Abstract
Background
Previous evidence have shown that long non-coding RNA (lncRNA) TMPO antisense RNA 1 (TMPO-AS1) is involved in the aggressiveness of several cancers. Nevertheless, the precise functions of TMOP-AS1 in hepatocellular carcinoma (HCC) are still unresolved.
Materials and Methods
The expressions of TMPO-AS1 and miR-320a were detected in HCC tissues and cells by qRT-RCR. The cell growth, migration and invasion were detected by colony formation, wound healing assay and Transwell assay, respectively. The targeting relation between miR-320a and TMPO-AS1 was predicted by bioinformatics analysis and identified by luciferase reporter gene as well as FISH assay. The expression of SERPINE1 MRNA Binding Protein 1 (SERBP1) was detected by Western blot. The growth of HCC cell was analyzed using transplanted tumor model.
Results
Currently, we revealed that TMPO-AS1 was overexpressed in clinical HCC samples and a panel of HCC cell lines. Clinically, a higher level of TMPO-AS1 was connected to the advanced stage of HCC and worse prognosis of patients. Depletion of TMPO-AS1 repressed HCC cell viability, migration ability and invasiveness. Nevertheless, upregulation of TMPO-AS1 caused opposite results. Further studies revealed that lncRNA TMPO-AS1 was largely located in the cytoplasm of HCC cell and sponge miR-320a, resulting in increasing the level of SERBP1 in HCC cell. Finally, TMPO-AS1 silencing suppressed tumor growth of HCC cell in vivo.
Conclusion
Collectively, our results suggested that TMPO-AS1 was a promoting factor for the aggressive behaviors of HCC cell.
Introduction
Hepatocellular carcinoma (HCC) is the most common type of liver cancers and the major cause for cancer-related death.Citation1 HCC is a highly heterogeneous disease that is prone to frequent recurrence and distant metastasis.Citation2,Citation3 Hence, a better understanding of mechanisms behind HCC cell metastasis is urgently needed, which will be helpful for exploring a novel biomarker and treatment target for combating HCC.
Recently, increasing evidence have indicated that long non-coding RNAs (lncRNAs), a kind of non-coding RNAs regulates the oncogenesis and metastasis in a variety of malignant cancers. For instance, lncRNA H19 promotes papillary thyroid carcinoma cell epithelial–mesenchymal transition (EMT).Citation4 LncRNA HOXD-AS1 induces the EMT process of breast cancer cell via serving as a competing endogenous RNA (ceRNA) of miR-421.Citation5 LncRNA ZNFX1-AS1 facilitates the progression and metastases of colon cancer cell through function as a ceRNA of miR-144 to regulating EZH2.Citation6 LncRNA TP73-AS1 targets miR-329-3p to regulate expression of SMAD2 in human cervical cancer tissue and cell lines.Citation7
Dysregulations of lncRNAs in HCC have been revealed in several previous investigations. Upregulation of lncRNA SNHG16 inhibits HCC cell proliferation and chemo-resistance via sponging miR-93.Citation8 LncRNA MIAT increases HCC cell proliferation and invasion by sponging miR-214.Citation9 Recently, TMPO-AS1 has been proven to induce the progression of cervical cancer by raising RAB14 by sponging miR-577.Citation10 Overexpression of TMPO-AS1 is connected to the progression of prostate cancer cell and poor clinical outcomes of patients.Citation11 In addition, lncRNA TMPO-AS1 accelerates the development of non-small-cell lung carcinoma (NSCLC) by modulating TMPO.Citation12 Despite the functional characteristics of lncRNA TMPO-AS1 in cancers that have been functionally characterized, the potential roles of TMPO-AS1 require further exploration.
Currently, we elaborated that lncRNA TMPO-AS1 was dysregulated in HCC samples and cell lines. Additionally, we elaborated that TMPO-AS1 exerted oncogenic roles in the growth and aggressive traits of HCC cell. Moreover, our findings suggested that TMPO-AS1 functioned as a competing endogenous RNA (ceRNA) to sponge miR-320a to modulate the level of SERBP1 in HCC cell.
Materials and Methods
HCC Tissues
Forty-two cases of HCC and corresponding non-cancerous specimens were collected from the International Zhuang Hospital District of Guangxi University of Chinese Medicine. The clinical characteristics of HCC patients are summarized in Supplementary Table 1. No patients received treatment, including chemotherapy and radiotherapy before operative treatment. Written informed consent was obtained and this research was approved by the Ethics Committee of the International Zhuang Hospital District of Guangxi University of Chinese Medicine.
Cell Lines and Transfections
HCC cells (HepG2, SNU-387, HCCLM3, SMMC-7721, Huh7) and normal human hepatic cell line, LO2 were purchased from Jiangsu KeyGEN BioTECH (Nanjing, Jiangsu, China) and were maintained in RPMI-1640 or DMEM containing 10% FBS at 37°C in a 5% CO2 incubator. Two shRNAs targeting TMPO-AS1 (sh-TMPO-AS1 #1, sh-TMPO-AS1 #2), miR-320a mimics, miR-320a inhibitor and negative controls, siRNA negative control (si-Con) and siRNA SERBP1 (si-SERBP1) were obtained from RiboBio (Guangzhou, Guangdong, China). Lentivirus plasmid was bought from RiboBio and was transfected sh-TMPO-AS1 #1. Virus-containing supernatant was collected 48 hours after lentivirus packaging, followed by its addition to the SNU-387 cell. After 24 hours, the stably infected SNU-387 cell was selected with 2 μg/mL of Puromycin 2HCL (Selleck). The TMPO-AS1 expression vector (pcDNA3.1-TMPO-AS1, named as pc-TMPO-AS1) and empty pcDNA3.1 vector (negative control, named as pc-vector) was bought from RiboBio. Cell transfections were performed using Lipofectamine 2000 (Thermo Fisher Scientific).
qRT-PCR
RNAs were prepared with Trizol reagent. RNA (1 μg) was used to reverse transcription with a Transcriptor First Strand cDNA Synthesis kit (Roche Diagnostics, Basel, Switzerland). The qRT-PCR was performed using LightCycler 480 instrument (Roche Diagnostics) with a SYBR Premix Ex Taq II kit (Takara, Shiga, Japan). The level of miR-320a was measured by One Step PrimeScript miRNA cDNA Synthesis kit (Takara). GAPDH or U6 was applied for normalized control. The relative expression was determined by utilizing the 2−ΔΔCt method. The primers are summarized in Supplementary Table 2.
