Abstract
Background
A large body of evidence has shown that circular RNAs (circRNAs) play a significant role in the progression of some malignant cancers, including hepatocellular carcinoma (HCC). However, the complex mechanism of circRNAs in hepatocellular carcinoma has not been clarified.
Methods
We identified circRNAs by microarray analysis and quantitative real-time polymerase chain reaction (RT-qPCR). We also carried out bioinformatics analysis, luciferase reporter assays, and RNA pull-down assays to define the relationship between microRNA (miR)-543 and circARPP21. Through silencing and overexpression of circARPP21, we investigated the effects of circARPP21 on proliferation, migration, and invasion abilities of HCC cells in vitro and in vivo.
Results
In this study, we found that a novel circRNA, circARPP21 (hsa_circ_0123629), exerts a strong effect on HCC progression. Reduced expression of circARPP21 in HCC patients is correlated with larger tumor size, higher tug-lymph node metastasis (TNM) stage, and poor prognosis as indicated by elevated levels of alpha-fetoprotein (AFP). Conversely, higher expression of circARPP21 can increase leukemia inhibitory factor receptor (LIFR) expression by sponging miR-543. Finally, overexpression of miR-543 can reverse the anti-proliferation and anti-metastasis effects of circARPP21.
Conclusion
The circARPP21/miR-543/LIFR axis suppresses the proliferation, invasion, and migration of hepatocellular carcinoma cells. In addition, circARPP21 can serve as a biomarker in HCC, thus offering a potential new approach to HCC therapy.
Introduction
With multiple causes, including HBV infection and liver cirrhosis, hepatocellular carcinoma ranks sixth in cancer incidence and fourth in cancer mortality worldwide.Citation1 Despite the development of numerous new alternative therapies, surgical resection still remains the mainstay of HCC treatment.Citation2 Unfortunately, due to the late appearance of clinical symptoms, the 5-year overall survival of hepatocellular carcinoma is still less than 20%.Citation3 Therefore, exploration of new potential biomarkers and clarification of their molecular mechanisms are of urgent importance to the quest for more effective therapies for this disease.
Circular RNAs (circRNAs) are a group of newly defined endogenous non-coding RNAs with covalently closed loops formed by the 5′ and 3′ ends of pre-mRNA transcripts.Citation4 CircRNAs derive mainly from exons or introns, while exonic circRNAs localize in the cytoplasm and show better stability than linear RNAs.Citation5 Recent studies have demonstrated that circRNAs have diverse functions. For instance, circRNAs can regulate transcription,Citation6 interact with RNA-binding proteins (RBPs),Citation7 act as sponges of miRNAsCitation8 and, in some cases, to have the ability to act as templates for protein translation.Citation9 Up to now, a large number of circRNAs have been shown to act as a sponge of miRNA to regulate cancer progression. Yu et al found that the circular RNA cSMARCA5 could suppress the growth and metastasis of hepatocellular carcinoma by sponging miR-17-3p and miR-181b-5p.Citation10 Wang et al indicated that circFOXP1 could enhance the Warburg effect in gallbladder cancer through circFOXP1/miR-370/PKLR axis.Citation11
MicroRNAs (miRNAs) are small, mostly non-coding RNAs containing approximately 22 nucleotides.Citation12 Dysregulation of miRNAs can have oncogenic or tumor-suppressive effects.Citation13 MiRNAs have been shown to affect mRNA levels by targeting their 3ʹ untranslated region (UTR).Citation14–Citation16 Thus, investigation of the upstream regulators of miRNAs is important in order to understand the mechanisms of miRNA effects in depth. It has been reported that miRNAs can be regulated by circRNAsCitation17 or lncRNAs.Citation18
Through microarray sequencing, we have identified dysregulated circRNAs in HCC tissues. Our microarray analysis showed that circARPP21 is only weakly expressed in HCC tissues. By determining circARPP21 levels in HCC cell lines and in 55 paired tumor and para-tumor normal tissues, we found that circARPP21 is stably downregulated in HCC. We also showed that circARPP21 can act as a sponge of miR-543 and indirectly regulate the leukemia inhibitory factor receptor (LIFR) to inhibit HCC progression. Our findings also offer a new promising biomarker to enhance current treatments for HCC.
