRT1 to migration as well as invasion capability of HCC cells. The migration ability of HepG2 cells was tested with a migration assay utilizing transwell chambers without coating. HepG2 received a supplement of conditioned medium from macrophages transfected with SIRT1 or vector. It was revealed via the migration assay that excessive SIRT1 expression significantly decreased the migration of cells (). Invasion capability of HepG2 cells was evaluated using an invasion assay that utilized polycarbonate transwell filters preliminarily coated with Matrigel. The number of invasive HepG2 cells that migrated to the bottom surface of the filter membrane was lower for HepG2 cells after SIRT1 transfection than that after transfection with the vector control (). These results indicated that SIRT1 promoted polarization of M1 macrophages which contributed to inhibitory effect on invasion as well as migration of HepG2 cells.
Figure 4 SIRT1 inhibited migration as well as invasion of HCC cells.
Notes: HepG2 cells received a supplement of conditioned medium from macrophages (MCM), which were transfected with SIRT1 or vector. (A) Images of HepG2 cells with migration ability on the bottom surface of transwell membranes. (B) Number of HepG2 cells with migration ability in five random fields under a microscope from various groups. (C) Images revealing HepG2 cells with invasion ability on the bottom surface of transwell membranes. (D) HepG2 cells with invasion ability in five random fields under a microscope from various groups. Results are displayed as mean ± SEM. **P<0.01 vs vector-MCM group.
Abbreviations: HCC, hepatocellular carcinoma; MCM, macrophage-conditioned medium; SEM, standard error of mean.
SIRT1 enhanced NF-κB pathway stimulation in macrophages
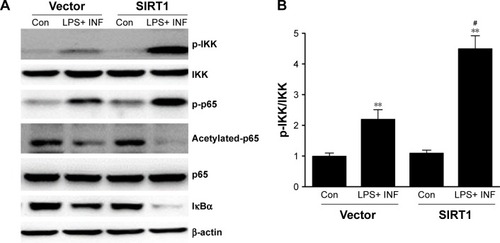
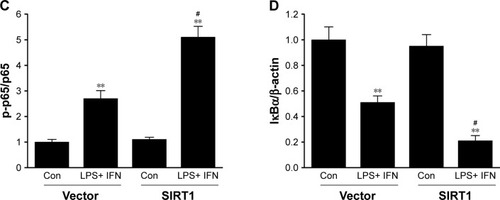
Because NF-κB pathways play an essential role in M1 macrophage polarization,Citation27 our research investigated its impact on NF-κB stimulation. Phosphorylation of p65 as well as IκB kinase (IKK) was noticeably increased in SIRT1-transfected macrophages compared to vector-transfected macrophages after LPS+ IFN-γ treatment (). Furthermore, IκB was remarkably downregulated in SIRT1-transfected macrophages compared to vector-transfected macrophages in reaction to LPS+ IFN-γ activation. The above-mentioned findings demonstrated that SIRT1 promoted macrophage NF-κB stimulation.
Figure 5 SIRT1 reinforced NF-κB pathway stimulation in macrophages.
Notes: RAW 264.7 macrophages were transfected with SIRT1 or vector and were subsequently activated utilizing 100 ng/mL LPS and 20 ng/mL IFN-γ for 24 hours. Representative immunoblots (A) and quantitative assessment of phosphorylation level of IKK (B), p65 (C), and IκBα (D) in RAW 264.7 macrophages. Results are displayed as mean ± SEM. **P<0.01 vs corresponding control; #P<0.05 vs LPS+ INF-γ-treated vector groups.
Abbreviations: LPS, lipopolysaccharide; IFN, interferon; IKK, IκB kinase; SEM, standard error of mean; Con, control.
