Abstract
Survivin is a critical regulator of mitosis, and an inhibitor of apoptosis which is overexpressed in almost all cancers. In the current study, cell cycle profiles of normal proliferating human umbilical vein endothelial cells, prostate cancer, and lung cancer cell lines expressing varying levels of survivin and its splice variants were compared using a novel functional complementation assay. Defects in chromosome segregation and cytokinesis that were observed after depletion of endogenous survivin were not complemented by any of the survivin splice variants: survivin-2B, survivin-3B, survivin-ΔEx3, or survivin-2A when expressed exogenously at a level comparable to endogenous full-length survivin. Survivin variants were not detectable at the endogenous protein level. Cancer cells with higher levels of full-length survivin and survivin-2B expression, exhibited reduced caspase-3 activation following doxorubicin treatment and radiation. Whereas earlier studies focused on function and expression levels of survivin specific to cancer cells, the current study brings forward the essential role of survivin in normal dividing cells. Full-length survivin was found to be associated with Aurora-B kinase in the chromosomal passenger complex and depletion of survivin mimics mitotic phenotypes observed after Aurora-B kinase inhibition, in cancer as well as normal proliferating cells. Thus, our study establishes survivin as a marker of proliferation, rather than a cancer specific marker. Therefore, systemic therapeutic interventions targeting survivin will affect cancer as well as normal proliferating cells.
Introduction
Survivin is highly expressed in human cancers but not detectable in non-proliferating normal adult tissues.Citation1 Considerable attention has been given to survivin as a target for cancer therapy because survivin expression levels in cancers are correlated with pro- survival and proliferative functions, preventing apoptosis and promoting mitosis. In cancer cell lines, survivin co-localizes with Aurora-B kinase, Borealin, and inner centromere protein (INCENP) which are components of the chromosomal passenger complex (CPC).Citation2,Citation3 Aurora-B kinase is the enzymatic core of the CPC. In cancer cells, survivin expresses at a higher level during mitosis, and seems to function as a structural component of the CPC. Survivin should therefore facilitate the timely deployment of Aurora-B kinase at specific locations during mitotic progression for mitotic spindle assembly.Citation4–Citation6 However, the role of survivin in mitotic progression in normal cells is a matter of debate.
Considerable attention has been given to survivin as a target for sensitization of cancer cells to radiation and chemotherapeutic agents.Citation7 Blocking the activation of caspases by direct binding is envisioned as a mechanism by which survivin confers resistance to apoptotic signals in cancers.Citation8,Citation9 The direct role of survivin in inhibition of apoptosis was originally proposed due to its homology to the family of inhibitors of apoptosis proteins (IAP) which have the capacity to bind to caspase-3 or caspase-7.Citation1,Citation3,Citation9,Citation10 Nonetheless, structurally, survivin is different from other mammalian IAPs, as it possesses only a single baculovirus IAP repeat domain and lacks the C-terminal RING finger and caspase recruitment domains.Citation1
In addition to the predominant 16 kDa full-length form of survivin, earlier studies have identified four low abundant transcripts generated by alternative splicing of survivin pre-mRNA. Citation11–Citation14 A number of studies have been published on the possible roles of these splice variants in various cancers.Citation15–Citation21 However, none have validated unequivocally these findings at the protein level with the use of specific antibodies in cancer cell lines or tissues. The variant, survivin-2B contains an additional 23 amino acid insertion from a cryptic exon, and at least two studies have proposed the possible role of this variant in promoting apoptosis.Citation22–Citation24 Survivin-ΔEx3 has been proposed as an anti-apoptotic protein.Citation24,Citation25 However, the relevance of each variant in the physiological context of mitosis or regulation of apoptosis has not been studied and is not understood. We have therefore developed a synthetic gene-based functional complementation assay to dissect the function of survivin and its variants at varying levels of expression, and investigated their function in the mitosis and regulation of apoptosis.
There are several ongoing efforts investigating the therapeutic potential of survivin, which include development of small molecule inhibitors and monoclonal antibody therapies. However, several critical and fundamental questions on survivin biology remain unanswered, which include: (1) Is survivin essential for mitosis in both cancer as well as normal proliferating cells? (2) Do survivin splice variants play essential roles in cell division at physiological levels? and (3) What are the relative effects of survivin overexpression in mitosis versus inhibition of apoptosis? The development of a functional complementation assay and epitope tagging has allowed us to elucidate the physiological functions of survivin and variants as a regulator of cell division and apoptosis. In addition to its established roles in cancer cell proliferation, our studies clearly show that survivin is essential for proliferation of normal cells. An increase in apoptosis following survivin depletion became evident only at time points following mitotic catastrophe suggesting that the primary role of survivin is mitosis rather than inhibition of apoptosis.
Materials and methods
Cell lines
Prostate cancer cell line PC3, and lung cancer cell line H466 were purchased from ATCC (Manassas, VA) and grown at 37°C with 5% CO2 in RPMI medium (Life Technologies, Carlsbad, CA) containing 10% fetal bovine serum (FBS) with 100u/mL penicillin and streptomycin. The HUVEC cells were purchased from ScienCell (Carlsbad, CA), and cultured in ECM medium (ScienCell). Early passage cells (P2 to P6) were used in the experiments.