Materials and Methods
Tissue Specimens
Tissue specimens (55 HCC tissues and 55 para-tumor normal tissues) were obtained from patients diagnosed with HCC between 2016 and 2019 in the Hepatobiliary Center of The First Affiliated Hospital, Nanjing Medical University. Every participant involved in this study provided written informed consent that was approved by the Ethics Committee of The First Affiliated Hospital, Nanjing Medical University and were carried out in accordance with The Code of Ethics of the World Medical Association (Declaration of Helsinki) for experiments involving humans. The tissue samples were frozen at −80°C before use. The circRNA microarray of three paired human HCC tissues was constructed and completed at Beijing CapitalBio Technology (Beijing, China).
Cell Culture
HCC cell lines (Huh7, Hep3B, HepG2, SMMC-7721, MHCC97H, HCCLM3) and normal liver cell lines (LO2) were purchased from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China). These cells were incubated in Dulbecco’s modified Eagle’s medium (DMEM) (Gibco, Life Technologies, Carlsbad, CA, USA) with 10% fetal bovine serum (FBS) (Gibco) and antibiotics (1% penicillin/streptomycin, Gibco, USA) at 37°C with 5% CO2.
Oligonucleotide Transfection
Si-circARPP21, miRNA-543 mimic, miRNA-543 inhibitor and their related control oligonucleotides were designed and synthesized by RiboBio (Guangzhou, China). The sequence of siRNAs has been shown as follows: siRNA1: CTGTCCAAAGCAGTCAGGA, siRNA2: AGCAGTCAGGAGCAGGAAA, siRNA3: CCAAAGCAGTCAGGAGCAG. A lipofectamine 3000 kit (Invitrogen) was used as transfection medium.
Stable Cell Transfection
To establish stable overexpression of circARPP21 in human HCC cell lines, we used lentivirus (HanBio, Shanghai, China) for transduction and the transfection was performed according to the supplier’s instructions. And all the cell lines were selected with 3μg/mL puromycin for 14 days.
RNase R Treatment
Total RNA extracted from HCC cell lines was mixed with RNase R(4U/mg) at 37°C for 30 minutes. RT-qPCR was then used to test the stability of circARPP21.
Real-Time Quantitative PCR
RNA was extracted from tissues and cell lines with TRIzol (Invitrogen, Carlsbad, CA). A PrimeScript RT reagent kit (Taka-ra Bio Inc, China) was used for reverse transcription of RNA to cDNA. Alternatively, the total RNA of miRNA was reversed by a RiboBio reverse transcription kit (Guangzhou, China). Next, RT-qPCR was conducted using the SYBR PremixEx Taq (Vazyme, China). The primer sequences were as follows: CircARPP21, Forward: CAATCTAAAGCCCGCCATGA and Reverse: CCTTTTCCTGCTCCTGACTG; ARPP21, Forward: CCCATCTGGTCAGTACCCTACC and Reverse: CACGCTTTGCACCGGAGTT;miR-543, Forward: ATGAGAAGTTGCCCGTGTT and Reverse: GCGAATGTTTCGTCACAAAT; LIFR, Forward: CATCATCAGCGTAGTGGCTAAA and Reverse: TTCCGACCGAGACGAGTTA; GADPH, Forward: CACCCACTCCTCCACCTTTG and Reverse: CCACCACCCTGTTGCTGTAG; U6, Forward: CTCGCTTCGCCAGCACA and Reverse: AACGCTTCACGAATTTGCGT.
Western Blotting Assay
Total protein was extracted from the HCC cell lines with ice-cold radioimmunoprecipitation-assay lysis buffer, then separated using sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE). The proteins were transferred to polyvinylidene difluoride (PVDF) membranes which were blocked in 5% non-fat powdered milk for 2h. Next, the membranes were incubated in primary detection antibodies overnight at 4°C and subsequently incubated with secondary detection antibodies for 2h at room temperature. The following primary antibodies were used to incubate membranes: LIFR (sc-659, Santa Cruz Biotechnology); GAPDH (#ab181602, Abcam). Protein expression levels were visualized using an enhanced chemiluminescence (ECL) detection system after washing the membranes three times with Tris-buffered saline-Tween (TBS-T) buffer.
Bio-Coupled Probe RNA Pull-Down Assay
HCCLM3 and Hep3B cells transfected with miR-543 mimics were washed in cold PBS and then lysed. The lysate was then incubated with a Biotin-coupled CircARPP21probe. The biotin-coupled probes were bounded magnetic beads. After a 2-hour incubation, the target RNA was isolated by a RNeasy Mini Kit (Qiagen, Germany). The RNA mixture was then washed, extracted with Trizol, and prepared for PCR.
Luciferase Reporter Assay
HCC cells were seeded onto a 24-well plate. Cells were co-transfected with wild-type or mutated circARPP21 or LIFR 3’-UTR reporter plasmids, and with miR-543 mimics or negative controls. After 48h, luciferase activity in co-transfected cells was determined by the dual-luciferase reporter assay system (Promega, WI, USA).