Suppressive effects of SIRT1 on M1 macrophages and tumors were counteracted via inhibition of NF-κB pathway
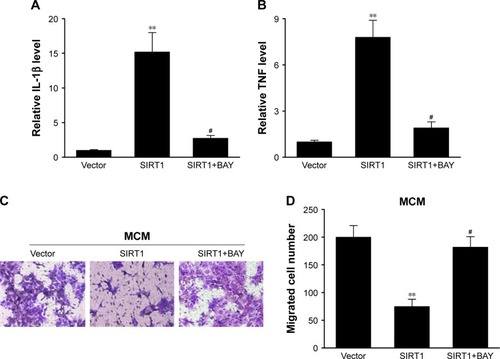
Our research has shown that SIRT1 reinforced polarization of M1 microglia as well as NF-κB stimulation. Bay-11-7082, an inhibitor specific to NF-κB, was supplemented in macrophages to examine whether reinforced NF-κB stimulation assisted polarization of M1 microglia. It was demonstrated via q-PCR that SIRT1-transfected macrophages displayed upregulation of genes associated with M1 compared to vector-transfected macrophages. However, the phenotype was counteracted via Bay-11-7082 (). Furthermore, conditioned medium from SIRT1-transfected macrophages significantly inhibited the migration of HepG2 cells, and this inhibitory effect was also counteracted via Bay-11-7082 (). These data indicated that SIRT1 reinforced M1 macrophage polarization and inhibited HCC cell migration via the NF-κB pathway.
Figure 6 The repressing impact of SIRT1 on M1 macrophages as well as cancer cells was counteracted via inhibition of NF-κB signaling.
Notes: RAW 264.7 macrophages were transfected with SIRT1 or vector and were subsequently activated utilizing 100 ng/mL LPS and 20 ng/mL IFN-γ and BAY for 24 hours. Expression of L-1β (A) and TNF-α (B) was evaluated via qPCR in RAW 264.7 macrophages. RAW 264.7 macrophages were transfected with SIRT1 or vector and were subsequently activated utilizing 100 ng/mL LPS, 20 ng/mL IFN-γ, and BAY for 24 hours. HepG2 cells were then treated with abovementioned conditioned medium from RAW 264.7 macrophages (MCM). (C) Images of HepG2 cells with migration ability on the bottom surface of transwell membranes. (D) Number of HepG2 cells with migration ability in five random fields under a microscope from various groups. Results are displayed as mean ± SEM, **P<0.01 vs vector groups; #P<0.05 vs SIRT1 groups.
Abbreviations: BAY, Bay-11-7082; LPS, lipopolysaccharide; IFN, interferon; MCM, macrophage-conditioned medium; SEM, standard error of mean; TNF, tumor necrosis factor.
Discussion
As a conserved protein deacetylase relying on NAD+, SIRT1 contributes to various cellular reactions.Citation28–Citation30 SIRT1 is able to deacetylate histones as well as emerging non-histone substrates that contribute to various signaling pathways.Citation31–Citation33 A growing number of studies have suggested that SIRT1 serves as an essential modulator of DNA injury, inflammation, survival extension, metabolism stress, and tumor progression.Citation34–Citation36 In terms of inflammation, SIRT1 is able to deacetylate some transcription factors as well as modulate immune cell reactions.Citation37 As for cancer, studies have reported a seemingly contradicting impact of SIRT1 under various circumstances, ranging from an oncoprotein to a tumor suppressor.Citation38 SIRT1 has an impact on not only infiltrating immunocytes but also cancer cells in the tumor microenvironment. Shedding light upon the activities of SIRT1 associated with cancer in both systems could offer possible avenues of treatment targeting SIRT1. In particular, Al-Bahrani et al found that an expression of SIRT1 was found in eleven of 16 (68.75%) HCC and in five of 14 (35.71%) cholangiocarcinoma cases. Western blot analysis showed the expression of SIRT1 in all cell lines studied, although a stronger signal was seen in the HCC cell line. Our research demonstrated that SIRT1 upregulation was correlated with an elevation in the density of M1-like macrophages in HCC tissues. SIRT1 reinforced polarization of M1 macrophages and inhibited metastasis of HCC.