Cell Counting Kit 8 (CCK-8) Assay
HCC cells (2 × 103) were cultured into 96 well plates and cultured for 1 day, 2 days, 3 days, 4 days or 5 days, respectively. After each time point, 10 μL of CCK-8 (Solarbio, Beijing, China) reagent was added into each well. Cells in plates were incubated at 37 °C for 2 hours. Finally, the OD value was examined using a microplate reader at 450 nm.
Colony Formation
One thousand HCC cells were added into six-well plates and maintained for 2 weeks. Cell colonies were fixed using 4% formaldehyde and stained by 1% crystal violet (Sigma). The number of visible cell colonies in plates was calculated.
Wound-Healing Assay
Cells (5 × 105) were grown into 6-well plates. After overnight, cell monolayers were scraped using a 100 μL pipette tip. The wound was photographed at 0 hour, 24 hours and the percentage of migration was calculated using ImageJ software.
Invasion Assay
Cells (1 × 105) were suspended in FBS-free medium and plated into the upper chamber of Transwell chamber (Millipore, Braunschweig, Germany), which was pre-coated with Matrigel (BD Bioscience). The lower chamber was filled with 500 μL of culture medium (20% FBS). After staining with 1% crystal violet, the number of invaded cells was counted.
Luciferase Reporter Assay
The wild-type (wt) 3′-UTR TMPO-AS1 was amplified and cloned into pmirGLO reporter (Promega) to construct TMPO-AS1-wt plasmid. TMPO-AS1 mutant 3′-UTR plasmid (TMPO-AS1-mut) was constructed by using a TaKaRa MutanBEST kit (TaKaRa). Then, the report vector and miR-320a were cotransfected into HCCLM3 and SNU-387 cell by utilizing Lipofectamine 2000 (Thermo Fisher Scientific). The wild-type (wt) 3′-UTR SERBP1 was amplified and cloned into pmirGLO reporter (Promega) to generate SERBP1-wt plasmid. SERBP1 mutant 3′-UTR plasmid (SERBP1-mut) was constructed by utilizing a TaKaRa MutanBEST kit (TaKaRa). Then, the report vector and miR-320a were cotransfected into HCCLM3 and SNU-387 cell using Lipofectamine 2000 (Thermo Fisher Scientific). After 48 h, the luciferase activities in cells were detected by Luciferase reporter gene system (Promega).
Isolation of Cytoplasmic and Nuclear RNA
Cytoplasm and nuclear RNA in HCC cells were separated using a Cytoplasmic & Nuclear RNA purification kit (Norgen, Belmont, USA). Then, the expression proportions of designated RNA molecules in the nucleus and cytoplasm fractions were measured using qRT-PCR test. U6 was selected as the nucleus control and GAPDH was the cytoplasm control.
RNA Immunoprecipitation (RIP)
RIP assay was conducted using Magna RNA immunoprecipitation (RIP) kit (Millipore, Braunschweig, Germany). Cell lysates were incubated with immunoprecipitation buffer containing magnetic bead conjugated with anti-Argonaute2 (Ago2) antibody (Abcam) or negative control anti-IgG. The Co-precipitated RNAs were collected and measured using qRT-PCR.
Fluorescence in-situ Hybridization (FISH)
HCCLM3 and SNU-387 cells were fixed in 4% formaldehyde for 15minutes and followed by washes with PBS. Fixed cells were treated with pepsin (1% in 10 mmol/L HCl) and subsequent dehydrated through 70%, 90%, and 100% ethanol. The air-dried cells were further incubated with 40 nmol/L TMPO-AS and miR-320a probe in hybridization buffer (100 mg/mL dextran sulfate, 10% formamide in 2xSSC) at 80°C for 2minutes. Hybridization was performed at 55°C for 2 hours, and the slide was washed and dehydrated. The air-dried slide was stained with 1 mg/mL DAPI for 10 min. RNA FISH probes were designed and synthesized by Bogu Co, Ltd. Specimens were analyzed on a Nikon inverted fluorescence microscope.
Immunoblotting
Total proteins in cells were prepared using RIPA and separated using 8% SDS-PAGE. After separation, proteins were transferred onto PVDF membranes (Millipore). After blocking, the PVDF membrane was incubated with SERBP1 or GAPDH antibody for overnight. After incubating with HRP-conjugated secondary antibody, the target bands were measured by using an ECL kit (Bioworld).
Xenograft Tumor Model
This animal experiment was approved by the Institutional Animal Care and Use Committee of the International Zhuang Hospital District of Guangxi University of Chinese Medicine and complied with the Guide for the Care and Use of Laboratory Animals (NIH publication No. 86–23, revised 1985). sh-NC or sh-TMPO-AS1 transfected SNU-387 cells were subcutaneously inoculated into BALB/c nude mice (n=5 in each group, Chinese Academy of Sciences, Shanghai, China). Tumor volume was measured weekly. After 5 weeks, nude mice were sacrificed, and tumor tissues were subjected for immunohistochemistry (IHC) staining with SERBP1.