Fluorescence in situ Hybridization (FISH)
We detected the presence of CircARPP21 and miR-543 with specific probes using a FISH (fluorescence in situ hybridization) kit (RiboBio, Guangzhou, China). The circARPP21 probe and the miR-543 probe were hybridized at 37°C overnight after pre-hybridization. Nuclei were stained with 4’,6-diamidino-2-phenylindole (DAPI). Images were obtained with confocal microscopy.
Wound Healing Assay
Transfected and controlled cells (1 × 106 cells/well) were seeded onto 6-well plates and cultured at 37°C. After the cells were fully grown, a vertical scratch was made in the middle of the well with a sterile 200-μL pipet tip. Images were taken immediately after the scratch was made, and compared to images of the same location 24 hours later.
Transwell Invasion and Migration Assay
Following the manufacturer’s protocol, HCC cells were introduced into the upper chamber (8-μm pore size; Millipore) with 200μL serum-free medium. The bottom chamber was filed with 500μL medium with 10% serum as a chemoattractant. The transwell chamber was coated with the Matrigel mix (BD Biosciences, San Jose, CA, USA) for invasion assay. After incubation for 48h, the cells were fixed with methanol for 20 minutes and stained with crystal violet for 20 minutes at normal temperature. Five randomly selected microscopic fields were chosen to count the cells for each group.
Colony Formation Assay
In this assay, Hep3B and HCCLM3 cells were plated into a 6-well culture dish at a density of 1000 cells per well. After being incubated for 2 weeks, the cells were fixed by methanol and stained by crystal violet. The cell colony which contained >50 cells under the microscope were counted as valid.
Cell Counting Kit-8 Assay (CCK8)
For the CCK8 assay, transfected HCC cells were seeded into a 96-well plate at a density of 1000 cells/well for 5 replicates. The Cell counting kit-8 (CCK8, Dojindo, Japan) was added into the plate to assess the viability of HCC cells every 24h for 5 days. After a 2-hour incubation with 10μL CCK8, the optical density of the cells was measured at 450 nm.
5-Ethynyl-2’-Deoxyuridine (EdU) Assay
An EdU measurement was carried out with a Cell-Light EdU Apollo567 in vitro Kit (RiboBio, Guangzhou, China). Following the manufacture’s protocol, HCC cells (1 × 104) were plated onto 96-well plates. After cell adherence, a 50 μM EdU solution was added onto plates and incubated for 2h. The cell lines were then fixed in paraformaldehyde and stained by Apollo Dye Solution and Hoechst. Stained images were examined under a fluorescence microscope and the ratio of cell proliferation was calculated.
Tumor Xenografts
We purchased BALB/c nude mice (4–5 weeks old, five mice per group) from the National Laboratory Animal Center (Shanghai, China). Huh7 cells were transfected with siRNA of circARPP21 or NC vector. The transfected cells were used for subcutaneous injection of the nude mice (1.5× 106 cells per mouse). After 30 days, the mice were sacrificed and the tumors excised. We measured tumor size every five days in vivo. All the animal experiments in this study were approved by Animal Experimental Ethics Committee of Nanjing Medical University. And we manipulated and cared the animals following National Institutes of Health Guide for the Care and Use of Laboratory Animals.
Statistics Analysis
All data are presented as the mean ± standard deviation (SD) of at least three independent experiments. Student’s t test (two-tailed) or ANOVA with GraphPad Prism 8 was used to analyze the experimental data. The relationship between circARPP21 expression and clinicopathological characteristics of HCC patients was calculated and analyzed with a chi-square test (χ2) or Fisher’s exact test. Differences were considered statistically significant with *p < 0.05, **p < 0.01, ***p < 0.001.
Results
CircARPP21 is Downregulated in HCC Tissues and Cell Lines
To investigate the dysregulated circRNAs, we carried out a microarray assay using three paired sets of tumor and adjacent non-cancerous tissues. Differentially expressed circRNAs were defined as those exhibiting at least a two-fold change in expression and a P < 0.05. The heatmap of the top 10 up- and down-regulated circRNAs is shown in (). We found that a novel circRNA called circARPP21, which has not previously been reported in earlier work, was expressed at significantly lower levels in HCC. For this reason, we selected circARPP21as a candidate circRNA for further investigation. According to the circbase database (http://www.circbase.org/), circARPP21, which is located at chr3:35725217–35763326, is derived from its parental gene ARPP21 (cAMP-regulated phosphoprotein, 21kDa), and its spliced length is 1054bp (). The stability of circARPP21 was demonstrated by RNase R treatment: CircARPP21 showed resistance to RNase R digestion due to its loop structure (), while linear mRNA ARPP21 lacked resistance to RNase ().
