Abstract
Purpose
Non-small cell lung cancer (NSCLC) is the first leading cause of cancer-related death globally. Long noncoding RNA KCNQ1 overlapping transcript 1 (KCNQ1OT1) was involved in the progression of multiple cancers by sponging target miRNA. We aimed to explore the pathological mechanism of KCNQ1OT1 in NSCLC progression.
Methods
The expression of KCNQ1OT1, miR-204-5p and autophagy-related gene 3 (ATG3) was measured by quantitative real-time polymerase chain reaction (qRT-PCR). 3-(4, 5-Dimethyl-2-thiazolyl)-2, 5-diphenyl-2-H-tetrazolium bromide (MTT) assay and flow cytometry assay were conducted for the detection of cell proliferation and apoptosis, respectively. Western blot assay was performed to examine the protein levels of B-cell lymphoma-2 (BCL-2), BCL2-Associated X (Bax), cleaved caspase-3, cleaved caspase-9 and LC3 II /LC3 I and P62. The interaction between miR-204-5p and KCNQ1OT1 or ATG3 was validated by dual-luciferase reporter system and RNA immunoprecipitation (RIP) assay. Murine xenograft assay was conducted to explore the function of KCNQ1OT1 in vivo. Immunohistochemistry (IHC) staining assay was used for the analysis of ki67-positive cell percentage.
Results
The expression of KCNQ1OT1 and ATG3 was up-regulated whereas miR-204-5p was down-regulated in NSCLC tumors and cells. MiR-204-5p was inversely correlated with KCNQ1OT1 or ATG3. In addition, KCNQ1OT1 knockdown facilitated apoptosis, inhibited autophagy and proliferation of NSCLC cells in vitro and blocked tumor growth in vivo. However, the miR-204-5p inhibitor reversed the effects. More importantly, ATG3 was a target gene of miR-204-5p and ATG3 overexpression restored the effect of miR-204-5p on NSCLC cell progression.
Conclusion
KCNQ1OT1 promotes cell proliferation and autophagy and inhibits cell apoptosis via regulating miR-204-5p/ATG3 axis, providing a promising target for NSCLC therapy.
Keywords:
Introduction
Non-small cell lung cancer (NSCLC) which accounts for more than 80% of lung cancer is the first leading cause of cancer-related death globally.Citation1,Citation2 Despite advanced treatment strategies, including surgical resection, chemotherapy, radiotherapy, immunotherapy and combined therapy, have palliated the symptoms, the 5-year overall survival rate is still less than 20% in patients with NSCLC.Citation3–Citation5 In addition, chemo-resistance and high metastasis of NSCLC vitiated the therapy outcomes.Citation6 Therefore, it is urgently needed to elucidate the physiological and pathological mechanism of NSCLC.
Long non-coding RNAs (lncRNAs) are fundamental modulators of cell cycle, proliferation, migration, differentiation, invasion, metastasis, apoptosis, inflammation and autophagy in multiple cancers.Citation7–Citation9 LncRNA KCNQ1 overlapping transcript 1 (KCNQ1OT1), mapped in human KCNQ1 locus with a length of 91 kb, plays essential roles in tumor proliferation, metabolism, epithelial–mesenchymal transition (EMT) and growth via functioning as competing endogenous RNA (ceRNA).Citation10,Citation11 KCNQ1OT1 mainly acts as an oncogene in different cancers. For instance, KCNQ1OT1 facilitated proliferation, migration and invasion of cholangiocarcinoma cells by interacting with miR-140-5p.Citation12 Besides, KCNQ1OT1 was reported to improve chemoresistance of oxaliplatin against hepatocellular carcinoma by up-regulating ABCC1 via sponging miR-7-5p.Citation13 Though a previous study has demonstrated that KCNQ1OT1 could be an oncogene in NSCLC,Citation14 the molecular mechanism of KCNQ1OT1 in NSCLC remains obscure.
MicroRNAs (miRNAs) are evolutionarily conserved small non-coding RNAs with 19–23 nucleotides in length.Citation15,Citation16 Despite without protein coding capacity, miRNAs are widely involved in the regulation of gene expression at the post-transcriptional level by complementarily binding with messenger RNAs (mRNAs).Citation15–Citation19 Ectopic expression of miRNA is closely associated with tumorigenesis and development.Citation20 As a tumor suppressor, miR-204-5p is a pivotal regulator in multiple cancers. For example, miR-204-5p suppressed cell invasion and metastasis by targeting forkhead box C1 in laryngeal squamous cell carcinoma.Citation21 Similarly, miR-204-5p attenuated proliferation, migration, invasion and metastasis of hepatocellular carcinoma and oral squamous cell carcinoma though repressing CXCR4 and SIRT1 expression, respectively.Citation22,Citation2Citation3, Autophagy-related gene 3 (ATG3) could be targeted by several miRNAs to participate in the regulation of NSCLCCitation24,Citation25 However, whether miR-204-5p can target ATG3 in NSCLC requires deep investigation.
In the present study, we attempted to investigate the underlying mechanism of the KCNQ1OT1/miR-204-5p/ATG3 axis in NSCLC cell progression. All the results indicated that KCNQ1OT1 promotes proliferation and autophagy while suppresses apoptosis by regulating the miR-204-5p/ATG3 axis. This study might provide promising biomarkers for the diagnosis and treatment of NSCLC.
Materials and Methods
Tissue Samples
Fresh NSCLC tumor tissues and the corresponding normal tissues were obtained from 35 NSCLC patients recruited from The First Affiliated Hospital of Zhengzhou University. The patients had not received preoperative treatment. All NSCLC patients have signed written informed consents and the experiment protocols were approved by the Ethics Committee of The First Affiliated Hospital of Zhengzhou University.