Generation of cell lines
The survivin constructs were generated by PCR and other conventional molecular biology techniques. For making lentiviral vector particles, 293T cells were transfected with survivin genes in lentiviral expression vector pCDH (System Biosciences, Mountain View, CA) together with plasmids encoding VSV-G protein (Stratagene, Santa Clara, CA), a packaging construct derived from HIV pΔR8. Supernatants were collected, filtered, and used for infection using polybrene as cation and 48 hours later infected cells were selected with 2 μg/mL puromycin for 72 hours.
Western blotting
50–70 μg protein from total extracts was used for probing with survivin (Cell Signaling, Danvers, MA [2808]; Novus, Littleton, CO [NB500-201]), Aurora-B kinase (Cell Signaling [3094]), Caspase-3 (R&D Systems, Minneapolis, MN [CPP32]), PARP (Cell Signaling [9542]), HA (Covance, Princeton, NJ [HA.11]) antibodies. Quantity One (v4.6.8; Bio-Rad, Hercules, CA) was used for quantitative relative signal intensity.
Apoptosis assays
Cells were treated with doxorubicin (0.5 μg or 1 μg/mL) irradiated with 6 to 8 gray x-rays from an RS-2000 Biological Irradiator (Rad Source, Suwanee, GA). Relative levels of apoptosis were compared using Annexin-V-FITC Apoptosis kit and APO-BrdU TUNEL assay kit (Life Technologies, Grand Island, NY).
Microscopy
Cells grown on coverslips were synchronized at G1/S phase by culturing in the presence of 2 mM thymidine for 16 hours. Cells were released from thymidine block by washing twice and culturing in fresh medium for 6–8 hours, fixed with 4% paraformaldehyde, permeabilized with 0.25% Triton X-100, and then stained with mouse monoclonal anti-HA antibody (Covance: HA.11 at 1:400 dilution in 5% BSA) and rabbit polyclonal anti-Aurora-B kinase (Cell Signaling: 3094 at 1:200 dilution in 5% BSA) antibody. Alexa Fluor 488 conjugated donkey anti-mouse and Alexa Fluor 594 conjugated anti-rabbit (1:400, Life Technologies) antibodies were used for detecting the respective primary antibodies. Slides were mounted in 4′,6-Diamidino-2-phenylindole (DAPI) containing medium and images were captured using a Zeiss CCD camera (AxioCam Mrm; New York, NY) attached to an ApoTome Microscope ([Axio Observer.Z1; Carl Zeiss MicroImaging GmbH, Jena, Germany] 20× or 63× objective) using Zeiss Axiovision Vs40 (v4.6.3.0) image acquisition software. Data were analyzed using Student’s t-test and were considered statistically significant if P < 0.05.
Small interference RNA and cell cycle analysis
Stealth siRNA oligonucletides targeting survivin was purchased from Life Technologies. RNAiMax oligofectamine reagent from Life Technologies was used for siRNA transfection according to the manufacturer’s protocol. Cells were collected at 48, 72, and 96 hours after siRNA, fixed with 70% methanol and prepared for flow cytometry in a BD (Franklin Lakes, NJ) FACSCalibur flow cytometer.
Results
Development of a functional complementation assay to evaluate survivin-variant specific effects on mitosis
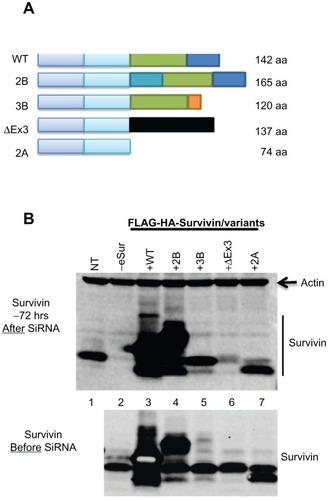
Cell lines expressing varying levels of survivin (full-length), survivin-2B, survivin-3B, survivin-ΔEx3, and survivin-2A () in prostate (PC3) and lung (H466) cancer cell lines, as well as normal human umbilical vein endothelial cells (HUVEC) were generated. Relative levels of expression of each variant and full-length survivin were compared by Western blot analysis using an antibody detecting a conserved N-terminal region. Over 38-fold overexpression of survivin and survivin-2B could be achieved by repeated infection with lentiviral particles. However, overexpression of survivin-3B, survivin-ΔEx3, and survivin-2A could not be achieved even after repeated infection (, Supplemental Figure 1C), potentially due to a dominant negative effect associated with these variants, which contain most of the dimerization domain. Similar levels of survivin and variant proteins were obtained following infections with viral particles for expression of untagged as well as an N-terminal FLAG-HA epitope tagged survivin ( and Supplemental Figure 1B and C).
