Abstract
Background
Lung cancer continues to be one of the most dangerous tumors around the world. It is an urgency to explore the molecular mechanism of non-small cell lung cancer (NSCLC) progression for developing novel therapeutic approaches. Circular RNA (circRNA) is a novel type of non-coding RNA with a stable closed loop structure. Abnormally expressed circRNAs have been found in many kinds of cancer including NSCLC.
Methods and Results
The expression of circGFRA1 and miR-188-3p was detected in NSCLC tissues by RT-qPCR and it was found that circGFRA1 was highly expressed and miR-183-3p was lowly expressed in NSCLC tissues. In NSCLC cell lines, we confirmed that circGFRA1 acted as an miR-188-3p sponge using dual-luciferase reporter assay and RNA immunoprecipitation (RIP) analysis. Overexpression of cirGFRA1 enhanced NSCLC progression while miR-188-3p overexpression inhibited it by CCK8 and colony formation analysis. In vivo tumor xenograft model, circGFRA1 and miR-188-3p synergistically regulated the proliferation of NSCLC tumors. Mechanistic study indicated that circGFRA1 and miR-188-3p regulated the proliferation of NSCLC cells at least through PI3K/AKT signaling pathway.
Conclusion
Our study elaborated a novel circGFRA-miR-188-3p-PI3K/AKT regulatory pathway, providing a potential diagnostic biomarker and therapeutic target for NSCLC.
Keywords:
Introduction
Lung cancer continues to be one of the most dangerous tumors around the world. According to biological characteristics, lung cancer can be divided into two categories: non-small cell lung cancer (NSCLC) and small cell lung cancer (SCLC).Citation1 NSCLC accounts for approximately 85% in lung cancer cases while the rest 15% belongs to SCLC.Citation2 Since it is asymptomatic at the early stages, up to 61% of lung cancers are diagnosed at late stages.Citation3 As cancer progresses from early stage to late stage, the survival rate decreases dramatically.Citation3 The 5-year relative survival rate for NSCLC for all stages is only about 20%, while the survival rate declines to 4% at late stage.Citation3 The molecular mechanism underlying NSCLC progression needs further exploration for developing novel therapeutic approaches.
Circular RNA is a novel type of non-coding RNA with a stable closed loop structure through a typical 5′ to 3′-phosphodiester bond.Citation4 Although circRNAs are originally thought to be abnormal splicing products of RNA, accumulating evidence indicated that circRNAs are involved in various biological pathways including acting as miRNA sponge to inhibit the expression of miRNA, regulating RNA binding protein and nuclear transcription.Citation5 Abnormally expressed circRNAs have been found in many kinds of cancer including NSCLC.Citation6,Citation7 Yao et al reported that the expression of circRNA 100876 was closely related to the carcinogenesis of NSCLC.Citation8 Tian et al reported that circABCB10 could facilitate the proliferation and migration of NSCLC through sponging miR-1252.Citation9 Wei et al found that circPTPRA could suppress epithelial-mesenchymal transformation and metastasis of NSCLC cells by sponging miR-96-5p.Citation10 CircGFRA1 is a recently identified cancer-related circRNA. He et al found that circGFRA1 level was significantly upregulated in triple negative breast cancer cell lines through a circRNA microarray analysis.Citation11 Liu et al reported that circGFRA1 promotes ovarian cancer progression by regulating miR-449a.Citation12 However, the role of circGFRA1 in NSCLC progression is unclear.
MiRNAs are another class of endogenous non-coding RNAs with a length of about 22 nucleotides, which can negatively regulate gene expression through targeting to the 3ʹ-untranslated region (UTR) of the target mRNAs.Citation13 Many miRNAs have been found to play important role in the development and progression of NSCLC. Qi et al found that miR-448 could target SIRT1 (Sirtuin 1) and promote progression of NSCLC.Citation14 Tu et al found that miR-34c could inhibit the development of NSCLC by inducing endoplasmic reticulum stress through targeting HMGB1 (High Mobility Group Box 1).Citation15 Xu et al reported that the expression of miR-155 and miR-21 was associated with the recurrence or metastasis of NSCLC.Citation16 MiR-188-3p has been reported to be a tumor suppressor in several cancers, including hepatocellular carcinoma,Citation17 breast cancer,Citation18 colorectal cancer,Citation19 and pancreatic cancer.Citation20 However, the function of miR-188-3p in NSCLC still remains unexplored.