Statistical Analysis
Data were shown as Mean ± SD. The differences were determined by utilizing one-way ANOVA or Student’s t-test. The prognostic value of TMPO-AS1 in HCC was determined using Kaplan–Meier plotter. P<0.05 was statistically significant.
Results
TMPO-AS1 Is Overexpressed in Human HCC
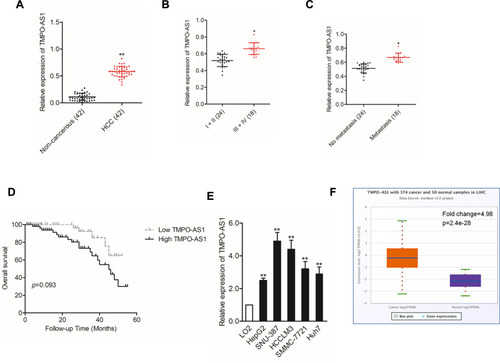
To analyze whether TMPO-AS1 was connected to HCC, we evaluated the expression pattern of TMPO-AS1 in 42 pairs of HCC and corresponding non-cancerous samples. As presented in , TMPO-AS1 was distinctly overexpressed in collected HCC samples compared with that in corresponding non-cancerous tissues. Clinically, a higher expression level of TMPO-AS1 was positively related to advanced tumor-node-metastases (TNM) stage and metastasis (). Moreover, HCC patients with a high level of TMPO-AS1 exhibited worse overall survival (OS) than patients with low TMPO-AS1 (). In addition, we also assessed the levels of TMPO-AS1 in a panel of HCC cells and LO2. The result of qRT-PCR test showed that lncRNA TMPO-AS1 was notably overexpressed in all HCC cell lines in contrast to in LO2 cell (). Finally, we examined the expression of TMPO-AS1 in TCGA Data Portal from starBASE v3.0 (http://starbase.sysu.edu.cn/panCancer.php) and found TMPO-AS1 exist higher expression in liver hepatocellular carcinoma (LIHC) than in normal tissue (). These data indicated that TMPO-AS1 was overregulated and might be a potential prognostic indicator in HCC.
Figure 1 High TMPO-AS1 expression is identified in HCC cells and tissues. (A) The expression levels of TMPO-AS1 in HCC tissues and non-cancerous tissues were detected using qRT-PCR assay. **P<0.01 compared with non-cancerous. (B) Expression levels of TMPO-AS1 in HCC tissues from stage I–II and stage III–IV. *P<0.01 compared with I–II. (C) Expression levels of TMPO-AS1 in HCC patients with metastasis and without metastasis. *P<0.01 compared with no metastasis. (D) Kaplan–Meier curves for HCC patients with higher expression of TMPO-AS1 or lower expression of TMPO-AS1. (E) The levels of TMPO-AS1 in HCC cell lines and LO2 cell were detected using qRT-PCR assay. **P<0.01 compared with LO2. (F) Expression of TMPO-AS1 in control tissues (n=50) and liver hepatocellular carcinoma (LIHC) tissues (n=374). TMPO-AS1 expression is significantly upregulated in LIHC tissues compared with control tissues based on the analysis of the high-throughput sequencing database of TCGA.
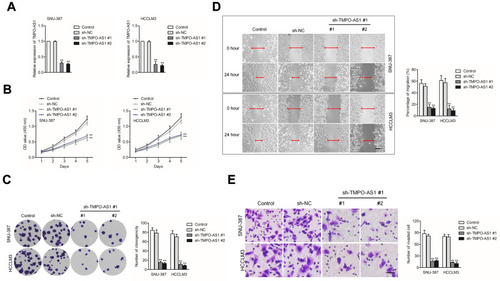
TMPO-AS1 Silencing Inhibits HCC Cell Proliferation, Migration and Invasion
Then, HCCLM3 and SNU-387 cells were transfected with sh-TMPO-AS1 #1 or sh-TMPO-AS1 #2 to reduce the endogenous expression of TMPO-AS1. We detected the knockdown efficiency of sh-TMPO-AS1 in HCC cell by using qRT-PCR test (). Next, CCK-8 assays were conducted, and we observed that TMPO-AS1 silencing distinctly reduced the cell proliferation of HCCLM3 and SNU-387 (). Consistently, depletion of TMPO-AS1 distinctly impaired the colony formation capacities of both HCCLM3 and SNU-387 cell in vitro (). Similarly, the migration abilities of HCCLM3 and SNU-387 cells were markedly repressed after TMPO-AS1 knockdown, as demonstrated by the wound healing assay (). Besides, TMPO-AS1 silencing inhibits the invasion capacity of HCC cells in vitro (). All these findings suggested that TMPO-AS1 silencing restrained aggressive phenotypes of HCC cells.
Figure 2 Knockdown of TMPO-AS1 inhibits HCC cell aggressive behavior. (A) qRT-PCR analysis of TMPO-AS1 in TMPO-AS1-silenced SNU-387 and HCCLM3 cell. (B) CCK-8 assay for testing the viability of SNU-387 and HCCLM3 cell after knockdown of TMPO-AS1. (C) The growth of SNU-387 and HCCLM3 cell after knockdown of TMPO-AS1 was detected by colony formation assay. (D) Wound healing assay for testing the migration of SNU-387 and HCCLM3 cell after knockdown of TMPO-AS1. (E) Transwell assay for testing the invasion of SNU-387 and HCCLM3 cell after knockdown of TMPO-AS1. **P<0.01 compared with control.