Figure 1 Development of the functional complementation assay and validation of expression of the survivin variants. (A) Diagrammatic representation of the organization of human survivin gene. Splice variant survivin-2B has an extra exon with 23 amino acids inserted after exon 2. Survivin-3B has seven amino acids introduced after exon 3 and exon 4 are deleted. Survivin-ΔEx3 has an altogether different sequence at the C-terminus due to a frame shift mutation. Survivin-2A has only the first two exons. (B) Western blots detecting expression of the full-length survivin and variants in PC3 cells, using an antibody against N-terminal region capable of detecting all variants and full-length survivin. The upper panels show the survivin levels 72 hours after transfection with siRNA oligonucleotide specifically targeting endogenous survivin, and the lower panel shows the relative levels of endogenous and the introduced survivin before transfection. Overexpression of FLAG-HA tagged full-length survivin WT (lane 3), surivin-2B (lane 4) and relatively lower levels of survivin-3B (lane 5), survivin-ΔEx3 (lane 6), and survivin-2A (lane 7). Smaller derivative products of survivin (less than 10%, were detected in cells with very high levels of expression) as reported from earlier studies that used overexpression approaches.
Abbreviations: NT, non-targeting control; –eSur, endogenous survivin.
To compare the effects specific to each of the variants, a functional complementation assay was developed. A region in exon 2 that is common to all variants was identified as a target for small interfering RNA (siRNA) to knockdown endogenous survivin. Modified genes encoding full-length survivin or variants with seven silent mutations at the siRNA targeting site were introduced using a lentiviral expression system. Following selection, endogenous survivin was specifically knocked down by siRNA. Over 95% knockdown of the endogenous survivin protein was observed 72 hours after siRNA, with no apparent reduction in the signal from the introduced gene ( and Supplemental Figure 1C). This allowed the evaluation of mitotic effects that are specific to the expression of individual variants. In addition, this has also allowed the comparative analysis of apoptotic regulatory effects of survivin overexpression following exposure to radiation or chemotherapeutic agent doxorubicin.
Full-length survivin is essential and sufficient for cell division
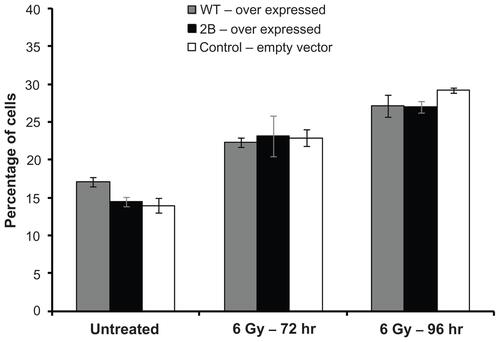
The cell cycle profile in cells with varying levels of expression was compared by flow cytometry () in normal cells as well as cancer cells. Even in cells with over 100-fold overexpression of full-length survivin, no significant difference in cell cycle profiles could be detected relative to control cells. Mitotic defects and alterations in cell cycle profiles due to the depletion of endogenous survivin were evident from the gradual increase in the percentage of cells with 4N (G2/mitosis) and 8N (multinucleated) DNA content at various time points after siRNA. In PC3 cells, comparison to the background level which had an average of 9% polyploidy in cells transfected with a non-targeting scrambled oligonucleotide control, an average of 25.3%, 43.6%, and 51.5% polyploidy was detected respectively at 48, 72, and 96 hours after siRNA transfection ( and Supplemental Figure 2). Similar cell cycle profile and levels of polyploidy were observed in cells expressing the variants survivin-3B, survivin-ΔEx3, and survivin-2A, ( and Supplemental Figure 2), suggesting that these variants are defective for mitotic function. All experiments were repeated independently at least three times. Consistent with the increase in polyploidy detected by flow cytometry, the nuclear volume and multilobed nuclear structure were evident from DAPI staining (). The chromosome segregation defects and mitotic catastrophe resulting from expression of each of variants alone were further quantified by counting the multilobed nuclei from DAPI stained cells after siRNA transfection (72 hours after siRNA: P-values: 3B: 0.00012; ΔEx3: 0.00001, 2A: 0. 012, WT: 0.28; 2B: 0.0735). Similar cell cycle profiles were obtained from early passage HUVEC cells expressing variants alone (), confirming that full-length survivin with intact C-terminal domain is essential for mitotic progression in normal cells as well. Thus, the internally controlled functional complementation assay conducted in cancer as well as normal proliferating cells confirmed that full-length survivin is essential for chromosome segregation and cytokinesis.
Figure 2 Effects of individual survivin variants on mitosis and cytokinesis after siRNA targeting the endogenous survivin. Representative FACS profile of PC3 (A) and human umbilical vein endothelial cells (B) showing survivin dependent effects as changes in cell cycle stages and polyploidy. Cell cycle stages, DNA content (2N, 4N, and ≤8N), percent polyploidy observed 72 hours after the siRNA targeting endogenous survivin, analyzed from 10,000 cells are marked. (C) Representative images showing nuclear morphology with increase in multilobed nuclei in PC3 cells as marked at 72 hours after siRNA detected by DAPI staining.