In this study, we detected the expression of circGFRA1 and miR-188-3p in NSCLC tissues and found that circGFRA1 expression was higher and miR-183-3p expression was lower in NSCLC tissues compared to that in normal adjacent tissues. In NSCLC cell lines, circGFRA1 and miR-183-3p could regulate the expression of each other. Overexpression of cirGFRA1 enhanced the development of NSCLC while miR-143-3p overexpression inhibited it both in vitro and in vivo. Further study indicated that circGFRA1 and miR-188-3p may act through PI3K/AKT signaling pathway to regulate the proliferation of NSCLC cells. Our study elaborated a novel circGFRA1-miR-188-3p-PI3K/AKT regulatory pathway, providing a potential diagnostic biomarker and therapeutic target for NSCLC.
Materials and Methods
Tissue Samples
NSCLC tumor tissues and normal adjacent tissues were obtained from 30 patients who have had surgery at Second Affiliated Hospital of Zhejiang University. A part of tissues was used for hematoxylin-eosin staining (HE staining), and the rest of tissues were rapidly frozen in liquid nitrogen and stored at −80°C preparing for RNA extraction. This study was approved by the Ethics Committee of Second Affiliated Hospital of Zhejiang University. Written informed consent was provided by all patients whose tissue samples were used. And the study was carried out in accordance with the principles of the Declaration of Helsinki.
Histology
Tissue samples were firstly fixed with 4% paraformaldehyde. Then, they were dehydrated, embedded, and sliced. The slices were stained using classical HE staining.
RT-qPCR Analysis
Total RNA of tissues or cells was extracted by TRNzol (TianGen, Beijing, China). For circRNA detection, total RNA was firstly digested by Rnase R to get rid of linear RNA, then cDNAs were synthesized with random primers. For miRNA, specific primer miRNA-RT was used for the reverse transcription of cDNA. Quantitative real-time PCR was conducted to detect the expression of circRNA and mRNA with GAPDH as an internal control. U6 was used as an internal control for detecting the expression of miRNA. The primer sequences were provided in Table S1.
Cell Culture
Human NSCLC cell lines (A549 and NCI-H838) were purchased from Tongpai biotechnology company (Shanghai, China). Both cell lines were cultured in RPMI 1640 medium (Gibco, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin at 37°C in an incubator with 5% CO2.
Cell Transfection
Negative control siRNA, si-GFRA1, miR-188-3p mimic and inhibitor were designed and synthesized by GenePharma (Shanghai, China). The sequences of circGFRA1 were cloned into the pcDNA3.1 by Sangon Biotech (Shang Hai, China). The pmirGLO luciferase vectors of circGFRA1 containing wild-type (WT) or mutated sequence in the putative miR-188-3p binding site (Mut) were also constructed by Sangon Biotech (Shang Hai, China), and named as pGLO-circGFRA1-WT and pGLO-circGFRA1-MUT separately. A549 or NCI-H838 cells were plated and transfected with siRNA, miRNA mimic and inhibitor, plasmids using lipofectamine 2000 (Invitrogen, USA) according to the manufacturer’s instruction. The sequences of siRNA were shown in Table S1.
Western Blot
Tissues or cells were harvested and lysed with RIPA Lysis Buffer (Beyotime Biotechnology, Beijing, China). The protein lysates were quantified and then separated by 8% SDS-PAGE (sodium dodecyl sulfate-polyacrylamide gel electrophoresis). Then, the proteins were transferred to polyvinylidenedifluoride membrane (Millipore, Billerica, USA). The membranes were blocked in 5% non-fat dry milk and incubated with primary antibodies at 4°C overnight, including Cyclin A1 (1:1000, ab53699, Abcam), Cyclin B1 (1:1000, ab72, Abcam), GAPDH (1:2000, 2118S, CST). Then, HRP-conjugated secondary antibodies were incubated for 1 h at room temperature. Finally, the bands were visualized by Tanon 5200.
Dual-Luciferase Reporter Assay
A549 and NCI-H838 cells were seeded and transfected with pGLO-circGFRA1-WT or pGLO-circGFRA1-MUT plasmid, together with miR-188-3p mimic or inhibitor. Cells were harvested 24 h later, luciferase activity was measured by the Dual-Luciferase Reporter Assay System (Promega Corp, Madison, WI, USA).
Cell Proliferation and Colony Formation Analysis
A549 or NCI-H838 cells were seeded in 24-well culture plates and transfected with relevant plasmids and siRNAs or treated with a PI3K pathway inhibitor-LY264002 for the indicated time-points. CCK-8 (Beyotime, Beijing, China) was used to perform the cell proliferation analysis according to the manufacturer’s instructions. For colony-forming analysis, circGFRA1 plasmid and miRNA mimic were transfected, respectively, or jointly into A549 or NCI-H838 cells. The transfected cells were seeded into twelve-well plates (600 cells/wells). Colonies were fixed by 4% paraformaldehyde 14 days later, and stained by 0.4% crystal violet solution. Colonies with diameter over 0.5 mm were counted under a light microscope.