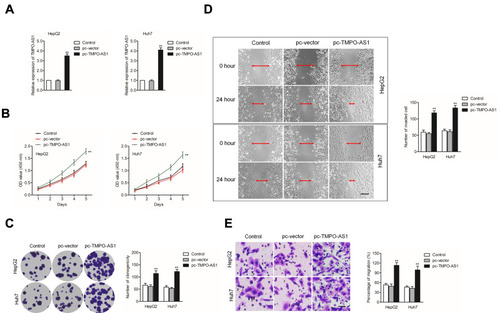
Upregulation of TMPO-AS1 Causes Opposite Trends in HCC Cell
Next, Huh7 and HepG2 cells were selected to construct TMPO-AS1-overexpressing cell lines. The qRT-PCR assay indicated that the levels of TMPO-AS1 were evidently raised in HepG2 and Huh7 cell after transfected with pc-TMPO-AS1 plasmid (). As presented in , upregulation of TMPO-AS1 significantly increased the cell viability of HepG2 and Huh7. Colony formation assay suggested that TMPO-AS1 overexpression enhanced colony formation ability of HepG2 and Huh7 cell in vitro (). Moreover, the results of wound healing and Transwell invasion displayed that overexpression of TMPO-AS1 markedly increased the aggressive phenotypes of HepG2 and Huh7 cells (). Thus, these observations further ascertained that upregulation of TMPO-AS1 promoted the aggressive phenotypes of HCC cells.
Figure 3 Overexpression of TMPO-AS1 promotes HCC cell aggressive behavior. (A) qRT-PCR analysis of TMPO-AS1 in TMPO-AS1-overexpressing HepG2 and Huh7 cells. (B) CCK-8 assay for testing the viability of HepG2 and Huh7 cell after overexpression of TMPO-AS1. (C) Colony formation assay for testing the growth of HepG2 and Huh7 cell after overexpression of TMPO-AS1. (D) Wound healing analysis for testing the migration of HepG2 and Huh7 cells after overexpression of TMPO-AS1. (E) Transwell assay for testing the invasion of HepG2 and Huh7 cell after overexpression of TMPO-AS1. **P<0.01 compared with control.
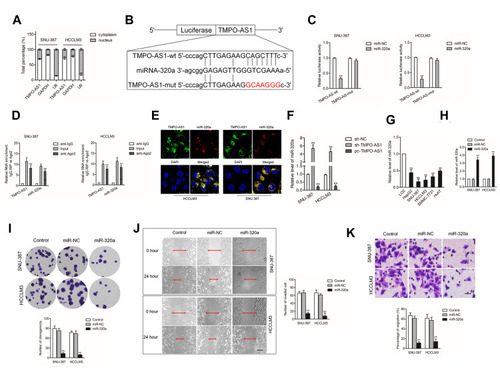
TMPO-AS1 Acts as a Sponge Role of miR-320a in HCC
In order to explore whether TMPO-AS1 acted as a sponge of miRNA, the nuclear and cytoplasmic fractionation was detected. As shown in , we found that lncRNA TMPO-AS1 was primarily located in the cytoplasm. starBASE v3.0 tool was used to predict the targeting relationship between miRNAs and TMPO-AS1. As shown in , TMPO-AS1 harbors conjectural binding sites of miR-320a. To prove whether lncRNA TMPO-AS1 interacted with miR-320a, the luciferase reporter test was carried out. As shown in , miR-320a transfection markedly reduced the luciferase activity in HCC cell transfected with pmirGLO reporter carrying TMPO-AS1-wt whereas had no significant inhibitory in a cell transfected with reporter carrying TMPO-AS1-mut. The RIP assay showed that miR-320a was significantly enriched by TMPO-AS1 probe in both HCCLM3 and SNU-387 cells (). RNA-FISH also showed that TMPO-AS1 co-localizes with miR-320a in both HCCLM3 and SNU-387 cells (). Moreover, we found that depletion of TMPO-AS1 raised the expression of miR-320a whereas TMPO-AS1 overexpressing reduced the expression of miR-320a in HCC cell (). More importantly, miR-320a was down-expressed in HCC compared with that in non-cancerous tissues (Supplementary Figure 1A). The Spearman correlation analysis revealed that the miR-320a level was inversely associated with TMPO-AS level in HCC tissues (Supplementary Figure 1B). To explore the function of miR-320a in HCC, qRT-PCR assay was conducted to measure the levels of miR-320a in HCC cell. As showed in , the level of miR-320a was lower in HCC cell when compared with that in LO2 cells. Then, miR-320a mimic was transfected into HCCLM3 and SNU-387 cell and the transfection efficiency were confirmed by qRT-PCR analysis (). The following colony formation assay also shows that the growth of HCC cell was dramatically declined in miR-320a transfected group (). As expected, the migratory capacity and invasion of HCC cell were also inhibited by miR-320a ().
Figure 4 TMPO-AS1 serves as a sponge of miR-320a. (A) The localization of TMPO-AS1 in cells (SNU-387 and HCCLM3) was confirmed by nuclear-cytoplasmic fractionation. (B) The potential binding sites between miR-320a and TMPO-AS1 were hypothesized using bioinformatics analysis starBase v.2.0 database. (C) The targeted binding effects between miR-320a and TMPO-AS1-wt or TMPO-AS1-mut in SNU-387 and HCCLM3 cells were detected using luciferase reporter assay. **P<0.01 compared with miR-NC. (D) RIP assay was conducted to confirm the binding ability between TMPO-AS1 and miR-320a. Both TMPO-AS1 and miR-320a were significantly enriched in Ago2 immunoprecipitate. **P<0.01 compared with anti-IgG. (E) RNA-FISH analysis showed that miR-320a co-localizes with TMPO-AS1 in SNU-387 and HCCLM3 cells. (F) The relative expression of miR-320a in SNU-387 and HCCLM3 cell transfected with sh-TMPO-AS1 or pc-TMPO-AS1 was measured by RT-qPCR. **P<0.01 compared with sh-NC. (G) The levels of miR-320a in HCC cells and LO2 cells were determined by qRT-PCR assay. **P<0.01 compared with LO2. (H) Cell viability of SNU-387 and HCCLM3 cell after miR-320a upregulation was conducted by performing CCK-8 assay. (I) The proliferation of miR-320a overexpressed SNU-387 and HCCLM3 cells was examined by colony formation assay. (J) The migration of miR-320a overexpressed SNU-387 and HCCLM3 cells was examined by wound healing assay. (K) The invasion of miR-320a overexpressed SNU-387 and HCCLM3 cell was examined by Transwell invasion assay. **P<0.01 compared with control.