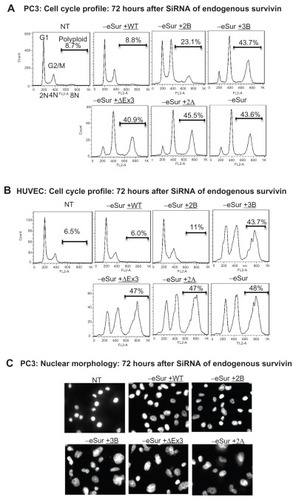
Survivin-2B, the only variant which contains C-terminal microtubule binding domain, was able to partially execute the mitotic function, when expressed at a very high and physiologically unachievable level (Supplemental Figure 2B). However, at a level equivalent to that of endogenous full-length survivin, survivin-2B was not sufficient for rescuing completely the mitotic defects resulting from depletion of endogenous survivin (). Thus, analysis of cells with varying levels of expression of survivin-2B confirmed that full-length wild type survivin is essential for mitotic function in the physiological context.
Expression of C-terminal truncation variants survivin-3B, survivin-ΔEx3, and survivin-2A individually at a level near to the endogenous full-length survivin () exhibited abnormal mitotic phenotypes in both PC3 as well as HUVEC cells. The cell division defects were similar to that observed in control knockdowns, with more than 40% of cells having 8 N or higher DNA content 72 hours post siRNA transfection (). Mitotic catastrophe and chromosome segregation defects were evident from the marked increase in multinucleated cells as compared by fluorescence activated cell sorting (FACS) analysis and DAPI staining and microscopy. An increase in the population of sub-G1 apoptotic cells was observed in these cells 96 hours after siRNA (Supplemental Figure 2). The increases in sub-G1 fraction were detected only at later time points (96 hours post siRNA), suggesting that the induction of apoptosis is primarily due to mitotic catastrophe, rather than the direct inhibition of caspases by survivin.
Association of survivin with the CPC is essential for mitosis in normal as well as cancer cells
To determine whether or not the association of survivin with the CPC is essential for cell division in non-cancerous, but proliferating cells, we used immunofluorescence (IF) and genetic approaches. Proliferating early passage HUVEC cells expressing epitope-tagged survivin were used to examine its localization during various stages of mitosis. Given our findings that N-terminal tags do not affect mitotic function, N-terminally FLAG-HA tagged proteins were used for localization studies.
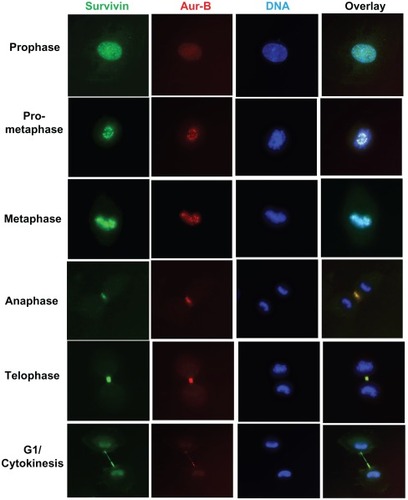
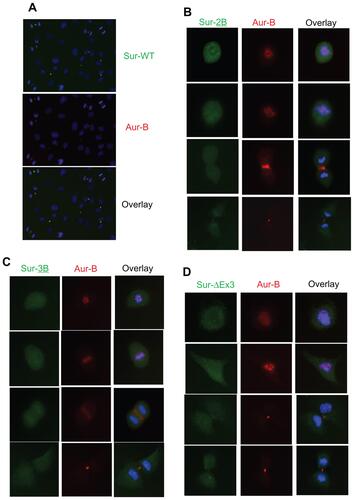
IF studies clearly showed that full-length survivin is associated with Aurora-B kinase, with a characteristic pattern of CPC movement during mitotic progression (). Survivin was found co-localized with Aurora-B kinase in the centromere during prometaphase and metaphase; spindle midzone during anaphase and at the midzone in telophase and cytokinesis. Survivin and Aurora-B kinase were co-localized at all stages in mitosis and cytokinesis in over 95% of the cells examined (Supplemental Figure 3A). Even under conditions of overexpression of survivin, signals were specifically localized with Aurora-B kinase. The CPC association of survivin during mitotic spindle assembly and progression in cancer as well as normal cells were confirmed by real time video microscopy using green fluorescence protein (GFP) tagged full-length survivin in PC3 as well as HUVEC cells (data not shown). This data confirmed that survivin and Aurora-B kinase are associated with the same machinery, essential for proliferation in cancer cells as well as normal cells.
Figure 3 Functional CPC association of survivin with Aurora-B kinase during mitosis in human umbilical vein endothelial cells. Cells expressing N-terminal FLAG-HA tagged full-length survivin, at various stages in mitosis were co-stained with anti-HA (green) and Aurora-B kinase (red) antibodies, and DNA counterstained with DAPI.