RNA Immunoprecipitation (RIP)
Magna RIPTM RNA Binding Protein Immunoprecipitation Kit (Millipore, Bedford, MA, USA) was used to perform the RIP experiments in SPCA-1 and A549 cells. Samples obtained by RIP were digested by protease K and RNA was isolated for the detection of circGFRA1.
In vivo Tumor Xenograft Model
The NCI-H838 cells were transfected with pcDNA/Mock, circGFRA1/miR-188-3p or pcDNA/miR-188-3p for 72 h, then 1× 107 transfected cells were injected subcutaneously into BALB/c nude mice (6–7 weeks old, Weitonglihua, Beijing, China). The tumor size in all nude mice was measured every 3 days. All mice were sacrificed 18 days later following inoculation. Tumors were weighed, photographed, and frozen for reserved for RT-qPCR and Western blot analysis. The experimental procedures were guaranteed by the Guidelines for Care and Use of Laboratory Animal with approval of Ethics Committee of Zhejiang University.
Statistical Analysis
All statistical analysis was performed using SPSS 22.0 (IBM, SPSS, Chicago, IL, USA) with Student’s t-test and one-way ANOVA to estimate the significant group differences. Data were expressed as the mean ± S.E.M. (standard error of mean). P values < 0.05 were considered statistically significant.
Results
circGFRA1 Was Highly Expressed and miR-188-3p Was Lowly Expressed in NSCLC Tissues
NSCLC and normal adjacent tissues were obtained from 30 patients. The pathological changes of lung tissue were detected by HE staining and shown in .
Figure 1 CircGFRA1 was highly expressed and miR-143-3p was lowly expressed in NSCLC tissues. (A) The pathological changes of NSCLC tissues were detected by HE staining. (B) The expression of circGFRA1 in NSCLC and adjacent tissues was detected by RT-qPCR, #p<0.01 compared with adjacent tissues. (C) The expression of miR-188-3p in NSCLC and adjacent tissues was detected by RT-qPCR, #p<0.01 compared with adjacent tissues. (D) The correlation analysis of circGFRA1 and miR-188-3p levels in 30 NSCLC tissues.
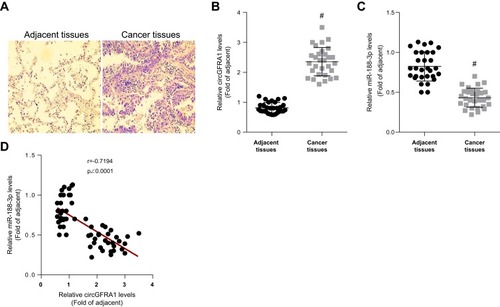
The expression of circGFRA1 and miR-188-3p in NSCLC and adjacent tissues was detected by RT-qPCR. As shown in , the expression of circGFRA1 was up-regulated and miR-188-3p expression was down-regulated in NSCLC tissues compared with adjacent tissues. Pearson correlation analysis was subsequently performed and the result displayed an inverse correlation between circGFRA1 and miR-188-3p (). These results indicated that circGFRA1 and miR-188-3p might play an important role in NSCLC pathogenesis.
circGFRA1 Acted as a ceRNA to Sponge miR-188-3p
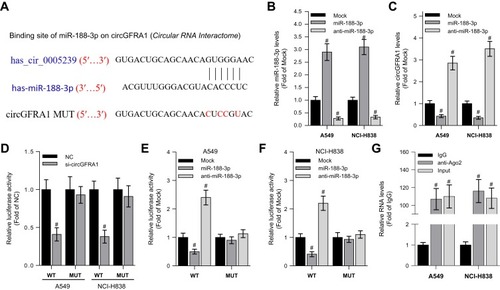
The “competing endogenous RNA (ceRNA) hypothesis” has been raised and validated by a plenty of studies.Citation21 Many circRNAs have been found to act as ceRNAs to sequestrate target miRNAs. To investigate whether circGFRA1 could sponge miR-188-3p, we first predicted the binding site for circGFRA1 in miR-188-3p using miRcode online website (). Then, we determined whether miR-188-3p overexpression or knockdown could affect the expression of circGFRA1. As shown in , miR-188-3p was successfully over-expressed or knocked down through transfecting miR-188-3p or anti-miR-188-3p mimics in A549 and NCI-H838 cells. The expression of circGFRA1 was also detected by RT-qPCR, and the results demonstrated that miR-188-3p overexpression inhibited the expression of circGFRA1 while miR-188-3p knockdown promoted the expression (). To further validate the association between circGFRA1 and miR-188-3p, circGFRA1 binding sites wild type or mutated luciferase plasmids (termed as pGLO-circGFRA1-WT or pGLO-circGFRA1-MUT) were constructed (the sequence information is shown in ), and luciferase activity was detected after pGLO-circGFRA1-WT or pGLO-circGFRA1-MUT was co-transfected with NC or si-circGFRA1 into A549 or NCI-H838 cells. As showed, compared with NC, si-circGFRA1 significantly reduced the luciferase activity when it was co-transfected with pGLO-circGFRA1-WT; however, the effects were absent when pGLO-circGFRA1-MUT plasmid was co-transfected with si-circGFRA1. The luciferase plasmids were also co-transfected with miR-188-3p or anti-miR-188-3p in A549 or NCI-H838 cells. As showed, miR-188-3p significantly reduced the luciferase activity when it was co-transfected with pGLO-circGFRA1-WT while anti-miR-188-3p promoted the activity. The effects were also absent when pGLO-circGFRA1-MUT plasmid was co-transfected. RIP analysis was performed to evaluate the enrichment of circGFRA1 and miR-188-3p by Ago2 antibody in A549 cells. The results further validated the binding between LINC00667 and miR-143-3p ().