SERBP1 Is a Target Gene of miR-320a
To predict the targets of miR-320a, five prediction tools (targetScan, picTar, RNA22, PITA and miRanda) were used and total six genes (E2F7, SOX4, NRP1, ZFP91, AP3M1 and SERBP1) were obtained from five bioinformatics analysis. SERBP1 was focus owing to its role in the progression of cancers, including HCC and the binding sites between SERBP1 miR-320a as shown in .Citation13–Citation15 Then, the result of luciferase reporter gene test suggested that transfection of markedly decreased the luciferase activity in HCC cell transfected with pmirGLO reporter carrying SERBP1-wt whereas had no significant inhibitory in a cell transfected with reporter carrying SERBP1-mut (). Besides, we observed that TMPO-AS1 silencing and overexpression of miR-320a conspicuously lessened the mRNA level and expression of SERBP1 in HCC cell (). In addition, SERBP1 expression was markedly higher in HCC tissue (). Furthermore, the level of SERBP1 was positively associated with TMPO-AS1 expression and negatively associated with the expression of miR-320a in HCC tissue (). The above findings manifested that SERBP1 was a target of miR-320a and its level was positively modulated by TMPO-AS1.
Figure 5 SERBP1 is a target gene of miR-320a. (A) Venn diagram depicted a common set of target genes of miR-320a in five prediction tools (targetScan, picTar, RNA22, PITA and miRanda). (B) The potential binding sites between miR-320a and SERBP1 were predicted by starBase v.3.0 database. The targeted binding effects between miR-320a and SERBP1-wt or SERBP1-mut in SNU-387 and HCCLM3 cells were detected using luciferase reporter assay. (C) The mRNA level of SERBP1 in SNU-387 and HCCLM3 cell transfected with miR-320a or sh-TMPO-AS1 was detected using qRT-PCR assay. **P<0.01 compared with miR-NC. (D) The protein expression of SERBP1 in SNU-387 and HCCLM3 cell transfected with miR-320a or sh-TMPO-AS1 was detected using immunofluorescence assay. (E) The expression levels of SERBP1 in HCC tissues and non-cancerous tissues were detected using qRT-PCR assay. **P<0.01 compared with non-cancerous. (F) Pearson’s correlation analysis of the relationship between TMOP-AS1 expression and SERBP1 expression level in HCC tissues. (G) Pearson’s correlation analysis of the relationship between miR-320a level and SERBP1 expression level in HCC tissues.
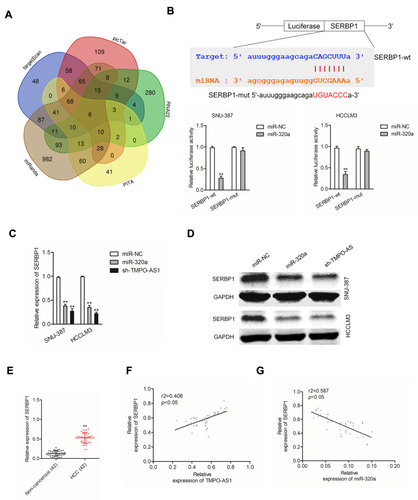
Downregulated miR-320a or SERBP1 Silencing Reverses the Effect of TMPO-AS1 in HCC Cell
Due to the sponge action of TMPO-AS1 on miR-320a, we thus inferred that TMPO-AS1 might regulate the aggressiveness of HCC via interacting with miR-320a. SNU-387 cell was divided into different groups, respectively (sh-NC, sh-TMPO-AS1, sh-TMPO-AS1 plus miR-320a inhibitor, pc-vector, pc-TMPO-AS1, pc-TMPO-AS1 plus si-SERBP1). The results of Western blot assay suggested that by contrast in the sh-NC group, the protein expression of SERBP1 was significantly reduced by sh-TMPO-AS1. However, the expression was restored in sh-TMPO-AS1 plus miR-320a inhibitor group (). By using the colony formation assay, we found that only in the cells transfected with sh-TMPO-AS1 alone, cell growth was repressed. In contrast, when cells were cotransfected with sh-TMPO-AS1 plus miR-320a inhibitor, the cell growth was remarkably reduced (). Consistently, the impacts of sh-TMPO-AS1 on the migratory and invasive abilities were abolished by cotransfection with miR-320a inhibitor (). Furthermore, the promoting effect of TMPO-AS1 overexpression on the growth, migration and invasion abilities of SNU-387 cell was abolished by si-SERBP1 (–). To confirm the sponge action of TMPO-AS1 on miR-320a, HCCLM3 cell was transfected with pc-TMPO-AS1 or transfected with pc-TMPO-AS1 plus miR-320a. The result of qRT-PCR assay suggested that by contrast in the pc-vector group, the level of miR-320 was significantly reduced by pc-TMPO-AS1. However, the expression of miR-320a was restored in pc-TMPO-AS1 plus miR-320a (). Meanwhile, the level of SERBP1 was significantly increased by pc-TMPO-AS1. However, the level of SERBP1 was decreased by miR-320a in the presence of pc-TMPO-AS1 (). By using the colony formation assay, we found that the growth of HCCLM3 cell transfected with pc-TMPO-AS1 was enhanced. In contrast, when the cell was cotransfected with pc-TMPO-AS1 plus miR-320a, the cell growth was not remarkably reduced (). Consistently, the promoting impacts of pc-TMPO-AS1 on the migratory and invasive abilities were abolished by cotransfection with miR-320a (). These data suggested that downregulation of TMPO-AS1 regulated the aggressiveness of HCC cell through modulating the miR-320a/SERBP1 axis.