Figure 1 CircARPP21 is downregulated in HCC specimens and cell lines. (A) Cluster heatmap illustrating the top 10 up- and down-regulated circRNAs in microarray. (B) The spliced mature sequence length of circARPP21 derived from the ARPP21 gene is 1054 bp and the head-to-tail splicing of circARPP21 was confirmed by Sanger sequencing. (C) RT-qPCR data documenting circARPP21 levels in HCC cells treated with RNase R. (D) RT-qPCR data on ARPP21 levels in HCC cells treated with RNase R. (E) Assessment of circARPP21 expression in cell lines using RT‐qPCR. (F) Evaluation of circARPP21 expression in 55 paired HCC tissues and adjacent normal tissues using RT-qPCR. (G) Evaluation of circARPP21 expression in 55 HCC tissues including early-stage group and late-stage group. (H) The diagnostic value of circARPP21 for HCC was evaluated by measuring the area under ROC curve. Values are shown as the mean ±SD of three independent experiments. **p < 0.01, ***p < 0.001.
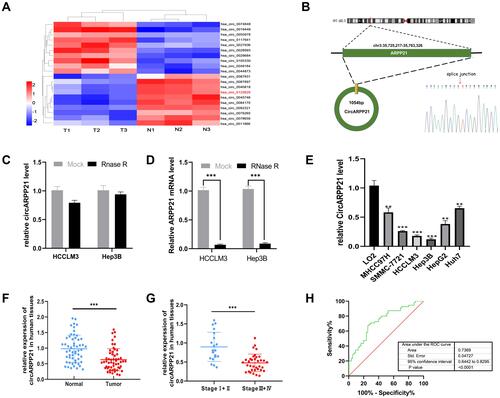
To further investigate the properties of this circRNA, we measured circARPP21 expression in HCC cell lines () and fresh human HCC tissue (), and found that circARPP21 was downregulated both in cell lines and in tumor tissue compared to the control group. Additionally, circARPP21 expression was markedly reduced in advanced tumors compared to that of early tumor stages (). Clinical statistics revealed that weak expression of circARPP21 was correlated with tumor size (p=0.0018), AFP level (p=0.0351), and tug-lymph node metastasis (TNM) stage (p<0.001)(Supplementary Table 1). However, this analysis did not show a significant association between circARPP21 and t patient gender, age, HBsAg level, or number of tumors. To further confirm the potential of circARPP21 as a therapeutic target for HCC patients, we determined the receiver operating characteristic (ROC) curve using the expression level of circARPP21 (); the area under curve (AUC) was 0.7369.
CircARPP21 Suppresses the Proliferation and Metastasis of HCC Cells in vitro
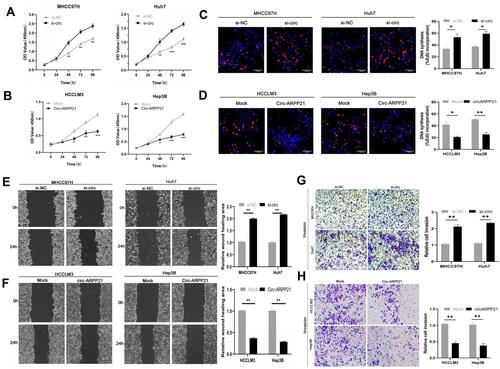
As is shown in , MHCC97H and Huh7 cells showed relatively higher circARPP21 levels than other HCC cells, and HCCLM3 and Hep3B cells showed the lowest circARPP21 expression. Thus, we chose HCCLM3 and Hep3B cells to investigate the phenotypic changes of HCC cells following overexpression of circARPP21. MHCC97H and Huh7 cells were also selected for loss-of-function experiments. After transfection with the circARPP21 overexpression vector, overexpression efficiency was detected by RT-qPCR (Figure S1A). Knock-down efficiency was also examined after siRNA transfection into MHCC97H and Huh7 cells (Figure S1C). None of the transfected cells influenced expression of ARPP21, which is the linear host gene of circARPP21 (Figure S1B and S1D).