Figure 2 CircGFRA1 acted as a ceRNA to sponge miR-188-3p. (A) The predicted binding site and mutant site for circGFRA1 in miR-188-3p. (B) RT-qPCR was performed to detect overexpression and knockdown efficiency of miR-188-3p in A549 or NCI-H838 cells, #P < 0.01 compared with Mock. (C) The expression of circGFRA1 was detected by RT-qPCR after miR-188-3p was overexpressed or knocked down in A549 or NCI-H838 cells. #P < 0.01 compared with Mock. (D) Luciferase activity was detected after pGLO-circGFRA1-WT or pGLO-circGFRA1-MUT was co-transfected with NC or si-circGFRA1 in A549 or NCI-H838 cells. #P < 0.01 compared with NC. (E and F) Luciferase activity was detected after pGLO-circGFRA1-WT or pGLO-circGFRA1-MUT was co-transfected with miR-188-3p or anti-miR-188-3p in A549 or NCI-H838 cells. #P < 0.01 compared with NC. (G) RIP analysis was used to evaluate the enrichment degree of circGFRA1 and miR-188-3p by Ago2 antibody in A549 cells. #P < 0.01 compared with IgG.
circGFRA1 and miR-188-3p Synergistically Regulated the Proliferation of NSCLC Cells
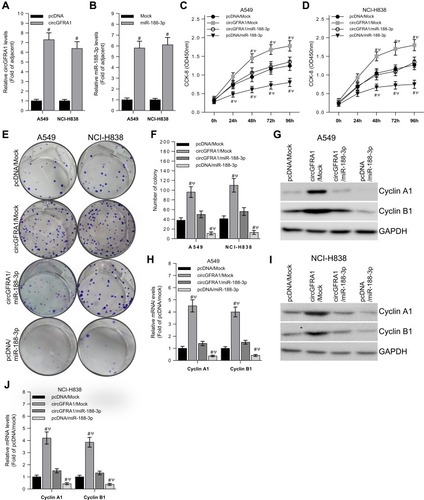
To study the function of circGFRA1 and miR-188-3p in NSCLC cells, circGFRA1 and miR-188-3p overexpression plasmids were constructed. The expression of circGFRA1 or miR-188-3p was significantly increased after being transfected into A549 or NCI-H838 cells, respectively, which indicated the overexpression efficiency (). Then, CCK8 assays were performed to determine the proliferation ability when circGFRA1 and miR-188-3p were transfected individually or jointly. As shown in , circGFRA1 overexpression significantly promoted the growth of A549 and NCI-H838 cells while miR-188-3p overexpression significantly inhibited it, and the pro- or anti-proliferation effects were absent when both circGFRA1 and miR-188-3p were overexpressed. circGFRA1 overexpression also increased the colony formation ability of A549 and NCI-H838 cells while miR-188-3p overexpression significantly inhibited it, and the pro- or anti-proliferation effect was absent when both circGFRA1 and miR-188-3p were overexpressed (). The expression of proliferation-related genes including Cyclin A1, Cyclin B1 was detected by western-blot. The results indicated that these genes were up-regulated when circGFRA1 was overexpressed; however, miR-188-3p overexpression inhibited the expression of these genes (–). These results indicate that the circGFRA1/miR-188-3p axis regulates the growth of NSCLC cells, cooperatively.