Figure 6 SERBP1 is regulated by TMPO-AS1. (A) SNU-387 cell was transfected with sh-NC, or sh-TMPO-AS1, or sh-TMPO-AS1 plus miR-320a inhibitor, respectively. SNU-387 cell was transfected with pc-vector, or pc-TMPO-AS1, or pc-TMPO-AS1 plus si-SERBP1, respectively. The expression of SERBP1 was measured by Western blotting assay. (B) The growth of SNU-387 cell was detected by colony formation assay. (C) The migration of SNU-387 cell was detected by wound healing assay. (D) The invasion of SNU-387 cell was detected by Transwell invasion assay. **P<0.01 compared with sh-NC or pc-vector, ##P<0.01 compared with sh-TMPO-AS1 or pc-TMPO-AS1. (E) HCCLM3 cell was transfected with pc-vector, or pc-TMPO-AS1, or pc-TMPO-AS1 plus miR-320a, respectively. The level of miR-320a was measured by qRT-PCR assay. (F) HCCLM3 cell was transfected with pc-vector, or pc-TMPO-AS1, or pc-TMPO-AS1 plus miR-320a, respectively. The level of SERBP1 was measured by qRT-PCR assay. (G) The growth of HCCLM3 cell was detected by colony formation assay. (H) The migration of HCCLM3 cell was detected by wound healing assay. (I) The invasion of HCCLM3 cell was detected by Transwell invasion assay. **P<0.01 compared with pc-vector, ##P<0.01 compared with pc-TMPO-AS1.
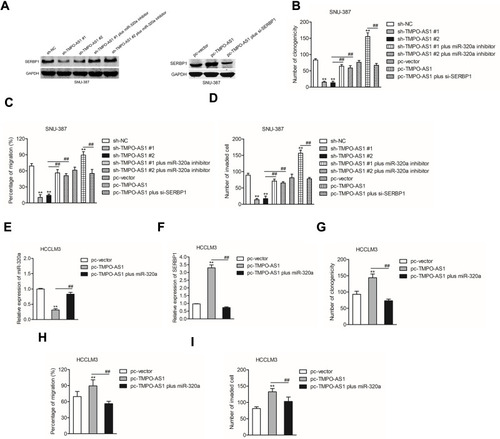
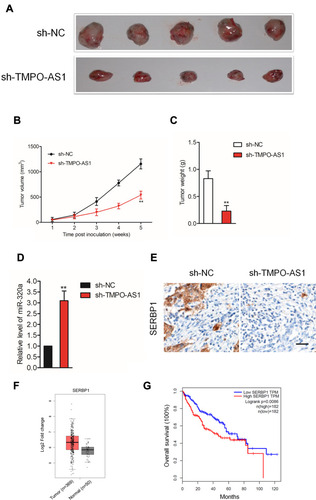
TMPO-AS1 Silencing Suppresses HCC Cell Growth in vivo
Finally, to illuminate the inhibitory effect of TMPO-AS1 silencing in vivo, xenograft tumor model was established by subcutaneous inoculation of sh-NC or sh-TMPO-AS1 stable transfected SNU-387 cell into nude mice. After 5 weeks, nude mice were sacrificed and the tumor tissues in each group were weighted. As shown in –, the tumor volume and weight in sh-TMPO-AS1 group were markedly reduced compared with those in sh-NC group. Meanwhile, qRT-PCR assay confirmed that depletion of TMPO-AS1 raised the level of miR-320a in tumor tissue (). However, the expression of the result of IHC staining using SERBP1 antibody further suggested that downregulation of TMPO-AS1 reduced the expression of SERBP1 in vivo (). These observations indicated that TMPO-AS1 silencing suppressed HCC cell growth in vivo. Finally, by using GEPIA analysis (http://gepia.cancer-pku.cn/index.html), we observed that SERBP1 was markedly overexpressed in HCC tissues (n=369) compared with that in normal tissues (n=50) (). Moreover, the higher level of SERBP1 was associated with poor OS of patients with HCC ().
Figure 7 Downregulation of TMPO-AS1 inhibits SNU-387 cell growth in vivo. (A) The appearances of the tumors harvested from euthanized SNU-387 tumor-bearing nude mice. (B, C) The tumor volume and weight of nude mice subcutaneously injected with SNU-387 cell. (D) qRT-PCR analysis of miR-320a level in tumor tissue formed by sh-NC or sh-TMPO-AS1 transfected SNU-387 cell. **P<0.01 compared with sh-NC. (E) IHC staining for SERBP1 in tumor tissue formed by sh-NC or sh-TMPO-AS1 transfected SNU-387 cell. (F) The expression of SERBP1 in HCC tissues (n=369) and normal tissues (n =50) from GEPIA analysis. Red box, HCC tissues (T); black box, normal tissue (N). (G) The correlation between expression of SERBP1 and overall survival of HCC patients from GEPIA analysis. Red, patients with high level of SERBP1; blue, patients with low level of SERBP1.
Discussion
LncRNAs, a kind of expressed non-coding RNAs are related to diverse diseases, especially the many types of cancers, including SBF2-AS1, HOTTIP and FAM201A.Citation16–Citation18 Increasing investigations have revealed the regulatory functions of lncRNAs during the development of HCC.Citation9,Citation19,Citation20 In the current study, we investigated the potential functions of TMPO-AS1 in human HCC. We observed that TMPO-AS1 was significantly overexpressed in HCC cell lines and clinical tissues. Moreover, higher expression level of lncRNA TMPO-AS1 was positively associated with advanced TNM stage and poor prognosis of patients, which indicated TMPO-AS1 could be used as a potential prognostic indicator in HCC.