Cell counting kit-8 (CCK8) assays showed that silencing of circARPP21 can promote the viability of HCC cells (), while circARPP21 overexpression had the opposite effect (). Next, we performed EdU essays, which revealed that silencing of circARPP21 enhanced DNA synthesis (); the overexpression group was able to suppress tumor proliferation in vitro (). Wound-healing assays and transwell invasion assays both revealed that mobility of HCC cells was increased in the si-circARPP21 group ( and ), and inhibited in the overexpression group ( and ).
Figure 2 CircARPP21 suppresses cancer progression in HCC cell lines. (A) The growth of HCC cells transfected with si‐circARPP21 was evaluated using the CCK‐8 assay. (B) The growth of HCC cells transfected with the circARPP21 vector was evaluated using the CCK‐8 assay. (C) EdU assays of HCC cells transfected with circARPP21 siRNAs compared to control. (D) EdU assays of HCC cells transfected with circARPP21 vector or control. (E) Wound healing assay performed in cells transfected with circARPP21 siRNAs or control. (F) Wound healing assay performed in cells transfected with circARPP21 vector or control. (G) Cell invasion assays in cells transfected with si-circARPP21 or control. (H) Cell invasion assays in cells transfected with circARPP21 vector or control. Values are shown as the mean ±SD of three independent experiments. *p < 0.05, **p < 0.01, ***p < 0.001.
CircARPP21 Functions as a Sponge of miR-543 in HCC Cells
Because many circRNAs play an important role in cancers through acting as a sponge of miRNA, we hypothesized that circARPP21 similarly affects progression of HCC. To determine which miRNA binds to circARPP21, we searched the database CircInteractome (https://circinteractome.irp.nia.nih.gov/). As shown in , the seed-binding region of miR-543 has a complementary sequence to circARPP21 and obtained one of the highest predicting score (context+ score percentile). The Kaplan–Meier Plotter database (http://kmplot.com/analysis/index.php) revealed that the survival time of patients with higher miR-543 levels was of shorter duration than that of patients with lower miR-543 expression, and the Starbase database (http://starbase.sysu.edu.cn/) showed a 1.86-fold difference in miR-543 levels in cancer samples compared to normal tissues (p=0.013) (). These results further raised the possibility of oncogenic miR-543 as a target of the anti-oncogene circARPP21.
Figure 3 CircARPP21 acts as a sponge of miR-543. (A) A schematic model shows potential binding sites for miR-543 and 3’UTR of circARPP21. (B) The survival curve and relative miR-543 expression in patients analyzed by database. (C) Expression level of miR-543 in HCC cells transfected with si-circARPP21 or control and circARPP21 vector or mock using RT-qPCR. (D) Schematic representation of potential binding sites of miR-543 with WT or MUT circARPP21. (E) Relative luciferase activity after co-transfection of miR-543 mimics, miR-NC, circARPP21-Mut and circARPP21-WT into 293T cells. (F) RT-qPCR analysis of circARPP21 expression in lysates of HCCLM3 and Hep3B cells with circARPP21 overexpression following biotinylated-circARPP21 pull-down assay. (G) MiR-543 was pulled down by circARPP21 in HCCLM3 and Hep3B cells. (H) CircARPP21 and miR-543 were co-localized in cytoplasm using FISH assays. Values are shown as the mean ±SD of three independent experiments. **p < 0.01, ***p < 0.001.
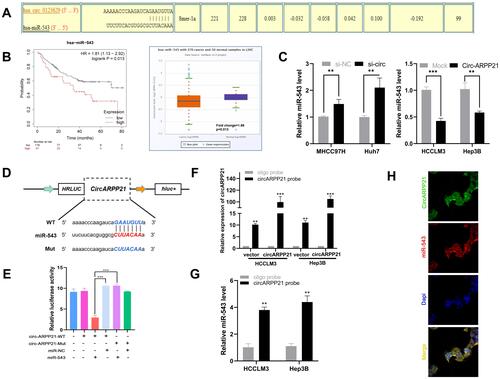
To test this hypothesis, we then conducted several experiments to confirm the relationship between circARPP21 and miR-543. First, RT-qPCR was carried out to determine whether knock down of circARPP21 upregulates miR-543 levels and whether overexpression of circARPP21 can reduce miR-543 expression (). Second, we constructed a 3′‐untranslated region (3′‐UTR) luciferase reporter containing a mutated variant (Mut) and wild-type variant (WT) of circARPP21 fragments () to determine whether circARPP21 can bind directly to miR-543. The results of luciferase assays definitively demonstrated that overexpression of miR-543 reduces luciferase activity in WT more strongly than in Mut variants (). Next, we designed a biotin-coupled probe to pull down circARPP21 in HCCLM3 and in Hep3B cells with overexpression of circARPP21 to increase pull-down efficiency (). shows that miR-543 was significantly pulled down compared to the control group. Last but not least, FISH assays further confirmed that both circARPP21 and miR-543 are located in cytoplasm of HCC cells (). In short, miR-543 is a direct target downstream of circARPP21.