Figure 3 The proliferation of NSCLC cells is regulated by circGFRA1 and miR-188-3p. (A and B) RT-qPCR was used to detect transfection efficiency of circGFRA1 or miR-188-3p in A549 and NCI-H838 cells. (C and D) Cell proliferation was determined by CCK-8 when circGFRA1 and miR-188-3p was transfected individually or jointly, #P < 0.01 compared with pcDNA/Mock, ΨP < 0.01 compared with circGFRA1/miR-188-3p. (E and F) Colony formation ability was evaluated in A549 and NCI-H838 cells when circGFRA1 and miR-188-3p was transfected individually or jointly, #P < 0.01 compared with pcDNA/Mock, ΨP < 0.01 compared with circGFRA1/miR-188-3p. (G, H, I, J) The expression of proliferation-related genes including Cyclin A1, Cyclin B1 was detected by western-blot and RT-qPCR in A549 and NCI-H838 cells. #P < 0.01 compared with pcDNA/Mock, ΨP < 0.01 compared with circGFRA1/miR-188-3p.
CircGFRA1 and miR-188-3p Synergistically Regulated the Proliferation of NSCLC Tumors in vivo
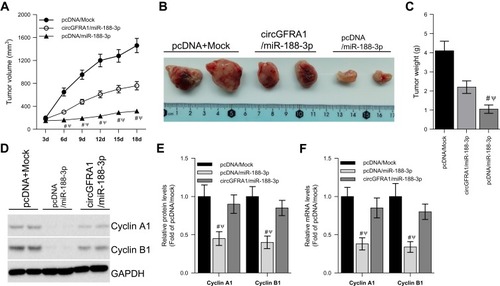
To explore the impact of the circGFRA1/miR-188-3p axis on tumorigenesis of NSCLC in vivo, NCI-H838 cells were transfected with pcDNA/Mock, circGFRA1/miR-188-3p or pcDNA/miR-188-3p and injected subcutaneously into BALB/c nude mice. Tumor volume was markedly decreased in pcDNA/miR-188-3p group when compared to pcDNA/Mock or circGFRA1/miR-188-3p group (). Representative images of resected tumor masses in each group were shown in . Tumor weight was also markedly decreased in pcDNA/miR-188-3p group when compared to pcDNA/Mock or circGFRA1/miR-188-3p group (). The expression of proliferation-related genes including Cyclin A1, Cyclin B1 was detected by Western blot. As shown in , miR-188-3p overexpression significantly inhibited the expression of Cyclin A1 and Cyclin B1, while circGFRA1 overexpression inhibited it. The expression of Cyclin A1 and Cyclin B1 was also detected by RT-qPCR, and a consistent result was obtained (). We also explored whether circGFRA1 knockdown could inhibit the proliferation of NSCLC tumors in vivo. Compared with the control, sh-circGFRA1 transfection significantly inhibited the growth of tumor. Tumor volume and weight were markedly decreased (Supplementary Figure 1A–C). The mRNA expression of Cyclin A1 and Cyclin B1 was significantly inhibited when circGFRA1 expression was inhibited (Supplementary Figure 1D).
Figure 4 circGFRA1 and miR-188-3p synergistically regulated the proliferation of NSCLC cells in vivo. The NCI-H838 cells were transfected with pcDNA/Mock, circGFRA1/miR-188-3p or pcDNA/miR-188-3p for 72 h, then the cells were inocubated subcutaneously into nude mice. (A) Tumor volume was markedly decreased in pcDNA/miR-188-3p when compared to pcDNA/Mock or circGFRA1/miR-188-3p group. #P < 0.01 compared with pcDNA/Mock, ΨP < 0.01 compared with circGFRA1/miR-188-3p. (B) Representative images of resected tumor masses in pcDNA/Mock, circGFRA1/miR-188-3p or pcDNA/miR-188-3p group. (C) Tumor weight was markedly decreased in pcDNA/miR-188-3p when compared to pcDNA/Mock or circGFRA1/miR-188-3p group. #P < 0.01 compared with pcDNA/Mock, ΨP < 0.01 compared with circGFRA1/miR-188-3p. (D and E) The expression of proliferation-related genes including Cyclin A1, Cyclin B1 was detected by western-blot, #P < 0.01 compared with pcDNA/Mock, ΨP < 0.01 compared with circGFRA1/miR-188-3p. (F) The expression of proliferation-related genes including Cyclin A1, Cyclin B1 was detected by RT-qPCR, #P < 0.01 compared with pcDNA/Mock, ΨP < 0.01 compared with circGFRA1/miR-188-3p.