Survivin variants are detected primarily in the cytoplasm during mitosis
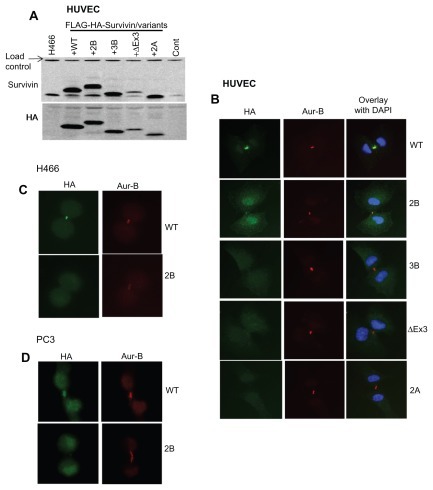
The subcellular localization of survivin and its splice variants has been extremely difficult because the available antibodies cross-reacted with a number of nuclear as well as cytoplasmic proteins (Supplemental Figure 1A, and data not shown). Therefore, cells expressing epitope tagged survivin variants were generated to circumvent possible errors in detection and quantification. Protein expression levels of the respective variants were confirmed by Western blotting using HA as well as survivin antibody (). Subcellular localization of each of the variants during mitosis was determined by indirect IF using HA and Aurora-B kinase antibodies. IF has shown that, during the mitotic phase, full-length survivin localizes in the nucleus; survivin-2B in the nucleus as well as in cytoplasm; while survivin-3B, survivin-ΔEx3, and survivin-2A are primarily in the cytoplasm ( and Supplemental Figure 3B–D). A small fraction of survivin-2B was found associated with CPC, which was apparent from partial co-localization with Aurora-B kinase at the midbody during cytokinesis. This was consistent with the findings from flow cytometry analysis of survivin-2B expressing cells (). A similar localization pattern for variants were observed in PC3 and H466 cancer cells (). Thus, flow cytometry data and microscopy following epitope tagging shows that at the physiological level only full-length survivin is associated with Aurora-B kinase in CPC. Functional association of survivin variants with any specific cellular compartment such as mitochondria could not be established.
Figure 4 Subcellular localization of survivin variants. (A) Western blot showing the expression of FLAG-HA tagged survivin in human umbilical vein endothelial cells detected using survivin and HA antibodies. (B) Localization of survivin variants in human umbilical vein endothelial cells at stages in mitosis co-stained with Aurora-B kinase and counterstained with DAPI. Immunofluorescence of full-length survivin and survivin-2B stained with Aurora-B kinase and DAPI in H466 (C) and PC3 (D) cell lines.
Full-length survivin and survivin-2B overexpression inhibits apoptosis in cancer cells
Since the inhibition of apoptosis is proposed to contribute to therapeutic resistance in cancer cells, the effect of overexpression of full-length survivin and survivin-2B on apoptosis was compared in cancer cells. PC3 cells stably expressing higher levels of survivin and survivin-2B (104- and 38-fold respectively, ), and an empty vector control were used to investigate their effects on the inhibition of apoptosis. In this experiment, cell lines expressing C-terminal truncation variants could not be included because of the dominant negative effect of these variants. To compare the expression level dependent apoptosis inhibition, cells over expressing survivin and survivin-2B were treated in parallel with a single dose of radiation, collected at various time points, and relative levels of cleaved PARP and cleaved caspase-3 signals were compared. A nominal decrease in radiation induced apoptosis was observed in cells over expressing full-length survivin and to a lesser extent even in survivin-2B cells as seen by the decrease in cleaved PARP signal (). The effect of over-expression of survivin and survivin-2B in doxorubicin induced apoptosis was also compared using the Annexin-V/ propidium iodide-based apoptosis assay. However, the apoptotic inhibitory effect detected in survivin or survivin-2B overexpression cells was not substantial (Supplemental Figure 4, and data not shown).
Figure 5 Effect of full-length survivin and survivin-2B overexpression on apoptosis in PC3 cells. (A) Western blots showing the overexpression of survivin and survivin-2B. (B) Reduction in cleaved caspase-3 (cCasp3) and cleaved PARP (cPARP) with overexpression of survivin in irradiated and doxorubicin treated cells and control. Signals were normalized using an internal load control and the relative levels of uncleaved to cleaved PARP were quantified. A 2.3-fold reduction in cleaved PARP was detected in cells with survivin overexpression, treated with a suboptimal dose of doxorubicin (0.5 μM, 72 hours). Variations found with radiation (8 Gy, 80 hours) were not significant. A 0.7-fold reduction in cleaved Caspase-3 signal was detected in cells over expressing full-length survivin treated with doxorubicin (0.5 μM for 72 hours). (C) Relative levels of DNA fragmentation detected by TUNEL assay 72 hours after 1 μM doxorubicin treatment.
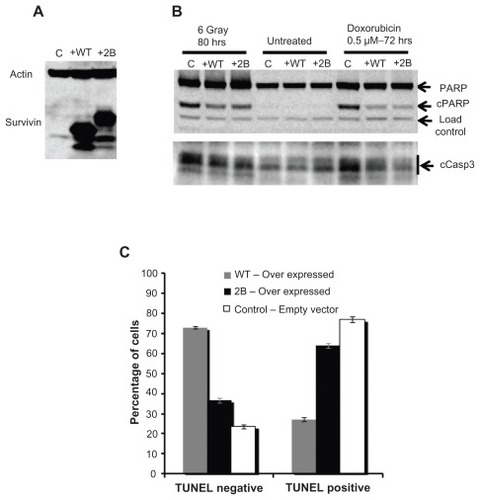
Since the inhibitory effect of survivin on radiation induced apoptosis detected from two different assays was not substantial, we compared the effects of the topoisomerase II inhibitor, doxorubicin. Doxorubicin treatment provides a consistent level of damage over the course of the experiment and hence is expected to cause higher levels of apoptosis. Reduction in cleaved caspase-3 (0.7-fold) and cleaved PARP signals (2.3-fold) in full-length survivin over expressing cells treated with a suboptimal dose of doxorubicin (0.5 μM, 72 hours) were observed (). Consistent with this, a significant reduction (2.4-fold) in DNA fragmentation was detected from a TUNEL-based assay (APO BrdU kit, Life Technologies) in cells with full-length survivin overexpression, when assayed 72 hours after treatment with 1 μM doxorubicin (). The apoptosis inhibitory effect of survivin and survivin-2B were similar. Of note, contrary to the previous reports,Citation16,Citation23 no pro-apoptotic effects of survivin-2B were detected in our internally controlled experiments ().