Cell Transfection
NSCLC cell lines HCC827, H1299, A549, H460 and human bronchial epithelial cell BEAS-2B were purchased from the American Type Culture Collection (ATCC, Manassas, VA, USA). All the cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM; Gibco, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum (FBS; Gibco) and 0.05% penicillin/streptomycin (Gibco). Small hairpin RNA (shRNA) targeting KCNQ1OT1 (sh-KCNQ1OT1), shRNA negative control (sh-NC), pcDNA, and pcDNA-ATG3 overexpression vector (ATG3) were synthesized by Genepharma (Shanghai, China). The miRNA mimics (miR-204-5p), miR-204-5p inhibitor (anti-miR-204-5p), miRNA negative control (miR-NC) and miRNA negative control inhibitor (anti-miR-NC) were purchased from RIBOBIO (Guangzhou, China). The synthetic oligonucleotides or plasmids were transfected in A549 and H460 cells using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA).
Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
NSCLC tissues and cells were incubated with TRIzol reagent (Invitrogen) to extract total RNA. The cDNA for KCNQ1OT1, miR-204-5p and ATG3 was synthesized by All-in-One™ First-Strand cDNA Synthesis Kit (FulenGen, Guangzhou, China). Then, qRT-PCR was performed using SYBR green (Applied Biosystems, Foster City, CA, USA) following the standard procedures. The primers for KCNQ1OT1, miR-204-5p, ATG3, GAPDH and U6 were listed as follows: KCNQ1OT1 (Forward, 5ʹ-TTGGTAGGATTTTGTTGAGG-3ʹ; Reverse, 5ʹ-CAACCTTCCCCTACTACC-3ʹ); miR-204-5p (Forward, 5ʹ-CGAAGTTCCCTTTGTCATCCT-3ʹ; Reverse, 5ʹ-GTGCAGGGTCCGAGGTATTC-3ʹ); ATG3 (Forward, 5ʹ-GCAAACAAGAACCTATGACCTG-3ʹ; Reverse, 5ʹ-GTCTTCATACATGTGCTCAACTG-3ʹ); GAPDH, (Forward, 5ʹ-AGGTCGGTGTGAACGGATTTG-3ʹ; Reverse, 5ʹ-GGGGTCGTTGATGGCAACA-3ʹ); U6, (Forward, 5ʹ-ACCCTGAGAAATACCCTCACAT-3ʹ; Reverse, 5ʹ-GACGACTGAGCCCCTGATG-3ʹ).
3-(4, 5-Dimethyl-2-Thiazolyl)-2, 5-Diphenyl-2-H-Tetrazolium Bromide (MTT) Assay and Flow Cytometry Assay
Cell viability and apoptosis were evaluated by MTT assay and flow cytometry assay, respectively. For MTT assay, transfected A549 and H460 cells (5000 cells/well) were seeded on 96-well plates for 24 h, 48 h, 72 h and 96 h. Then, the cells were reacted with 10 μL MTT reagent (Beyotime, Shanghai, China) and kept for an additional 4 h. The OD value at 490 nm was measured by a spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA). For flow cytometry assay, transfected A549 and H460 cells were collected, stained using Annexin V-FITC/PI Apoptosis Detection Kit (Vazyme, Nanjing, China) and analyzed by BD FACS Canto II flow cytometry (BD Biosciences, Franklin Lakes, NJ, USA).
Dual-Luciferase Reporter Assay
The sequences of KCNQ1OT1 and 3ʹ-UTR of ATG3 including the wild-type or mutant binding sequences of miR-204-5p were cloned and introduced into pmiR-REPORTTM vectors (RiboBio, Guangzhou, China) to generate luciferase reporter vectors WT-KCNQ1OT1, MUT-KCNQ1OT1, ATG3 3ʹUTR-WT and ATG3 3ʹUTR-MUT, respectively. Then, miR-204-5p or miR-NC was transfected into A549 and H460 cells together with indicated luciferase reporter vector using Lipofectamine 2000 transfection reagent (Invitrogen). Dual-Luciferase Reporter Assay Kit (Vazyme) was utilized for the detection of luciferase activity after 48 h.
RNA Immunoprecipitation (RIP) Assay
Magna RNA immunoprecipitation kit (Millipore, Bedford, MA, USA) was employed for RIP assay. In brief, A549 and H460 cells transfected with KCNQ1OT1 and negative control (NC) were lysed by RIP buffer. Then, cell lysis was incubated with magnetic beads coated with Anti-Argonaute2 (Anti-Ago2; Abcam, Cambridge, MA, USA) or immunoglobulin G (Anti-IgG; Abcam). The enrichment of miR-204-5p was subjected to qRT-PCR after RNAs in the magnetic beads were purified.
Western Blot Assay
Western blot assay was performed following the standard protocol. In brief, after total protein was isolated and quantified, the proteins were separated by sodium dodecyl sulfonate-polyacrylamide gel (SDS-PAGE; Solarbio, Beijing, China) and then transferred onto polyvinylidene difluoride membranes (PVDF; Pall Corporation, New York, NYC, USA). Next, the membranes were blocked in slim milk for 2 h and then incubated with primary antibodies against ATG3 (ab108251; 1:2000; Abcam, Cambridge, MA, USA), LC3 II /LC3 I (ab128025; 1:200; Abcam), P62 (ab56416; 1:500; Abcam), B-cell lymphoma-2 (BCL-2; ab196495; 1:1000; Abcam), cleaved caspase-3 (ab49822; 1:500; Abcam), cleaved caspase-9 (ab2324; 1:2000; Abcam), BCL2-Associated X (Bax; ab182733; 1:2000; Abcam) or GAPDH (ab181602; 1:5000; Abcam) at 4°C overnight followed by incubation with HRP-conjugated secondary antibody (D110150; 1:5000; Sangon, Shanghai, China) for 2 h. The protein bands were analyzed using an enhanced chemiluminescence reagent (Beyotime).