MiR-543 Targets LIFR and Promotes the Proliferation, Invasion and Migration of Hepatocellular Carcinoma Cells
Because miRNAs can directly silence downstream coding mRNAs to regulate tumor progression,Citation19 we searched the miRNA target prediction database miRDB (http://mirdb.org/),Citation20 TargetScan (http://www.targetscan.org/vert_71/),Citation21 and RNA22 (https://cm.jefferson.edu/rna22/)Citation22 to determine the target mRNA of miR-543. shows that the genes common to all three databases are leukemia inhibitory factor receptor (LIFR), musashi RNA-binding protein 2 (MSI2), and armadillo repeat containing 8 (ARMC8). Among the three genes, LIFR shows lower expression in tumors than in normal tissues and high levels of LIFR are correlated with longer survival times in HCC patients () according to data from Kaplan–Meier Plotter and Gepia databases (http://gepia.cancer-pku.cn/). We therefore hypothesized that LIFR might be a direct target of miR-543. The binding site between miR-543 and LIFR is shown in . For further confirmation, we carried out real-time quantitative PCR and Western blotting assays, which showed that both mRNA levels and protein expression of LIFR was reduced after transfection of cells with miR-543 mimics ( and ). In addition, we carried out a luciferase assay in cells with mutations in the binding site in the 3’ UTR of LIFR (). As expected, the luciferase activity of LIFR 3’ UTR-WT was markedly suppressed in cells transfected with miR-543 mimics and the luciferase activity of mutated group showed no obvious change ().
Figure 4 Mir-543 can promote HCC progression by targeting LIFR. (A) TargetScan, miRDB, RNA22 databases were used to find the target mRNA of miR-543. (B) Survival curve and expression of LIFR in HCC obtained from TCGA database. (C) Predicted binding site between miR-543 and 3’UTR of LIFR obtained from TargetScan. (D) RT-qPCR analysis of LIFR mRNA expression in cells transfected with miR-543 mimics and controls. (E) Relative LIFR protein level in cells transfected with miR-543 mimics and control using Western blotting assays. (F) Schematic representation of potential binding sites of miR-543 with WT or MUT LIFR. (G) Relative luciferase activity detected after co-transfecting miR-543 mimics, miR-NC, LIFR-Mut and LIFR-WT into 293T cells. (H) Colony forming assays using cells transfected with miR-543 mimics or control. (I) The growth curve of cells transfected with miR-543 mimics or miR-NC and miR-543 inhibitor or inh-NC using CCK8 assays. (J) Mobility of cells transfected with miR-543 or miR-NC, assessed by wound healing assay. (K) Transwell invasion and migration assays showing the metastasis ability of cells transfected with miR-543 mimics or control. Values are shown as the mean ±SD of three independent experiments. *p < 0.05, **p < 0.01, ***p < 0.001.
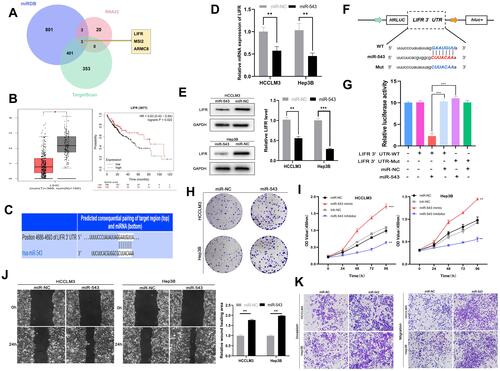
Because the oncogenic role of miR-543 appears to be linked to silencing of LIFR expression, we carried out colony forming assays and found that high miR-543 levels can enhance the proliferation of HCC cells (). Cell viability as measured by the CCK8 assay was also reduced in the miR-543 inhibited group compared to that of the miR-543 overexpressed group (). Subsequent wound healing assay and transwell invasion and migration assays demonstrated that the HCCLM3 and Hep3B cells with miR-543 mimics exhibit higher metastatic ability compared to that of the control group ( and ).