TAMs are able to convey oncogenic signals in chronic inflammatory reactions and eliminate precancerous senescent liver cells in order to avoid HCC.Citation39 Selectively steering or reeducating macrophages to anticancer phenotypes could serve as a promising treatment strategy.Citation40 Furthermore, programmed cell death protein 1 (PD-1) and programmed cell death ligand 1 (PD-L1) could be an innovative regulating “checkpoint” of macrophages, if the macrophages display PD-1 expression in the tumor microenvironment.Citation41 As C-C chemokine ligand 2 and colony-stimulating factor 1 have an essential impact on recruitment of macrophages to tumor tissues, targeting these ligands and/or corresponding receptors in order to eliminate the cancer-promoting capability of macrophages has attracted significant attention.Citation42 These treatment methods allow for improved outcomes in multiple preclinical models, but adding additional targets could have a complementary effect. Targeting liver macrophages could reduce HCC in the background of chronic hepatic illness or promote novel treatments in diagnosed HCC. Despite these promising findings, any HCC treatment that targets macrophages should take into consideration the versatile activity of liver macrophages in fibrosis, chronic inflammatory reactions, and tumor progression.
IKKβ inhibition has been shown to reinforce tumor-repressing polarization of macrophages, while sustained IKKβ stimulation propels macrophages in a malignancy-enhancing direction.Citation43 Nevertheless, in established fibrosarcomas, inhibition of NF-κB stimulation assists in steering M2-like TAMs in a cancer-promoting direction. Additionally, NF-κB restimulation is able to transform an M2 phenotype to M1.Citation44,Citation45 Above-mentioned findings indicate that NF-κB stimulation of macrophages could have an essential impact on various cancers. It has been shown in our research that reinforced NF-κB stimulation in macrophages with excessive SIRT1 expression propels TAM polarization to a tumor-repressing phenotype. IκBα phosphorylation inhibition eliminates the tumor-repressing activity of macrophages with excessive SIRT1 expression in vitro.
Conclusion
This study has shown that M1 macrophages have an essential impact on HCC, and SIRT enhances this effect by regulating polarization of macrophages to a tumor-repressing phenotype via reinforcing NF-κB stimulation. These findings suggest that macrophage SIRT1 could serve as an innovative target to treat HCC.
Ethics approval and informed consent
Written informed consent was obtained from all participants prior to the study. For any participant under 18 years of age, a parent or guardian provided written informed consent. This research was approved by the School of Basic Medical Sciences, Xinxiang Medical University First Affiliated Hospital of Xinxiang Medical College, and conducted in accordance with the Declaration of Helsinki.
Acknowledgments
This work was supported by the Xinxiang Medical University (grant number 505120).
Disclosure
The authors report no conflicts of interest in this work.
References
- ChangjunLFeizhouHDezhenPZhaoLXianhaiMMiR-545-3p/MT1M axis regulates cell proliferation, invasion and migration in hepatocellular carcinomaBiomed Pharmacother201810834735430227328
- YueDSunXIdelalisib promotes Bim-dependent apoptosis through AKT/FOXO3a in hepatocellular carcinomaCell Death Dis201891093530224718
- CaoJDongYMaoFWangWDynamic three-dimensional contrast-enhanced ultrasound to predict therapeutic response of radiofrequency ablation in hepatocellular carcinoma: preliminary findingsBioMed Res Int20182018518
- QiuS-QWaaijerSJHZwagerMCde VriesEGEvan der VegtBSchröderCPTumor-associated Macrophages in breast cancer: innocent bystander or important player?Cancer Treat Rev20187017818930227299