The apoptotic inhibitory effect of survivin overexpression following radiation damage was also investigated by comparing protein markers (cleaved caspase-3 and cleaved PARP), Annexin V, and TUNEL assays in cells collected at various time points (36 hours to 96 hours) following varying doses of radiation (6 gray to 10 gray). However, the inhibition of radiation induced apoptosis detected in survivin or survivin-2B overexpression observed were not significant (data not shown). The differences in the levels of apoptotic inhibition from radiation versus doxorubicin treated cells were predicted due to the intensity of the damage, timing, and mechanism of induction of apoptosis. While many of the DNA breaks incurred by single dose radiation damage get promptly repaired, continuous DNA damage is expected from doxorubicin. Moreover, cell death by radiation is primarily mediated through mitotic catastrophe rather than due to DNA breaks, as most breaks are usually repaired within hours. Apoptosis following radiation treatment in these cells are likely the result of mitotic catastrophe as cells attempt to undergo mitosis with misrepaired lesions. Thus, variations in the levels, timing, and detection of these signals were expected. Taken together, these data confirm that, while full-length survivin is essential for mitotic progression, the projected apoptotic regulatory effects of survivin or survivin-2B are nominal.
Discussion
The genetic studies presented in this paper shed light on several outstanding questions on the biology of survivin. Our studies clearly show that survivin splice variants, even when present at higher levels, do not contribute significantly to pro-survival or proliferative functions in cancer cells such as promoting mitosis and inhibition of apoptosis. Thus, the significance and relevance of survivin variants as a prognostic or predictive marker are questioned.Citation13,Citation19,Citation21,Citation26–Citation37 Splicing is a posttranscriptional event and a proportional increase in variants is expected when the survivin promoter is activated. Altered levels of splice variants are expected if mutations and polymorphism in the regulatory elements that regulate splicing are present.
The current genetic studies establish that the C-terminal microtubule binding region of survivin is essential for chromosome segregation and completion of cell division. Survivin functions as a dimer with a tie-knot like structure in the CPC, and C-terminal coiled-coiled structure of both molecules are likely essential for microtubule binding. The relative abundance of variants that can dimerize with full-length survivin and lack the C-terminal domain are predicted to modulate CPC function.
The exact mechanism by which higher levels of survivin promote proliferation and survival of cancer cells, conferring resistance to therapy is still not fully clear. Studies in model systems have shown that survivin-like IAP molecules predominantly participate in cytokinesis, associated with Aurora-B kinase and mitotic machinery.Citation38,Citation39 Even when overexpressed, survivin signals were found localized to Aurora-B kinase and hence survivin is likely a limiting factor for promoting cell growth. Higher levels of survivin likely provide an enhanced structural integrity to mitotic machinery, potentially helping to bypass the mitotic spindle checkpoints common in cancers. Enhanced structural support provided by higher levels of survivin could be contributing to better centromere and kinetochore attachment, bipolar spindle formation, and cytokinesis. Survivin overexpression likely provides better integrity and structural support to the CPC and this helps to deliver Aurora-B kinase at its destined locations during mitotic events. Aurora-B kinase is required for phosphorylation of mitotic regulators, which include histone H3 at serine 10 and centromere protein CENP-A which are critical for chromatin condensation. The other targets include micro-tubule destabilizing Kin I kinesin and MCAK, important for spindle assembly.
The present study further established that survivin is essential for cell division in cancers as well as normal cells. Identifying survivin as a marker for proliferation is consistent with reports showing elevated levels of survivin during liver regeneration following hepatectomy.Citation40 In normal cells, the expression of survivin from the endogenous promoter is restricted to the G2/M phase of the cell cycle. Given that most adult normal tissues are not proliferating, it has been difficult to detect survivin in normal adult tissues. The systemic approaches targeting survivin in cancers may affect proliferating normal cells as well and future studies should focus on a more targeted approach specific to cancer cells.
Acknowledgments
This work was supported by the NIH/NCI awards R01CA108633; RC2CA148190 and Institutional Research Funds.