circGFRA1/miR-188-3p May Regulate the Proliferation of NSCLC Cells Through the PI3K/AKT Signaling Pathway
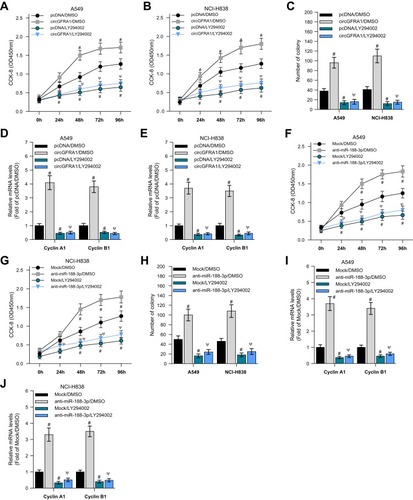
PI3K/AKT signalling pathway plays an important role in tumor progression.Citation22 We explored whether the circGFRA1/miR-188-3p axis regulated the development of NSCLC through PI3K/AKT signaling pathway. A549 and NCI-H838 cells transfected with circGFRA1 or pcDNA were treated with a classical PI3K inhibitor-LY264002 or DMSO, and the cell proliferation was detected by CCK8 assay. As shown in , LY264002 reversed the proliferative effect of circGFRA1. And LY264002 reversed the enhanced colony formation ability induced by circGFRA1 overexpression (). The expression of proliferation-related genes including Cyclin A1, Cyclin B1 was also detected by RT-qPCR in A549 and NCI-H838 cells. The result indicated that LY264002 reversed the upregulated expression of Cyclin A1 and Cyclin B1 caused by circGFRA1 overexpression (). A549 and NCI-H838 cells were also treated with anti-miR-188-3p and LY264002 individually or jointly. CCK8 analysis was also performed to evaluate the proliferation. As showed, anti-miR-188-3p significantly promoted the growth of A549 and NCI-H838 cells, this effect was reversed by LY264002. Additionally, LY264002 reversed the enhanced colony formation ability mediated by anti-miR-188-3p (). LY264002 rescued the upregulated expression of Cyclin A1 and Cyclin B1 caused by anti-miR-188-3p (). We also detected the phosphorylation level of AKT which plays a central role in the activation of PI3K/AKT signal. As shown in supplementary Figure 2, overexpression of circGFRA1 significantly increases the phosphorylation level of AKT. However, PI3K/AKT inhibitor LY294002 reduced the increased phosphorylation level of AKT caused by circGFRA1 overexpression. The results further revealed that circGFRA1/miR-188-3p may regulate the proliferation of NSCLC cells through PI3K/AKT signal. These results indicate that the circGFRA1/miR-188-3p axis may regulate the proliferation of NSCLC cells through the PI3K/AKT signaling pathway.
Figure 5 circGFRA1 and miR-188-3p synergistically regulated the proliferation of NSCLC cells through the PI3K/AKT signaling pathway. (A and B) A549 and NCI-H838 cell proliferation was detected by CCK-8 assay after circGFRA1 and LY264002 was given individually or jointly. #P < 0.01 compared with pcDNA/DMSO, ΨP < 0.01 compared with circGFRA1/DMSO. (C) Colony formation ability was evaluated in A549 and NCI-H838 cells after circGFRA1 and LY264002 was given individually or jointly. #P < 0.01 compared with pcDNA/DMSO, ΨP < 0.01 compared with circGFRA1/DMSO. (D and E) The expression of proliferation-related genes including Cyclin A1, Cyclin B1 was detected by RT-qPCR, #P < 0.01 compared with pcDNA/DMSO, ΨP < 0.01 compared with circGFRA1/DMSO. (F and G) A549 and NCI-H838 cell proliferation was detected by CCK-8 assay after anti-miR-188-3p and LY264002 was given individually or jointly. #P < 0.01 compared with pcDNA/DMSO, ΨP < 0.01 compared with circGFRA1/DMSO. (H) Colony formation ability was evaluated in A549 and NCI-H838 cells after anti-miR-188-3p and LY264002 was given individually or jointly. #P < 0.01 compared with pcDNA/DMSO, ΨP < 0.01 compared with circGFRA1/DMSO. (I and J) The expression of proliferation-related genes including Cyclin A1, Cyclin B1 was detected by RT-qPCR, #P < 0.01 compared with pcDNA/DMSO, ΨP < 0.01 compared with circGFRA1/DMSO.