LncRNAs function as oncogenic or tumor-suppressive factors in cancers. Previous report has proven that upregulation of TMPO-AS1 promotes the aggressive traits of NSCLC cell in vitro.Citation12 In addition, overexpression of TMPO-AS1 increases cell proliferation through inducing cell cycle progression and promotes migratory of prostate cancer cell.Citation11 Likewise, loss-of-functional assay in this work indicated that TMPO-AS1 knockdown repressed the proliferation, colony formation and migration as well as invasion abilities of HCC cell. Nevertheless, upregulation of TMPO-AS1 resulted in opposite trends. Furthermore, downregulation of TMPO-AS1 remarkably inhibited the growth of HCC cell in vivo. Altogether, our findings uncovered an oncogenic action of TMPO-AS1 in the cell growth and metastatic abilities of HCC cell.
Functional lncRNAs were reported to act as a sponge for miRNAs and modulate the expression of target genes at the post-transcriptional levels.Citation21,Citation22 Several lncRNAs have been found to serve as ceRNAs during the development of HCC, such as ANRIL and CDKN2B-AS1.Citation23,Citation24 In our study, by using bioinformatics analysis and luciferase reporter assay, we demonstrated that TMPO-AS1 interacted with miR-320a in HCC cell. The RIP assay further proved that TMPO-AS1 contained the potential binding sites of miR-320a and the level of miR-320a was negatively regulated by TMPO-AS1 as demonstrated by qRT-PCR assay. MiR-320a has been reported to be a suppressor in tumor cell proliferation and metastasis via targeting oncogenes or cancer suppressors.Citation25,Citation26 Consistently, we revealed that miR-320a was downregulated in HCC and cell lines. Upregulation of miR-320a remarkably weakened the growth and decreased the migration, invasion abilities of HCC cell. These observations indicated that miR-320a had an inhibitory action in the aggressive capabilities of HCC cell.
MiRNAs modulate their target genes in a sequence-specific manner and their roles in cancers have been largely investigated.Citation27–Citation29 Of note, SERBP1 was ascertained as a target of miR-320a. Importantly, TMPO-AS1 silencing and overexpression of miR-320a conspicuously lessened the mRNA level and protein expression of SERBP1 in HCC cell. The dysregulation of ceRNA regulatory axis is involved in the malignant progression of cancers. Herein, we proved that TMPO-AS1 was also a ceRNA that concurrently sponges miR-320a and upregulate its target gene, SERBP1. Importantly, we further demonstrated that lncRNA TMPO-AS1 exerts oncogenic roles in HCC through regulating miR-320a/SERBP1 axis.
Conclusions
Altogether, our research revealed the oncogenic function of TMPO-AS1 in the aggressiveness of HCC cell. Furthermore, our findings illuminated that TMPO-AS1 acted as a ceRNA to regulate the level of SERBP1 via sponging miR-320a. This study provided new insights into the mechanism of TMPO-AS1-miR-320a-SERBP1 axis in the progression of human HCC.
Disclosure
The authors report no conflicts of interest in this work.
References
- Han TS, Ban HS, Hur K, Cho HS. The epigenetic regulation of HCC metastasis. Int J Mol Sci. 2018;19(12):3978. doi:10.3390/ijms19123978
- Sun L, Wang L, Chen T, et al. LncRNA RUNX1-IT1 which is downregulated by hypoxia-driven histone deacetylase 3 represses proliferation and cancer stem-like properties in hepatocellular carcinoma cells. Cell Death Dis. 2020;11(2):95. doi:10.1038/s41419-020-2274-x32024815
- Chen X, Tang FR, Arfuso F, et al. The Emerging role of long non-coding RNAs in the metastasis of hepatocellular carcinoma. Biomolecules. 2019;10(1):66. doi:10.3390/biom10010066
- Liang WQ, Zeng CCF, Sun SM, Lu XF, Peng CY, Lin HY. Long noncoding RNA H19 is a critical oncogenic driver and contributes to epithelial-mesenchymal transition in papillary thyroid carcinoma. Cancer Manag Res. 2019;11:2059–2072. doi:10.2147/CMAR.S19590630881130
- Li Y, Han X, Li Q, et al. Long noncoding RNA HOXD-AS1 induces epithelial-mesenchymal transition in breast cancer by acting as a competing endogenous RNA of miR-421. J Cell Biochem. 2019;120(6):10633–10642. doi:10.1002/jcb.2835330730081
- Shi L, Hong X, Ba L, et al. Long non-coding RNA ZNFX1-AS1 promotes the tumor progression and metastasis of colorectal cancer by acting as a competing endogenous RNA of miR-144 to regulate EZH2 expression. Cell Death Dis. 2019;10(3):150. doi:10.1038/s41419-019-1332-830770796
- Guan MM, Rao QX, Huang ML, et al. Long noncoding RNA TP73-AS1 targets MicroRNA-329-3p to regulate expression of the SMAD2 Gene in human cervical cancer tissue and cell lines. Med Sci Monit. 2019;25:8131–8141. doi:10.12659/MSM.91629231663517