Murine Xenograft Assay
Male nude mice of 5 weeks old were purchased from Vital River Laboratory Animal Technology (Beijing, China). Our animal experiment protocols were approved by the Ethics Committee of The First Affiliated Hospital of Zhengzhou University and performed according to the NIH guidelines for the care and use of laboratory animals (NIH Publication No. 85–23 Rev.1985). In brief, A549 and H460 cells stably transfected with sh-KCNQ1OT1 and sh-NC were subcutaneously injected in mice to establish xenograft mice model. Tumor volume was measured every 4 d from day 8 and calculated using the formula: (length×width2)/2. Tumor weight was measured after 28 d and tumors were harvested and preserved at −80°C.
Immunohistochemistry (IHC) Staining Assay
The tissue sections were deparaffinized, rehydrated using a graded ethanol series and heated in citrate buffer for antigen retrieval. Then, the sections were washed, blocked and incubated with normal goat serum. Then, the samples were incubated with a primary antibody anti-Ki67 (ab15580; 1:500; Abcam) at 4°C overnight and secondary antibody (ab205718; 1:5000; Abcam). The sections were stained with diaminobenzidine (DAB) and then counterstained with hematoxylin, dehydrated and mounted. At last, the section was photographed using a digital microscope camera (Nikon, Tokyo, Japan).
Statistical Analysis
All the experiments were conducted at least in triplicate and data were presented as means ± standard deviation (SD). Statistical analysis was carried out using SPSS 13.0 software (SPSS, Chicago, IL, USA) and GraphPad Prism 7 (GraphPad Inc., San Diego, CA, USA). The correlation between miR-204-5p and KCNQ1OT1 or ATG3 was analyzed by Pearson’s correlation coefficient analysis. P value less than 0.05 (P<0.05) was considered as statistically significant.
Results
KCNQ1OT1 Depletion Induces Apoptosis and Suppresses Proliferation and Autophagy in NSCLC Cells
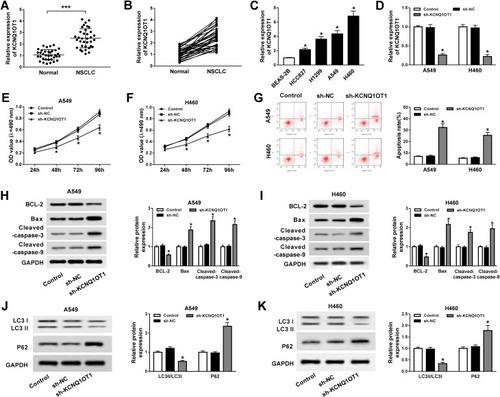
The functions of KCNQ1OT1 on NSCLC cell proliferation, apoptosis and autophagy were assessed by MTT assay, flow cytometry analysis and Western blot assay, respectively. As illustrated in , KCNQ1OT1 expression was extremely higher in NSCLC tumor tissues than that in the corresponding normal tissues. Likewise, KCNQ1OT1 expression was up-regulated in NSCLC cell lines (HCC827, H1299, A549, H460) compared with human bronchial epithelial cell BEAS-2B (). Furthermore, loss-of-function experiments were employed by knocking down KCNQ1OT1 to explore the regulatory effects of KCNQ1OT1 on NSCLC cell progression. An obvious reduction of KCNQ1OT1 expression was noticed in A549 and H460 cells stably transfected with sh-KCNQ1OT1, indicating the transfection efficiency was relatively high (). Moreover, cell growth was inhibited apparently in NSCLC cells after KCNQ1OT1 silencing (). On the contrary, the cell apoptosis rate was enhanced in the sh-KCNQ1OT1 transfected group compared with sh-NC group (). Hence, the expression of apoptosis-related proteins was measured. We found that Bax, cleaved caspase-3 and cleaved caspase-9 were dramatically elevated whereas anti-apoptosis protein BCL-2 was decreased in both A549 and H460 cells stably transfected with sh-KCNQ1OT1 (). We also analyzed the expression of autophagy markers LC3 and P62 and observed that deficiency of KCNQ1OT1 repressed LC3II/LC3I expression and boosted P62 expression (). Collectively, KCNQ1OT1 knockdown induced apoptosis and suppressed proliferation and autophagy in NSCLC cells.
Figure 1 KCNQ1OT1 knockdown repressed proliferation and autophagy and induced apoptosis in NSCLC. (A, B) KCNQ1OT1 expression in 35 pairs of NSCLC tumor tissues and normal tissues. (C) KCNQ1OT1 expression in NSCLC cell lines (HCC827, H1299, A549, H460) and human bronchial epithelial cell BEAS-2B. (D–K) A549 and H460 cells were stably transfected with sh-KCNQ1OT1 or sh-NC. (D) KCNQ1OT1 expression in stably transfected A549 and H460 cells. (E, F) Cell viability of transfected A549 (E) and H460 cells (F). (G) Cell apoptosis of transfected A549 and H460 cells. (H, I) The expression of apoptosis-related protein cleaved caspase-3, cleaved caspase-9, Bax and anti-apoptosis protein BCL-2 in transfected A549 (H) and H460 cells (I). (J, K) Protein expression of autophagy markers LC3 and P62 in transfected A549 (J) and H460 cells (K). *P<0.05, ***P<0.001.