CircARPP21 Regulates LIFR Expression by Sponging miR-543
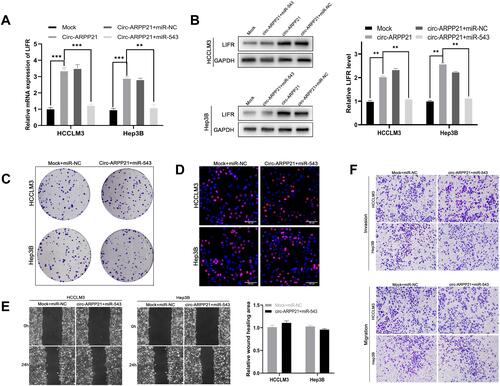
In order to clarify the relationship between circARPP21, miR-543 and LIFR, we then conducted rescue experiments to find if overexpression of circARPP21 could offset the oncogenic function of miR-543. RT-qPCR assays demonstrated that miR-543 can reverse the upregulation of LIFR caused by overexpression of circARPP21 (). Western blotting assay also showed that co-transfection of circARPP21 and miR-543 had no effect on levels of protein expression of LIFR (). The results of functional experiments of circARPP21 in HCC, mentioned above, confirm the tumor-suppressing effect of circARPP21 in HCC. Nevertheless, the effect of circARPP21 t in HCC can be significantly counteracted by miR-543 after co-transfection (-).
Figure 5 CircARPP21 regulates LIFR by targeting miR-543. (A) mRNA expression levels of LIFR detected by RT-qPCR in cells co-transfected with circARPP21 vector or mock and miR-543 mimics or miR-NC. (B) Western blot showing LIFR protein levels in HCC cells co-transfected with vectors overexpressing circARPP21 and miR‐543 mimics and their respective controls. (C) Colony formation assays were conducted to reveal cell proliferation of cells co-transfected with circARPP21 vectors and miR-543 mimics. (D) EdU assays showing the proliferation of cells co-transfected with circARPP21 vectors and miR-543 mimics. (E) Wound healing assays showing the mobility of cells co-transfected with circARPP21 vectors and miR-543 mimics. (F) Transwell assays were performed to examine the ability of migration and invasion of HCC cells co-transfected with circARPP21 vectors and miR-543 mimics compared to the control group which was co-transfected with miR-NC and Mock. Values are shown as the mean ±SD of three independent experiments. **p < 0.01, ***p < 0.001.
CircARPP21 Silencing Promotes the Growth of HCC Cells in vivo
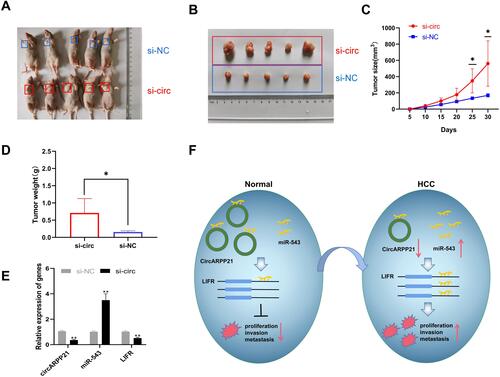
To investigate whether circARPP21 can affect HCC progression in vivo, we constructed five paired xenograft models. Huh7 cells transfected with circARPP21 silencing siRNA were injected into nude mice and the xenograft tumors were excised after 30 days. The tumor size of the group containing cells transfected with si-circARPP21 appeared to be larger than the control group ( and ). The growth curve of tumors knocked down with circARPP21 tended to be significantly higher than that of the control non-cancer group (). The weight of xenograft tumors also showed the same trend (). Moreover, we used RT-qPCR to measure the expression of circARPP21, miR-543 and LIFR in the tumors and found that circARPP21 and LIFR were significantly reduced while miR-543 was upregulated in the si-circARPP21 group (). All of these findings indicate that circARPP21 is able to sponge miR-543 and indirectly regulate LIFR to suppress the progression of HCC ().
Figure 6 Knockdown of circARPP21 promotes HCC cell growth in vivo. (A) Representative images of the HCC tumor bearing BALB/c nude mice. (B) Comparison of the tumor size of xenograft tumors induced by circARPP21 silencing and control in transfected Huh7 cells. (C) The growth curve of xenograft tumors, which were measured every five days in vivo. (D) Comparison of tumor weight between si-NC and si-circARPP21 transfected cells. (E) The expression of circARPP21, miR-543 and LIFR in xenograft tumors measured by RT-qPCR. (F) Schematic representation of the circARPP21 mechanism in HCC cells. *p < 0.05, **p < 0.01.