Discussion
NSCLC is one of the most common cancers with a low cure rate. Despite great efforts, the underlying mechanism of NSCLC tumorigenesis still remains to be fully elucidated.Citation3 According to the research in recent years, ncRNAs including circRNA and miRNA are extensively involved in various biological processes.Citation23 Increased evidence suggested that an aberrant expression of ncRNAs was associated with human diseases including cancer.Citation24 In the past couple of years, several circRNAs have been found to act as important tumor suppressors or promote tumorigenesis in NSCLC.Citation9,Citation10 In the present study, we detected the expression of circGFRA1 and miR-188-3p in NSCLC tissues and found that the expression of circGFRA1 was negatively correlated with the expression of miR-188-3p. The expression of circGFRA1 was relatively high while miR-188-3p expression was low in NSCLC tissues. The increased expression of circGFRA1 in NSCLC tissues was consistent with the recently reported data in ovarian cancer and breast cancer.Citation11,Citation12 miR-188-3p has been found to be down-regulated in many kinds of tumors such as pancreatic cancer, hepatocellular carcinoma, and so on.Citation17,Citation20
Emerging evidence reveals that circRNAs can act as a ceRNA to sponge miRNAs. We then predicted that circGFRA1 could bind with miR-188-3p. The expression of circGFRA1 was down-regulated when miR-188-3p was overexpressed. The association between cirGFRA1 and miR-188-3p was further validated by dual-luciferase reporter and RIP analysis. These results revealed that circGFRA1 acts as an miR-188-3p sponge.
We next explore the function of cicGFRA1 and miR-188-3p in the tumor tumorigenesis. In vitro, we found circGFRA1 overexpression significantly promoted the growth of A549 and NCI-H838 cells and increased the expression of proliferation-related genes; however, miR-188-3p overexpression reversed these effect led by circGFRA1. These results revealed that the function of circGFRA1 and microRNA are antagonistic to each other. In vivo, we constructed tumor xenograft model and found a same trend with that in vitro. circGFRA1 overexpression promoted the development of NSCLC tumor, which was attenuated by miR-188-3p overexpression.
The PI3K/AKT signalling pathway is a classical oncogenic signalling pathway. Several ncRNAs have been found to regulate tumorigenesis through regulating the PI3K/AKT signalling pathway.Citation25,Citation26 In the present study, we used an inhibitor of PI3K/AKT and found that inhibition of PI3K/AKT signalling pathway could reverse the effects of cirGFRA1 and anti-miR-188-3p. These results imply that the cirGFRA1/miR-188-3p axis may act through PI3K/AKT signalling pathway to inhibit NSCLC progression. It is interesting to further explore the direct target that is regulated by the cirGFRA1/miR-188-3p axis in PI3K/AKT signalling pathway.
In conclusion, our study revealed that a novel cirGFRA1-miR-188-3p-PI3K/AKT axis playing an important role in the tumorigenesis of NSCLC. cirGFRA1 acts as a ceRNA to regulate the PI3K/AKT signalling pathway through sponging miR-188-3p. cirGFRA1 and miR-188-3p might be potential diagnostic biomarker and therapeutic targets for NSCLC.
Acknowledgments
This work is financially supported by research grants from the Natural Science Foundation of Zhejiang Province (LQ16H020005), and the National Science Foundation for Young Scientists of China (81500203).
Disclosure
The authors report no conflicts of interest in this work.
References
- Brambilla E. Lung cancer: multidisciplinary approach for management: cell and molecular biology assembly contribution to the celebration of 20 years of the ERS. Eur Respir J. 2010;35:717–720. doi:10.1183/09031936.0001881020356981
- Tsim S, O’Dowd CA, Milroy R, Davidson S. Staging of non-small cell lung cancer (NSCLC): a review. Respir Med. 2010;104:1767–1774. doi:10.1016/j.rmed.2010.08.00520833010
- Miller KD, Nogueira L, Mariotto AB, et al. Cancer treatment and survivorship statistics, 2019. CA Cancer J Clin. 2019.
- Ebbesen KK, Kjems J, Hansen TB. Circular RNAs: identification, biogenesis and function. Biochim Biophys Acta. 2016;1859:163–168. doi:10.1016/j.bbagrm.2015.07.00726171810
- Hsiao KY, Sun HS, Tsai SJ. Circular RNA - new member of noncoding RNA with novel functions. Exp Biol Med. 2017;242:1136–1141. doi:10.1177/1535370217708978
- Zhu LP, He YJ, Hou JC, et al. The role of circRNAs in cancers. Biosci Rep. 2017;37. doi:10.1042/BSR20170750
- Qian L, Yu SL, Chen Z, Meng ZQ, Huang SL, Wang P. The emerging role of circRNAs and their clinical significance in-human cancers. Bba-Rev Cancer. 2018;1870:247–260.