KCNQ1OT1 is a Sponge of miR-204-5p
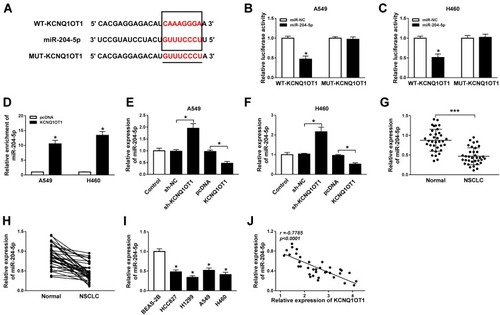
Growing evidence has validated that lncRNA KCNQ1OT1 exerts its function by sponging the target miRNA. As searched by the online prediction tool StarBase v2.0, miR-204-5p includes the binding sites of KCNQ1OT1 (). To confirm the prediction, wild type (WT-KCNQ1OT1) and mutant type (MUT-KCNQ1OT1) vectors were constructed and co-transfected into A549 and H460 cells with miR-204-5p or miR-NC to establish dual-luciferase reporter system. Luciferase activity was reduced evidently in NSCLC cells co-transfected with WT-KCNQ1OT1 and miR-204-5p compared to WT-KCNQ1OT1 and miR-NC co-transfected cells, while luciferase activity in MUT-KCNQ1OT1 transfection group remained unchanged (). Additionally, the RIP assay indicated that the enrichment of miR-204-5p in the Anti-Ago2 RIP group was elevated in NSCLC cells transfected with KCNQ1OT1 (). More importantly, overexpression of KCNQ1OT1 repressed miR-204-5p expression whereas depletion of KCNQ1OT1 facilitated miR-204-5p expression (). Subsequently, we discovered that the expression of miR-204-5p was down-regulated in NSCLC tumor tissues and cell lines compared with normal tissues () and cells (). Pearson’s correlation coefficient analysis exhibited that KCNQ1OT1 was negatively correlated with miR-204-5p (r=−0.7785, P<0.0001) in NSCLC tissues (). These findings demonstrated that KCNQ1OT1 acted as a sponge of miR-204-5p.
Figure 2 KCNQ1OT1 directly interacted with miR-204-5p. (A) The putative binding sites between KCNQ1OT1 and miR-204-5p. (B, C) Luciferase activity of A549 (B) and H460 cells (C) co-transfected with WT-KCNQ1OT1 or MUT-KCNQ1OT1 and miR-204-5p or miR-NC. (D) The enrichment of miR-204-5p in Anti-Ago2 immunoprecipitation complex in A549 and H460 cells transfected with KCNQ1OT1 or pcDNA. (E, F) The expression of miR-204-5p in A549 (E) and H460 cells (F) transfected with KCNQ1OT1, sh-KCNQ1OT1, pcDNA or sh-NC. (G, H) The expression of miR-204-5p in NSCLC tumor tissues and normal tissues. (I) The expression of miR-204-5p in NSCLC cell lines (HCC827, H1299, A549, H460) and human bronchial epithelial cell BEAS-2B. (J) The correlation between expression of KCNQ1OT1 and miR-204-5p (r=−0.7785, P<0.0001). *P<0.05, ***P<0.001.
KCNQ1OT1 Regulates Proliferation, Apoptosis and Autophagy by Binding to miR-204-5p in NSCLC Cells
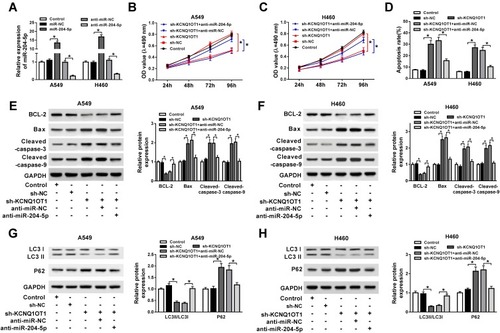
To explore whether KCNQ1OT1 could regulate NSCLC progression by binding to miR-204-5p, A549 and H460 cells were transfected with sh-KCNQ1OT1+anti-miR-204-5p, sh-KCNQ1OT1+anti-miR-NC, sh-KCNQ1OT1 or sh-NC. As exhibited in , all the vectors were transfected into A549 and H460 cells successfully with high transfection efficiency. MTT results revealed that miR-204-5p inhibitor could abrogate KCNQ1OT1 silencing induced inhibition on cell proliferation in NSCLC (). Conversely, the KCNQ1OT1 knockdown promoted the apoptosis of A549 and H460 cells and miR-204-5p inhibitor rescued the effect (). Consistently, KCNQ1OT1 knockdown enhanced the expression of apoptosis-related protein Bax, cleaved caspase-3 and cleaved caspase-9, and reduced the expression of anti-apoptosis protein BCL-2. However, the miR-204-5p inhibitor reversed the trends (). Furthermore, the level of LC3II/LC3I was decreased whereas P62 was increased after KCNQ1OT1 silencing; however, the miR-204-5p inhibitor transfection group overturned the effects (). All the data implicated that KCNQ1OT1 regulated proliferation, apoptosis and autophagy by interacting with miR-204-5p in NSCLC cells.
Figure 3 miR-204-5p inhibitor attenuated the effects of KCNQ1OT1 silencing on cell proliferation, apoptosis and autophagy in NSCLC. A549 and H460 cells were transfected with sh-KCNQ1OT1+anti-miR-204-5p, sh-KCNQ1OT1, sh-KCNQ1OT1+anti-miR-NC or sh-NC. (A) The expression of miR-204-5p in transfected A549 and H460 cells. (B, C) Cell viability of transfected A549 (B) and H460 cells (C). (D) Cell apoptosis of transfected A549 and H460 cells. (E, F) The expression of apoptosis-related protein cleaved caspase-3, cleaved caspase-9, Bax and anti-apoptosis protein BCL-2 in transfected A549 (E) and H460 cells (F). (G, H) Protein expression of autophagy markers LC3 and P62 in transfected A549 (G) and H460 cells (H). *P<0.05.