- LoyherPLHamonPLavironMCombadiere C and Boissonnas A macrophages of distinct origins contribute to tumor development in the lungJ Exp Med2018215102536255330201786
- CarasITucureanuCLerescuLInfluence of tumor cell culture supernatants on macrophage functional polarization: in vitro models of macrophage-tumor environment interactionTumori201197564765422158498
- AlsaabHOSauSAlzhraniRMTumor hypoxia directed multimodal nanotherapy for overcoming drug resistance in renal cell carcinoma and reprogramming macrophagesBiomaterials201818328029430179778
- LacalPMGrazianiGTherapeutic implication of vascular endothelial growth factor receptor-1 (VEGFR-1) targeting in cancer cells and tumor microenvironment by competitive and non-competitive inhibitorsPharmacol Res20181369710730170190
- RoySBagAKDuttaSMacrophage-derived neuropilin-2 exhibits novel tumor-promoting functionsCancer Res201878195600561730111533
- ChenPZhouJLiJZhangQZuoQTIPE1 suppresses osteosarcoma tumor growth by regulating macrophage infiltrationClin Transl Oncol201921333434130062520
- CookKLSoto-PantojaDR“UPRegulation” of CD47 by the endoplasmic reticulum stress pathway controls anti-tumor immune responsesBiomark Res2017512628815041
- ZhangXWangSNanYInhibition of autophagy potentiated the anti-tumor effects of VEGF and CD47 bispecific therapy in glioblastomaAppl Microbiol Biotechnol Epub2018512
- SakakuraKTakahashiHKairaKRelationship between tumor-associated macrophage subsets and CD47 expression in squamous cell carcinoma of the head and neck in the tumor microenvironmentLab Invest2016969994100327322955
- WeiWHu-JieCAssociation of the infiltration of tumor-associated macrophages, expression of Smad7 protein and prognosis in oral squamous cell carcinomaArch Oral Biol201895222930036733
- PilchZToneckaKBraniewskaAAntitumor activity of TLR7 is potentiated by CD200R antibody leading to changes in the tumor microenvironmentCancer Immunol Res20186893094030021725
- ZinovkinDAPranjolMZIBilskyIAZmushkoVATumor-associated T-lymphocytes and macrophages are decreased in endometrioid endometrial carcinoma with MELF-Pattern stromal changesCancer Microenviron2018112–310711430008066
- LiuQTongDLiuGMetformin inhibits prostate cancer progression by targeting tumor-associated inflammatory infiltrationClin Cancer Res201824225622563430012567
- TsukamotoHFujiedaKMiyashitaACombined blockade of IL6 and PD-1/PD-L1 signaling abrogates mutual regulation of their immunosuppressive effects in the tumor microenvironmentCancer Res201878175011502229967259
- Salazar-GonzálezRATuriján-EspinozaEHeinDWMilán-SegoviaRCUresti-RiveraEEPortales-PérezDPExpression and genotype-dependent catalytic activity of N-acetyltransferase 2 (NAT2) in human peripheral blood mononuclear cells and its modulation by sirtuin 1Biochem Pharmacol201815634034730149019
- ZhangXLuYWuQAstaxanthin mitigates subarachnoid hemorrhage injury primarily by increasing sirtuin 1 and inhibiting the Toll-like receptor 4 signaling pathwayFASEB J201933172273730048156
- GuoYXingLQianCDingZJinBInvolvement of Flavonoids from the Leaves of Carya cathayensis Sarg in Sirtuin 1 Expression in HUVEC SenescenceEvid Based Complementary Alternat Med2018201818
- HafezYMEl-DeebOSAtefMMThe emerging role of the epigenetic enzyme sirtuin-1 and high mobility group box 1 in patients with diabetic foot ulcerationDiabetes Metab Syndr20181261065107030168428
- WangYZhangLCheXLiWLiuZJiangJRoles of SIRT1/FoxO1/SREBP-1 in the development of progestin resistance in endometrial cancerArch Gynecol Obstet Epub2018911
- RifaïKJudesGIdrissouMSIRT1-dependent epigenetic regulation of H3 and H4 histone acetylation in human breast cancerOncotarget2018955306613067830093977