- Xu F, Zha G, Wu Y, Cai W, Ao J. Overexpressing lncRNA SNHG16 inhibited HCC proliferation and chemoresistance by functionally sponging hsa-miR-93. Onco Targets Ther. 2018;11:8855–8863. doi:10.2147/OTT.S18200530573973
- Huang X, Gao Y, Qin J, Lu S. lncRNA MIAT promotes proliferation and invasion of HCC cells via sponging miR-214. Am J Physiol Gastrointest Liver Physiol. 2018;314(5):G559–G565. doi:10.1152/ajpgi.00242.201729097358
- Yang J, Liang B, Hou S. TMPO-AS1 promotes cervical cancer progression by upregulating RAB14 via sponging miR-577. J Gene Med. 2019;21(11):e3125. doi:10.1002/jgm.312531483914
- Huang W, Su X, Yan W, et al. Overexpression of AR-regulated lncRNA TMPO-AS1 correlates with tumor progression and poor prognosis in prostate cancer. Prostate. 2018;78(16):1248–1261. doi:10.1002/pros.2370030105831
- Qin Z, Zheng X, Fang Y. Long noncoding RNA TMPO-AS1 promotes progression of non-small cell lung cancer through regulating its natural antisense transcript TMPO. Biochem Biophys Res Commun. 2019;516(2):486–493. doi:10.1016/j.bbrc.2019.06.08831230752
- Koensgen D, Mustea A, Klaman I, et al. Expression analysis and RNA localization of PAI-RBP1 (SERBP1) in epithelial ovarian cancer: association with tumor progression. Gynecol Oncol. 2007;107(2):266–273. doi:10.1016/j.ygyno.2007.06.02317698176
- Wang T, Xu L, Jia R, Wei J. MiR-218 suppresses the metastasis and EMT of HCC cells via targeting SERBP1. Acta Biochim Biophys Sin (Shanghai). 2017;49(5):383–391. doi:10.1093/abbs/gmx01728369267
- Mari Y, West GM, Scharager-Tapia C, et al. SERBP1 is a component of the liver receptor homologue-1 transcriptional complex. J Proteome Res. 2015;14(11):4571–4580. doi:10.1021/acs.jproteome.5b0037926398198
- Tang Y, Ji F. lncRNA HOTTIP facilitates osteosarcoma cell migration, invasion and epithelial-mesenchymal transition by forming a positive feedback loop with c-Myc. Oncol Lett. 2019;18(2):1649–1656. doi:10.3892/ol.2019.1046331423232
- Gao F, Feng J, Yao H, et al. LncRNA SBF2-AS1 promotes the progression of cervical cancer by regulating miR-361-5p/FOXM1 axis. Artif Cells Nanomed Biotechnol. 2019;47(1):776–782. doi:10.1080/21691401.2019.157788330856345
- He W, Qiao ZX, Ma B. Long noncoding RNA FAM201A mediates the metastasis of lung squamous cell cancer via regulating ABCE1 expression. Eur Rev Med Pharmacol Sci. 2019;23(23):10343–10353. doi:10.26355/eurrev_201912_1967231841188
- Zhang Y-T, Li B-P, Zhang B, et al. LncRNA SBF2-AS1 promotes hepatocellular carcinoma metastasis by regulating EMT and predicts unfavorable prognosis. Eur Rev Med Pharmacol Sci. 2018;22(19):6333–6341. doi:10.26355/eurrev_201810_1604430338801
- Wu Y, Zhou Y, Huan L, et al. LncRNA MIR22HG inhibits growth, migration and invasion through regulating the miR-10a-5p/NCOR2 axis in hepatocellular carcinoma cells. Cancer Sci. 2019;110(3):973–984. doi:10.1111/cas.1395030680848
- Peng WX, Koirala P, Mo YY. LncRNA-mediated regulation of cell signaling in cancer. Oncogene. 2017;36(41):5661–5667. doi:10.1038/onc.2017.18428604750
- Bhan A, Soleimani M, Mandal SS. Long noncoding RNA and cancer: a new paradigm. Cancer Res. 2017;77(15):3965–3981. doi:10.1158/0008-5472.CAN-16-263428701486
- Ma J, Li T, Han X, Yuan H. Knockdown of LncRNA ANRIL suppresses cell proliferation, metastasis, and invasion via regulating miR-122-5p expression in hepatocellular carcinoma. J Cancer Res Clin Oncol. 2018;144(2):205–214. doi:10.1007/s00432-017-2543-y29127494
- Huang Y, Xiang B, Liu Y, Wang Y, Kan H. LncRNA CDKN2B-AS1 promotes tumor growth and metastasis of human hepatocellular carcinoma by targeting let-7c-5p/NAP1L1 axis. Cancer Lett. 2018;437:56–66. doi:10.1016/j.canlet.2018.08.02430165194
- Li Y, Liu H, Shao J, Xing G. miR-320a serves as a negative regulator in the progression of gastric cancer by targeting RAB14. Mol Med Rep. 2017;16(3):2652–2658. doi:10.3892/mmr.2017.693728713899
- Zhao H, Dong T, Zhou H, et al. miR-320a suppresses colorectal cancer progression by targeting Rac1. Carcinogenesis. 2014;35(4):886–895. doi:10.1093/carcin/bgt37824265291
- Yin J, Zhuang G, Zhu Y, et al. MiR-615-3p inhibits the osteogenic differentiation of human lumbar ligamentum flavum cells via suppression of osteogenic regulators GDF5 and FOXO1. Cell Biol Int. 2017;41(7):779–786. doi:10.1002/cbin.1078028460412
- Mak CS, Yung MM, Hui LM, et al. MicroRNA-141 enhances anoikis resistance in metastatic progression of ovarian cancer through targeting KLF12/Sp1/survivin axis. Mol Cancer. 2017;16(1):11. doi:10.1186/s12943-017-0582-228095864
- Li C, Zhang Y, Zhao W, Cui S, Song Y. miR-153-3p regulates progression of ovarian carcinoma in vitro and in vivo by targeting MCL1 gene. J Cell Biochem. 2019;120(11):19147–19158. doi:10.1002/jcb.2924431297886