ATG3 is a Target of miR-204-5p
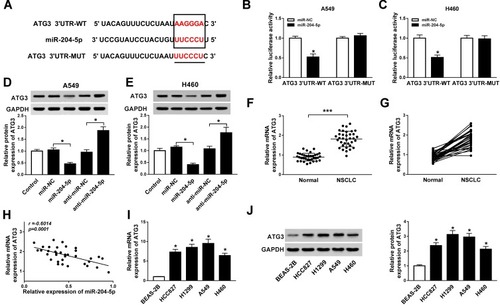
Based on bioinformatics analysis using StarBase v2.0, miR-204-5p comprised the binding sites of 3ʹUTR ATG3 (). Dual-luciferase reporter assay exhibited that luciferase activity was relatively lower in NSCLC cells co-transfected with ATG3 3ʹUTR-WT and miR-204-5p than ATG3 3ʹUTR-WT and miR-NC co-transfected group, confirming the interaction between ATG3 and miR-204-5p (). Besides, ATG3 protein expression was reduced by the introduction of miR-204-5p while enhanced by miR-204-5p inhibitor in A549 and H460 cells (). As expected, ATG3 expression was up-regulated in NSCLC tumor tissues compared with normal tissues (). Then, we found ATG3 expression was negatively correlated with miR-204-5p expression in NSCLC tissues, as illustrated by Person’s correlation coefficient analysis (r=−0.6014, P<0.0001) (). Similarly, the mRNA and protein levels of ATG3 were considerably higher in NSCLC cell lines (HCC827, H1299, A549, H460) than those in BEAS-2B cells (). Altogether, ATG3 was a target of miR-204-5p.
Figure 4 ATG3 is a target of miR-204-5p. (A) The putative binding sites between ATG3 and miR-204-5p. (B, C) Luciferase activity of A549 (B) and H460 cells (C) co-transfected with ATG3 3ʹUTR-WT or ATG3 3ʹUTR-MUT and miR-204-5p or miR-NC. (D, E) Protein expression of ATG3 in A549 (D) and H460 cells (E) transfected with miR-204-5p, anti-miR-204-5p, anti-miR-NC or miR-NC. (F, G) The expression of ATG3 in NSCLC tumor tissues and normal tissues. (H) The correlation between ATG3 and miR-204-5p (r=−0.6014, P<0.0001). (I, J) The expression of ATG3 mRNA (I) and protein (J) in NSCLC cell lines (HCC827, H1299, A549, H460) and human bronchial epithelial cells BEAS-2B. *P<0.05, ***P<0.001.
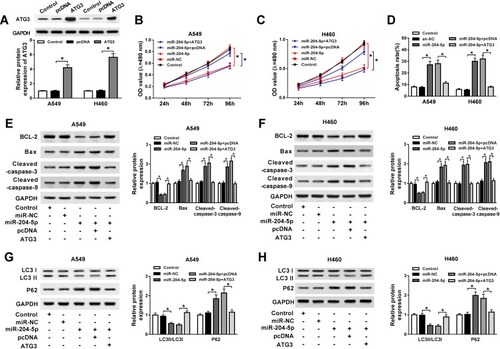
ATG3 Overexpression Reverses miR-204-5p Mediated Regulatory Effects on Proliferation, Apoptosis and Autophagy in NSCLC Cells
Following confirmation that ATG3 is a target of miR-204-5p, A549 and H460 cells were transfected with miR-204-5p+ATG3, miR-204-5p+pcDNA, miR-204-5p or miR-NC to illuminate the effects of miR-204-5p/ATG3 axis on NSCLC cell development. As presented in , the expression of ATG3 protein was raised in NSCLC cells transfected with ATG3 compared with the pcDNA group. Moreover, ATG3 restored the suppressive effect of miR-204-5p on the proliferation of A549 and H460 cells ( and C). Likewise, ATG3 elevation reversed miR-204-5p induced promotional effect on NSCLC cell apoptosis (). As expected, the expression levels of apoptosis-related protein Bax, cleaved caspase-3 and cleaved caspase-9 were enhanced by miR-204-5p and repressed by the overexpression of ATG3. However, the anti-apoptosis protein BCL-2 expression showed the opposite results (). In addition, the ratio of LC3II/LC3I was reduced while P62 protein expression was increased by miR-204-5p; however, the effects were reversed by ATG3 (). The results clarified that miR-204-5p could regulate NSCLC cell proliferation, apoptosis and autophagy through ATG3.
Figure 5 ATG3 abrogated miR-204-5p mediated acceleration on cell apoptosis and suppression on cell proliferation and autophagy in NSCLC. A549 and H460 cells were transfected with miR-204-5p+ATG3, miR-204-5p+pcDNA, miR-204-5p or miR-NC. (A) The expression of ATG3 protein in transfected A549 and H460 cells. (B, C) Cell viability of transfected A549 (B) and H460 cells (C). (D) Cell apoptosis of transfected A549 and H460 cells. (E, F) The expression of apoptosis-related protein caspase-3, caspase-9, Bax and anti-apoptosis protein BCL-2 in transfected A549 (E) and H460 cells (F). (G, H) Protein expression of autophagy markers LC3 and P62 in transfected A549 (G) and H460 cells (H). *P<0.05.