- CeballosMPDecándidoGQuirogaADInhibition of sirtuins 1 and 2 impairs cell survival and migration and modulates the expression of P-glycoprotein and MRP3 in hepatocellular carcinoma cell linesToxicol Lett2018289637429545174
- ZhouJZhouWKongFXiaoXKuangHZhuYmicroRNA-34a overexpression inhibits cell migration and invasion via regulating SIRT1 in hepatocellular carcinomaOncol Lett20171466950695429344126
- GaoX-RGeJLiW-YZhouW-CXuLGengD-QNF-κB/let-7f-5p/IL-10 pathway involves in wear particle-induced osteolysis by inducing M1 macrophage polarizationCell Cycle201817172134214530176162
- ShiXWeiWWangNTremella polysaccharides inhibit cellular apoptosis and autophagy induced by Pseudomonas aeruginosa lipopoly-saccharide in A549 cells through sirtuin 1 activationOncol Lett20181569609961629805682
- YouZLiuYLiuXNicotinamide N-methyltransferase enhances the progression of prostate cancer by stabilizing sirtuin 1Oncol Lett20181569195920129805651
- Bugyei-TwumAFordCCivitareseRSirtuin 1 activation attenuates cardiac fibrosis in a rodent pressure overload model by modifying Smad2/3 transactivationCardiovasc Res2018114121629164129800064
- ChunSKLeeSFlores-ToroJLoss of sirtuin 1 and mitofusin 2 contributes to enhanced ischemia/reperfusion injury in aged liversAging Cell2018174e12761
- KangJYKimJYKimKBKDM2B is a histone H3K79 demethylase and induces transcriptional repression via sirtuin-1-mediated chromatin silencingFASEB J201832105737575029763382
- YamaçAHKılıçÜEffect of statins on sirtuin 1 and endothelial nitric oxide synthase expression in young patients with a history of premature myocardial infarctionTurk Kardiyol Dern Ars201846320521529664427
- XiaBLiYLiREffect of sirtuin-1 on synaptic plasticity in nucleus accumbens in a rat model of heroin addictionMed Sci Monit2018243789380329870523
- KaurIRawalPRohillaSEndothelial progenitor cells from aged subjects display decreased expression of sirtuin 1, angiogenic functions, and increased senescenceCell Biol Int Epub2018531
- ShiJ-XHuangQGlucagon-like peptide-1 protects mouse podocytes against high glucose-induced apoptosis, and suppresses reactive oxygen species production and proinflammatory cytokine secretion, through sirtuin 1 activation in vitroMol Med Rep20181821789179729845208
- LiKLvGPanLSIRT1 alleviates LPS induced inflammation of periodontal ligament fibroblasts via downregulation of TLR4Int J Biol Macromol201811924925430026098
- HuLYHouYBYuLHMiYHZhangJWWangKExpression of SIRT1 gene in human lung cancer lines enhances their sensitivity to the anticancer effects of cisplatinEur Rev Med Pharmacol Sci201822144551455630058691
- LiuC-QXuJZhouZ-GExpression patterns of programmed death ligand 1 correlate with different microenvironments and patient prognosis in hepatocellular carcinomaBr J Cancer20181191808829921949
- XiaoPLongXZhangLNeurotensin/IL-8 pathway orchestrates local inflammatory response and tumor invasion by inducing M2 polarization of tumor-associated macrophages and epithelial-mesenchymal transition of hepatocellular carcinoma cellsOncoImmunology201877e144016629900041
- GordonSRMauteRLDulkenBWPD-1 expression by tumour-associated macrophages inhibits phagocytosis and tumour immunityNature2017545765549549928514441
- LiXYaoWYuanYTargeting of tumour-infiltrating macrophages via CCL2/CCR2 signalling as a therapeutic strategy against hepatocellular carcinomaGut201766115716726452628
- FongCHYBebienMDidierlaurentAAn antiinflammatory role for IKKβ through the inhibition of “classical” macrophage activationJ Exp Med200820561269127618490491
- SaccaniASchioppaTPortaCP50 nuclear factor-kappaB overexpression in tumor-associated macrophages inhibits M1 inflammatory responses and antitumor resistanceCancer Res20066623114321144017145890
- HagemannTBiswasSKLawrenceTSicaALewisCERegulation of macrophage function in tumors: the multifaceted role of NF-kappaBBlood2009113143139314619171876