Supplementary figures
Figure S1 (A) Western blot showing non-specific immunoreactivity of two commonly used commercial survivin antibodies, in extracts made from several cell lines as marked. Lanes 1, 2, 3: Total extract from human umbilical vein endothelial cells, H466, and PC3 cells probed with anti-survivin rabbit monoclonal antibody (Cell Signaling: 2808), Lane 4: PC3 extracts probed with anti-survivin polyclonal antibody from Novus Biological (NB-500-201). Endogenous survivin (lower band) and non-specific signals are marked; (B) Western blot detecting full-length survivin, survivin-2B and survivin-3B at a level comparable to the endogenous survivin and actin was used as loading control; (C) Western blot detecting expression of untagged survivin and all the variants in PC3 cells. The upper panel: survivin levels 72 hours after transfection with siRNA oligonucleotide specifically targeting the endogenous survivin (knock down efficiency >95%) and the lower panel shows the levels of endogenous and introduced survivin before transfection.
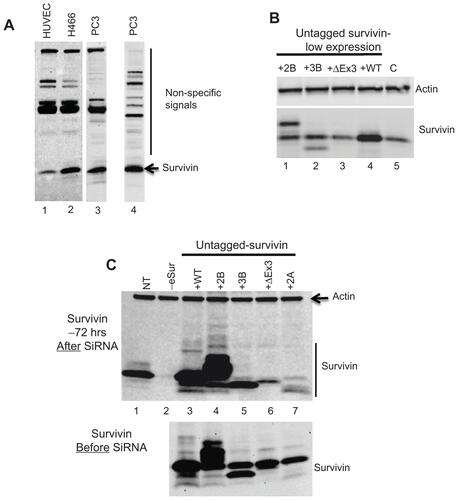
Figure S2 Flow cytometry profile of PC3 cells at 48 and 96 hours after siRNA targeting endogenous survivin showing increase in cells with polyploidy in the absence of functional endogenous survivin as marked.
Notes: (A) endogenous survivin depletion; (B) survivin 2B only; (C) survivin 3B only; (D) survivin ΔEx3 only; (E) survivin 2A only.
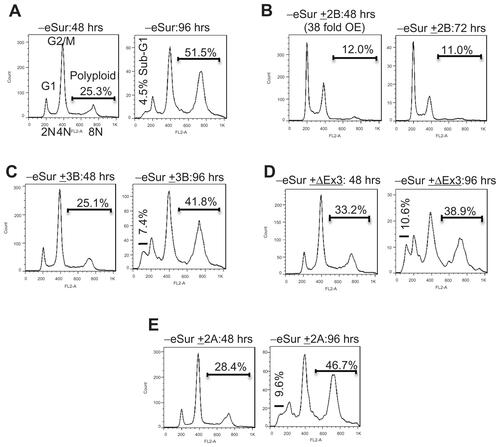
Figure S3 (A) A broader view of immunostaining showing co-localization of survivin with Aurora-B kinase in human umbilical vein endothelial cells during mitosis. Green: Survivin, Red: Aurora-B kinase, Blue: DAPI. (B–D) Immunofluorescence demonstrating predominant cytoplasmic localization of survivin variants. Human umbilical vein endothelial cells expressing respective FLAG-HA-tagged proteins were stained with HA, Aurora-B antibodies, and DAPI.
Disclosure
The authors declare no conflicts of interest in this work.
References
- AmbrosiniGAdidaCAltieriDCA novel anti-apoptosis gene, survivin, expressed in cancer and lymphomaNat Med1997389179219256286
- JeyaprakashAAKleinURLindnerDEbertJNiggEAContiEStructure of a Survivin-Borealin-INCENP core complex reveals how chromosomal passengers travel togetherCell2007131227128517956729
- RuchaudSCarmenaMEarnshawWCChromosomal passengers: conducting cell divisionNat Rev Mol Cell Biol200781079881217848966
- VaderGMedemaRHLensSMThe chromosomal passenger complex: guiding Aurora-B through mitosisJ Cell Biol2006173683383716769825
- NiggEAMitotic kinases as regulators of cell division and its check-pointsNat Rev Mol Cell Biol200121213211413462
- CarmenaMEarnshawWCThe cellular geography of aurora kinasesNat Rev Mol Cell Biol200341184285414625535
- DohiTBeltramiEWallNRPlesciaJAltieriDCMitochondrial survivin inhibits apoptosis and promotes tumorigenesisJ Clin Invest200411481117112715489959
- LiFAmbrosiniGChuEYControl of apoptosis and mitotic spindle checkpoint by survivinNature199839667115805849859993
- TammIWangYSausvilleEIAP-family protein survivin inhibits caspase activity and apoptosis induced by Fas (CD95), Bax, caspases, and anticancer drugsCancer Res19985823531553209850056
- AsanumaKKobayashiDFuruyaDTsujiNYagihashiAWatanabeNA role for survivin in radioresistance of pancreatic cancer cellsJpn J Cancer Res20029391057106212359060
- BadranAYoshidaAIshikawaKIdentification of a novel splice variant of the human anti-apoptopsis gene survivinBiochem Biophys Res Commun2004314390290714741722
- CaldasHHonseyLEAlturaRASurvivin 2alpha: a novel Survivin splice variant expressed in human malignanciesMol Cancer2005411115743529
- CaldasHJiangYHollowayMPSurvivin splice variants regulate the balance between proliferation and cell deathOncogene200524121994200715688031
- ConwayEMPollefeytSCornelissenJThree differentially expressed survivin cDNA variants encode proteins with distinct anti-apoptotic functionsBlood20009541435144210666222
- SampathJPelusLMAlternative splice variants of survivin as potential targets in cancerCurr Drug Discov Technol20074317419117986000
- ZhuNGuLFindleyHWLiFZhouMAn alternatively spliced survivin variant is positively regulated by p53 and sensitizes leukemia cells to chemotherapyOncogene200423457545755115334064