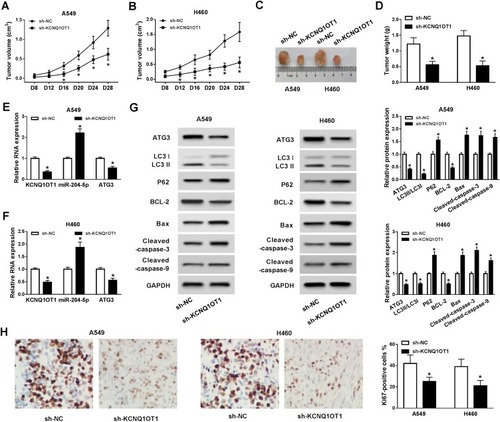
Silencing of KCNQ1OT1 Inhibits Tumor Growth in vivo
Subsequently, xenograft model was established by subcutaneously injecting sh-KCNQ1OT1 and sh-NC stably transfected A549 and H460 cells into the nude mice. We found that tumor growth was significantly repressed in the xenograft model stably transfected with sh-KCNQ1OT1 compared with sh-NC group (). Similarly, tumor weight was lower in sh-KCNQ1OT1 transfected mice than sh-NC group (). Besides, the expression of KCNQ1OT1 and ATG3 was down-regulated whereas miR-204-5p was up-regulated in the tissue samples collected from sh-KCNQ1OT1 transfected group (). More importantly, the levels of ATG3, LC3II/LC3I, BCL-2 were suppressed whereas P62, Bax, cleaved caspase-3, cleaved caspase-9 were promoted in sh-KCNQ1OT1 group (). IHC staining assay indicated that KCNQ1OT1 knockdown led to a significant decrease in the percentage of ki67-positive cells compared to sh-NC group (). Taken together, KCNQ1OT1 silencing suppressed tumor growth in vivo by accelerating apoptosis and suppressing autophagy in NSCLC.
Figure 6 KCNQ1OT1 silencing suppressed tumor growth in vivo. Sh-NC or sh-KCNQ1OT1 transfected A549 cells or H460 cells were injected into the nude mice. (A, B) Tumor volume of xenograft mice was measured every 4 d from day 8. (C, D) Tumor weight was measured when xenograft mice were sacrificed after 28 days. (E, F) The expression of KCNQ1OT1, miR-204-5p, ATG3 in xenograft mice tumor tissues. (G) Protein expression of ATG3, LC3, P62, BCL-2, cleaved caspase-3, cleaved caspase-9 and Bax in xenograft mice tumor tissues. (H) The percentage of KI67-positive cells in xenograft mice tumor tissues. *P<0.05.
Discussion
LncRNAs have been demonstrated to be associated with the progression of diverse human cancers, including NSCLC. In this study, we aimed to explore the function of lncRNA KCNQ1OT1 in NSCLC development. Previous studies have identified that lncRNA KCNQ1OT1 is an essential prognosis biomarker in a variety of diseases, including diabetic retinopathy, osteolysis and breast cancer,Citation26–Citation28 while the function and the molecular mechanism of KCNQ1OT1 in NSCLC progression were not fully validated. KCNQ1OT1 was up-regulated in breast cancer and overexpression of KCNQ1OT1 accelerated cell growth in vitro and repressed tumor growth in vivo by enhancing CCNE2 via sponging miR-145.Citation29 Upregulation of KCNQ1OT1 stimulated NSCLC cell progression by inhibiting miRNA-27b-3p and boosting HSP90AA1 expression.Citation30 Additionally, KCNQ1OT1 promoted cell development as well as cisplatin resistance against tongue cancer by targeting miR-211-5p to alter Ezrin/Fak/Src signaling pathway.Citation31 Consistently, KCNQ1OT1 enhanced oxaliplatin resistance by modulating the miR-34a/ATG4B axis in colon cancer.Citation32 Therefore, we guessed that KCNQ1OT1 exerted its functions by sponging the specific miRNA in NSCLC.
MiR-204-5p was identified to be a target of KCNQ1OT1 in this study. Interestingly, miR-204-5p was found to be an autophagy-associated gene in several tumor types, such as rhabdomyosarcoma, endometrial and colorectal cancer.Citation33–Citation35 In addition, Luan et al reported that miR-204-5p improved cell sensitivity to chemotherapy and inhibited melanoma growth by targeting MMP9 and B-cell lymphoma-2.Citation36 Similarly, miR-204-5p enhanced chemotherapeutic sensitivity and repressed invasion of colorectal cancer cells by interacting with RAB22A.Citation37 Moreover, miR-204-5p was clarified to weaken cell proliferation and induce cell apoptosis in papillary thyroid carcinoma as well as expedite apoptosis and repress cell metastasis in osteosarcoma by targeting IGFBP5 and EBF2, respectively.Citation38,Citation39 Therefore, we speculated that miR-204-5p functioned as a tumor suppressor to suppress proliferation, migration, invasion and autophagy as well as enhance apoptosis of NSCLC cells.
Wang et al declared that miR-16 could target the 3ʹ UTR of ATG to hamper TGF-β1-induced epithelial-to-mesenchymal transition (EMT) via the activation of autophagy in NSCLC.Citation24 Hua et al reported that miR-1/ATG3 participated in the regulation of chemoresistance in NSCLC.Citation25 Thus, we speculated that KCNQ1OT1 modulated proliferation, apoptosis and autophagy of NSCLC cells via modulating miR-204-5p/ATG3 axis. Initially, we noticed that the expression of KCNQ1OT1 and ATG3 was up-regulated whereas miR-204-5p was down-regulated in NSCLC tumors and cells compared with the corresponding normal tissues and cells. As expected, KCNQ1OT1 knockdown expedited apoptosis and repressed proliferation and autophagy of NSCLC cells in vitro and in vivo. However, the miR-204-5p inhibitor reversed the effects. The rescue experiments also displayed that ATG3 elevation abolished miR-204-5p mediated regulatory effects on cell proliferation, apoptosis and autophagy in NSCLC.