- Yao JT, Zhao SH, Liu QP, et al. Over-expression of CircRNA_100876 in non-small cell lung cancer and its prognostic value. Pathol Res Pract. 2017;213:453–456. doi:10.1016/j.prp.2017.02.01128343871
- Tian XF, Zhang L, Jiao Y, Chen JS, Shan Y and Yang WF. circABCB10 promotes nonsmall cell lung cancer cell proliferation and migration by regulating the miR-1252/FOXR2 axis. J Cell Biochem. 2019;120:3765–3772. doi:10.1002/jcb.2765730417418
- Wei S, Zheng Y, Jiang Y, et al. The circRNA circPTPRA suppresses epithelial-mesenchymal transitioning and metastasis of NSCLC cells by sponging miR-96-5p. EBioMedicine. 2019;44:182–193. doi:10.1016/j.ebiom.2019.05.03231160270
- He RF, Liu P, Xie XM, et al. circGFRA1 and GFRA1 act as ceRNAs in triple negative breast cancer by regulating miR-34a. J Exp Clin Cancer Res. 2017;36. doi:10.1186/s13046-017-0614-1
- Liu J, Yu F, Wang S, et al. circGFRA1 promotes ovarian cancer progression by sponging miR-449a. J Cancer. 2019;10:3908–3913. doi:10.7150/jca.3161531417634
- Friedman RC, Farh KKH, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi:10.1101/gr.082701.10818955434
- Qi H, Wang H, Pang D. miR-448 promotes progression of non-small-cell lung cancer via targeting SIRT1. Exp Ther Med. 2019;18:1907–1913. doi:10.3892/etm.2019.773831410153
- Tu L, Long X, Song WD, et al. MiR-34c acts as a tumor suppressor in non-small cell lung cancer by inducing endoplasmic reticulum stress through targeting HMGB1. Onco Targets Ther. 2019;12:5729–5739. doi:10.2147/OTT.S20693231410019
- Xu S, Shi L. High expression of miR-155 and miR-21 in the recurrence or metastasis of non-small cell lung cancer. Oncol Lett. 2019;18:758–763. doi:10.3892/ol.2019.1033731289551
- Meng FZ, Zhang SG, Song RP, et al. NCAPG2 overexpression promotes hepatocellular carcinoma proliferation and metastasis through activating the STAT3 and NF-kappa B/miR-188-3p pathways. EBioMedicine. 2019;44:237–249. doi:10.1016/j.ebiom.2019.05.05331176678
- Pei J, Zhang J, Yang XW, et al. TMED3 promotes cell proliferation and motility in breast cancer and is negatively modulated by miR-188-3p. Cancer Cell Int. 2019;19. doi:10.1186/s12935-019-0791-4
- Pichler M, Stiegelbauer V, Vychytilova-Faltejskova P, et al. Genome-wide miRNA analysis identifies miR-188-3p as a novel prognostic marker and molecular factor involved in colorectal carcinogenesis. Clin Cancer Res. 2017;23:1323–1333. doi:10.1158/1078-0432.CCR-16-049727601590
- Shi W, Zhang C, Ning Z, et al. Long non-coding RNA LINC00346 promotes pancreatic cancer growth and gemcitabine resistance by sponging miR-188-3p to derepress BRD4 expression. J Exp Clin Cancer Res. 2019;38:60. doi:10.1186/s13046-019-1055-930728036
- Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146:353–358. doi:10.1016/j.cell.2011.07.01421802130
- Dobbin ZC, Landen CN. The importance of the PI3K/AKT/mTOR pathway in the progression of ovarian cancer. Int J Mol Sci. 2013;14:8213–8227. doi:10.3390/ijms1404821323591839
- Novikova IV, Hennelly SP, Tung CS, Sanbonmatsu KY. Rise of the RNA machines: exploring the structure of long non-coding RNAs. J Mol Biol. 2013;425:3731–3746. doi:10.1016/j.jmb.2013.02.03023467124
- Lu W, Zhang H, Niu Y, et al. Long non-coding RNA linc00673 regulated non-small cell lung cancer proliferation, migration, invasion and epithelial mesenchymal transition by sponging miR-150-5p. Mol Cancer. 2017;16:118. doi:10.1186/s12943-017-0685-928697764
- Dong P, Konno Y, Watari H, Hosaka M, Noguchi M, Sakuragi N. The impact of microRNA-mediated PI3K/AKT signaling on epithelial-mesenchymal transition and cancer stemness in endometrial cancer. J Transl Med. 2014;12:231. doi:10.1186/s12967-014-0231-025141911
- Du W, Zhang X, Wan Z. miR-3691-5p promotes hepatocellular carcinoma cell migration and invasion through activating PI3K/Akt signaling by targeting PTEN. Onco Targets Ther. 2019;12:4897–4906. doi:10.2147/OTT.S20812731417285