- WuttigDKunzeDFuesselSAre overexpressed alternative survivin transcripts in human bladder cancer suitable targets for siRNA-mediated in vitro inhibition?Int J Oncol20073061317132417487351
- LingXYangJTanDDifferential expression of survivin-2B and survivin-DeltaEx3 is inversely associated with disease relapse and patient survival in non-small-cell lung cancer (NSCLC)Lung Cancer200549335336115936846
- SpanPNTjan-HeijnenVCHeuvelJJde KokJBFoekensJASweepFCDo the survivin (BIRC5) splice variants modulate or add to the prognostic value of total survivin in breast cancer?Clin Chem20065291693170016873289
- LiFRole of survivin and its splice variants in tumorigenesisBr J Cancer200592221221615611788
- FangusaroJRJiangYHollowayMPSurvivin, Survivin-2B, and Survivin-deItaEx3 expression in medulloblastoma: biologic markers of tumour morphology and clinical outcomeBr J Cancer200592235936515655550
- MirzaAMcGuirkMHockenberryTNHuman survivin is negatively regulated by wild-type p53 and participates in p53-dependent apoptotic pathwayOncogene200221172613262211965534
- LingXChengQBlackJDLiFForced expression of survivin-2B abrogates mitotic cells and induces mitochondria-dependent apoptosis by blockade of tubulin polymerization and modulation of Bcl-2, Bax, and survivinJ Biol Chem200728237272042721417656368
- MahotkaCWenzelMSpringerEGabbertHEGerharzCDSurvivin-deltaEx3 and survivin-2B: two novel splice variants of the apoptosis inhibitor survivin with different antiapoptotic propertiesCancer Res199959246097610210626797
- KnauerSKBierCSchlagPThe survivin isoform survivin-3B is cytoprotective and can function as a chromosomal passenger complex proteinCell Cycle20076121502150917582222
- KriegAMahotkaCKriegTExpression of different survivin variants in gastric carcinomas: first clues to a role of survivin-2B in tumour progressionBr J Cancer200286573774311875736
- MahotkaCKriegTKriegADistinct in vivo expression patterns of survivin splice variants in renal cell carcinomasInt J Cancer20021001303612115583
- ChengZHuLFuWZhangQLiaoXExpression of survivin and its splice variants in gastric cancerJ Huazhong Univ Sci Technolog Med Sci200727439339817828494
- YamadaYKuroiwaTNakagawaTTranscriptional expression of survivin and its splice variants in brain tumors in humansJ Neurosurg200399473874514567610
- AtlasiYMowlaSJZiaeeSADifferential expression of survivin and its splice variants, survivin-DeltaEx3 and survivin-2B, in bladder cancerCancer Detect Prev200932430831319186007
- BoidotRVegranFLizard-NacolSPredictive value of survivin alternative transcript expression in locally advanced breast cancer patients treated with neoadjuvant chemotherapyInt J Mol Med200923228529119148555
- De MariaSPannoneGBufoPSurvivin gene-expression and splicing isoforms in oral squamous cell carcinomaJ Cancer Res Clin Oncol2009135110711618642030
- FutakuchiHUedaMKandaKFujinoKYamaguchiHNodaSTranscriptional expression of survivin and its splice variants in cervical carcinomasInt J Gynecol Cancer20071751092109817877643
- HeXFWenDGHouJQHeJCenJNExpressions of survivin and the splice variants survivin-2B and survivin-DeltaEx3 in bladder cancer and their clinical significanceAi Zheng200928111209121319895744
- NakanoJHuangCLiuDThe clinical significance of splice variants and subcellular localisation of survivin in non-small cell lung cancersBr J Cancer20089861109111718283319
- NouraeeNMowlaSJOzhandAParvinMZiaeeSAHatefiNExpression of survivin and its spliced variants in bladder tumors as a potential prognostic markerUrol J20096210110819472128
- VegranFBoidotROudinCDef rainCRebucciMLizard-NacolSAssociation of p53 gene alterations with the expression of antiapoptotic survivin splice variants in breast cancerOncogene200726229029716847456
- UrenAGBeilharzTO’ConnellMJRole for yeast inhibitor of apoptosis (IAP)-like proteins in cell divisionProc Natl Acad Sci U S A19999618101701017510468581
- SpeliotesEKUrenAVauxDHorvitzHRThe survivin-like C. elegans BIR-1 protein acts with the Aurora-like kinase AIR-2 to affect chromosomes and the spindle midzoneMol Cell20006221122310983970
- DeguchiMShirakiKInoueHExpression of survivin during liver regenerationBiochem Biophys Res Commun20022971596412220508