In conclusion, we clarified that KCNQ1OT1 contributes to cell proliferation and autophagy and suppresses cell apoptosis by altering the miR-204-5p/ATG3 axis in NSCLC. Our study represented potential biomarkers for targeted therapy of NSCLC.
Disclosure
The authors report no conflicts of interest in this work.
References
- Ai X, Mao F, Shen S, et al. Bexarotene inhibits the viability of non-small cell lung cancer cells via slc10a2/PPARgamma/PTEN/mTOR signaling pathway. BMC Cancer. 2018;18:407. doi:10.1186/s12885-018-4224-x29642873
- You J, Cheng J, Yu B, et al. Baicalin, a Chinese herbal medicine, inhibits the proliferation and migration of human non-small cell lung carcinoma (NSCLC) cells, A549 and H1299, by activating the SIRT1/AMPK signaling pathway. Med Sci Monit. 2018;24:2126–2133. doi:10.12659/MSM.90962729632297
- Xie Y, Lv Y, Zhang Y, et al. LATS2 promotes apoptosis in non-small cell lung cancer A549 cells via triggering Mff-dependent mitochondrial fission and activating the JNK signaling pathway. Biomed Pharmacother. 2019;109:679–689. doi:10.1016/j.biopha.2018.10.09730551520
- Li Q, Yang Z, Chen M, et al. Downregulation of microRNA-196a enhances the sensitivity of non-small cell lung cancer cells to cisplatin treatment. Int J Mol Med. 2016;37:1067–1074. doi:10.3892/ijmm.2016.251326936095
- Tang Q, Li M, Chen L, et al. miR-200b/c targets the expression of RhoE and inhibits the proliferation and invasion of non-small cell lung cancer cells. Int J Oncol. 2018;53:1732–1742.30066855
- Moravcikova E, Krepela E, Donnenberg VS, et al. BOK displays cell death-independent tumor suppressor activity in non-small-cell lung carcinoma. Int J Cancer. 2017;141:2050–2061. doi:10.1002/ijc.3090628744854
- Cui M, Chen M, Shen Z, et al. LncRNA-UCA1 modulates progression of colon cancer through regulating the miR-28-5p/HOXB3 axis. J Cell Biochem. 2019.
- Li C, Lv Y, Shao C, et al. Tumor-derived exosomal lncRNA GAS5 as a biomarker for early-stage non-small-cell lung cancer diagnosis. J Cell Physiol. 2019.
- Li N, Zhan X, Zhan X. The lncRNA SNHG3 regulates energy metabolism of ovarian cancer by an analysis of mitochondrial proteomes. Gynecol Oncol. 2018;150:343–354. doi:10.1016/j.ygyno.2018.06.01329921511
- Wang CG, Liao Z, Xiao H, et al. LncRNA KCNQ1OT1 promoted BMP2 expression to regulate osteogenic differentiation by sponging miRNA-214. Exp Mol Pathol. 2019;107:77–84. doi:10.1016/j.yexmp.2019.01.01230703347
- Bian Y, Gao G, Zhang Q, et al. KCNQ1OT1/miR-217/ZEB1 feedback loop facilitates cell migration and epithelial-mesenchymal transition in colorectal cancer. Cancer Biol Ther. 2019;20:886–896. doi:10.1080/15384047.2019.157995930794031
- Sun H, Li Y, Kong H, et al. Dysregulation of KCNQ1OT1 promotes cholangiocarcinoma progression via miR-140-5p/SOX4 axis. Arch Biochem Biophys. 2018;658:7–15. doi:10.1016/j.abb.2018.09.01930243712
- Hu H, Yang L, Li L, et al. Long non-coding RNA KCNQ1OT1 modulates oxaliplatin resistance in hepatocellular carcinoma through miR-7-5p/ABCC1 axis. Biochem Biophys Res Commun. 2018;503:2400–2406. doi:10.1016/j.bbrc.2018.06.16829966655
- Zheng L, Zhang FX, Wang LL, et al. LncRNA KCNQ1OT1 is overexpressed in non-small cell lung cancer and its expression level is related to clinicopathology. Eur Rev Med Pharmacol Sci. 2019;23(16):6944–6950. doi:10.26355/eurrev_201908_1873431486494
- Chang RM, Yang H, Fang F, et al. MicroRNA-331-3p promotes proliferation and metastasis of hepatocellular carcinoma by targeting PH domain and leucine-rich repeat protein phosphatase. Hepatology. 2014;60:1251–1263. doi:10.1002/hep.2722124825302
- Luan X, Wang Y. LncRNA XLOC_006390 facilitates cervical cancer tumorigenesis and metastasis as a ceRNA against miR-331-3p and miR-338-3p. J Gynecol Oncol. 2018;29:e95. doi:10.3802/jgo.2018.29.e9530207103
- Ji H, Sang M, Liu F, et al. miR-124 regulates EMT based on ZEB2 target to inhibit invasion and metastasis in triple-negative breast cancer. Pathol Res Pract. 2019;215:697–704. doi:10.1016/j.prp.2018.12.03930611621
- Wang YX, Zhu HF, Zhang ZY, et al. MiR-384 inhibits the proliferation of colorectal cancer by targeting AKT3. Cancer Cell Int. 2018;18:124. doi:10.1186/s12935-018-0628-630186040
- Wang Y, Li J, Dai L, et al. MiR-17-5p may serve as a novel predictor for breast cancer recurrence. Cancer Biomark. 2018;22:721–726. doi:10.3233/CBM-18122829914008
- Liu T, Song Z, Gai Y. Circular RNA circ_0001649 acts as a prognostic biomarker and inhibits NSCLC progression via sponging miR-331-3p and miR-338-5p. Biochem Biophys Res Commun. 2018;503:1503–1509. doi:10.1016/j.bbrc.2018.07.07030029881
- Gao W, Wu Y, He X, et al. MicroRNA-204-5p inhibits invasion and metastasis of laryngeal squamous cell carcinoma by suppressing forkhead box C1. J Cancer. 2017;8:2356–2368. doi:10.7150/jca.1947028819440
- Wang X, Li F, Zhou X. miR-204-5p regulates cell proliferation and metastasis through inhibiting CXCR4 expression in OSCC. Biomed Pharmacother. 2016;82:202–207. doi:10.1016/j.biopha.2016.04.06027470356
- Jiang G, Wen L, Zheng H, et al. miR‐204‐5p targeting SIRT1 regulates hepatocellular carcinoma progression. Cell Biochem Funct. 2016;34:505–510. doi:10.1002/cbf.322327748572
- Wang H, Zhang Y, Wu Q, et al. MiR-16 mimics inhibit TGF-β1-induced epithelial-to-mesenchymal transition via activation of autophagy in non-small cell lung carcinoma cells. Oncol Rep. 2018;39:247–254.29138833
- Hua L, Zhu G, Wei J. MicroRNA‐1 overexpression increases chemosensitivity of non‐small cell lung cancer cells by inhibiting autophagy related 3‐mediated autophagy. Cell Biol Int. 2018;42(9):1240–1249. doi:10.1002/cbin.v42.929851226
- Sun X, Xin Y, Wang M, et al. Overexpression of long non-coding RNA KCNQ1OT1 is related to good prognosis via inhibiting cell proliferation in non-small cell lung cancer. Thorac Cancer. 2018;9:523–531. doi:10.1111/tca.2018.9.issue-529504267
- Shao J, Pan X, Yin X, et al. KCNQ1OT1 affects the progression of diabetic retinopathy by regulating miR-1470 and epidermal growth factor receptor. J Cell Physiol. 2019;234:17269–17279. doi:10.1002/jcp.2834430784065
- Gao X, Ge J, Li W, et al. LncRNA KCNQ1OT1 ameliorates particle-induced osteolysis through inducing macrophage polarization by inhibiting miR-21a-5p. Biol Chem. 2018;399:375–386. doi:10.1515/hsz-2017-021529252185
- Feng W, Wang C, Liang C, et al. The dysregulated expression of KCNQ1OT1 and its interaction with downstream factors miR-145/CCNE2 in breast cancer cells. Cell Physiol Biochem. 2018;49:432–446. doi:10.1159/00049297830157476
- Dong Z, Yang P, Qiu X, et al. KCNQ1OT1 facilitates progression of non-small-cell lung carcinoma via modulating miRNA-27b-3p/HSP90AA1 axis. J Cell Physiol. 2019;234:11304–11314. doi:10.1002/jcp.v234.730471108
- Zhang S, Ma H, Zhang D, et al. LncRNA KCNQ1OT1 regulates proliferation and cisplatin resistance in tongue cancer via miR-211-5p mediated Ezrin/Fak/Src signaling. Cell Death Dis. 2018;9:742. doi:10.1038/s41419-018-0793-529970910
- Li Y, Li C, Li D, et al. lncRNA KCNQ1OT1 enhances the chemoresistance of oxaliplatin in colon cancer by targeting the miR-34a/ATG4B pathway. Onco Targets Ther. 2019;12:2649–2660. doi:10.2147/OTT.S18805431040703
- Tk C, Ts L, Th C, et al. Dysregulation of microRNA-204 mediates migration and invasion of endometrial cancer by regulating FOXC1. Int J Cancer. 2012;130:1036–1045. doi:10.1002/ijc.2606021400511
- Imam JS, Plyler JR, Bansal H, et al. Genomic loss of tumor suppressor miRNA-204 promotes cancer cell migration and invasion by activating AKT/mTOR/Rac1 signaling and actin reorganization. PLoS ONE. 2012;7:e52397. doi:10.1371/journal.pone.005239723285024
- Sumbul AT, Gogebakan B, Ergun S, et al. miR-204-5p expression in colorectal cancer: an autophagy-associated gene. Tumour Biol. 2014;35:12713–12719. doi:10.1007/s13277-014-2596-325209181
- Luan W, Qian Y, Ni X, et al. miR-204-5p acts as a tumor suppressor by targeting matrix metalloproteinases-9 and B-cell lymphoma-2 in malignant melanoma. Onco Targets Ther. 2017;10:1237–1246. doi:10.2147/OTT28280358
- Yin Y, Zhang B, Wang W, et al. miR-204-5p inhibits proliferation and invasion and enhances chemotherapeutic sensitivity of colorectal cancer cells by downregulating RAB22A. Clin Cancer Res. 2014;20:6187–6199. doi:10.1158/1078-0432.CCR-14-103025294901
- Liu L, Wang J, Li X, et al. MiR-204-5p suppresses cell proliferation by inhibiting IGFBP5 in papillary thyroid carcinoma. Biochem Biophys Res Commun. 2015;457:621–626. doi:10.1016/j.bbrc.2015.01.03725603050
- Li M, Shen Y, Wang Q, et al. MiR-204-5p promotes apoptosis and inhibits migration of osteosarcoma via targeting EBF2. Biochimie. 2019;158:224–232. doi:10.1016/j.biochi.2018.12.00330529043