Discussion
In 2012, circular RNAs were shown to be universally expressed in eukaryotic genes, with circular RNA isoforms found in hundreds of human genes.Citation23 In the past decade, many studies have shed light on the biogenesis and potential mechanism of circRNAs in human diseases.Citation24 Current studies of the circular RNAs associated with HCC progression as prognostic biomarkers or functional molecules are in progress. Investigating the network of noncoding regulatory RNAs in HCC, Yanping Wei et al found that circ-CDYL, along with HDGF and HIF1AN, can serve as biomarkers for detection of early-stage HCC.Citation25 Our study had demonstrated that circular RNA circARPP21 is downregulated in HCC tissues and cells compared with circARPP21 levels in normal, non-cancer tissues. Moreover, low expression of circARPP21 is correlated with clinicopathologic data showing larger tumor size, late tumor stage, and AFP levels. Our functional experiments also revealed that overexpression of circARPP21 can inhibit the proliferation, viability, invasion, and migration of HCC cells, while the experimental group with silenced circARPP21 showed the opposite phenotype. Altogether, these results suggest the potential of circARPP21 as a new biomarker and tumor-suppressor molecule in hepatocellular carcinoma.
Following our initial investigation of circARPP21, we then determined that circARPP21 sponges its downstream microRNA miR-543. Current research indicates that miR-543 acts as an oncogene in many types of cancers. For example, in lung cancer, Wang et al reported that miR-543 promotes lung cancer cell growth and angiogenesis in vitro as well as tumor growth in vivo through silencing of MTA1.Citation26 Additionally, Shi et al showed that miR-543 is overexpressed by the Helicobacter pylori CagA protein, thus suppressing autophagy and promoting epithelial–mesenchymal transition in gastric cancer through targeting SIRT1.Citation27 miR-543 also plays an important oncogenic role in prostate cancer.Citation28 Yu et al confirmed the strong expression of miR-543 in hepatocellular carcinoma, and its ability to promote HCC cell growth and invasion,Citation29 a finding consistent with the results of our own study. Leukemia inhibitory factor receptor (LIFR), reported by Luo et al to function as a metastasis suppressor in HCC,Citation30 was predicted and confirmed in our own research as a downstream target that binds directly to miR-543. In short, our work suggests that circARPP21 might alleviate HCC progression via the circARPP21/miR-543/LIFR axis.
As far as we know, this is the first study to investigate the characteristics of circARPP21 in HCC. We have also described for the first time the relationship between miR-543 and LIFR. These findings offer a promising new therapeutic approach for treatment of HCC. Nevertheless, there are still several limitations to our study. First, we need a larger sample size to confirm the outcomes. Second, since the mode of action of circRNAs remains unclear, it is possible that additional miRNAs or other kinds of genes also interact with circARPP21 to regulate HCC development. Moreover, the downstream pathway of LIFR also needs to be investigated to clarify how it works in HCC progression. Hence, further research is essential to acquire a deep understanding of operation of molecular mechanisms in cancer biology.
Disclosure
The authors report no conflicts of interest related to this work.
References
- Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi:10.3322/caac.21492
- Bruix J, Gores GJ, Mazzaferro V. Hepatocellular carcinoma: clinical frontiers and perspectives. Gut. 2014;63(5):844–855. doi:10.1136/gutjnl-2013-306627
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69(1):7–34. doi:10.3322/caac.21551
- Szabo L, Morey R, Palpant N, et al. Statistically based splicing detection reveals neural enrichment and tissue-specific induction of circular RNA during human fetal development. 2015;16:126.
- Jeck W, Sharpless N. Detecting and characterizing circular RNAs. Nat Biotechnol. 2014;32(5):453–461. doi:10.1038/nbt.2890
- Conn V, Hugouvieux V, Nayak A, et al. A circRNA from SEPALLATA3 regulates splicing of its cognate mRNA through R-loop formation. Nat Plants. 2017;3(p):17053. doi:10.1038/nplants.2017.53
- Abdelmohsen K, Panda A, Munk R, et al. Identification of HuR target circular RNAs uncovers suppression of PABPN1 translation by CircPABPN1. RNA Biol. 2017;14(3):361–369. doi:10.1080/15476286.2017.1279788
- Hansen T, Jensen T, Clausen B, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495(7441):384–388. doi:10.1038/nature11993
- Legnini I, Di Timoteo G, Rossi F, et al. Circ-ZNF609 is a circular RNA that can be translated and functions in myogenesis. Mol Cell. 2017;66(1):22–37.e9. doi:10.1016/j.molcel.2017.02.017
- Yu J, Xu Q, Wang Z, et al. Circular RNA cSMARCA5 inhibits growth and metastasis in hepatocellular carcinoma. J Hepatol. 2018;68(6):1214–1227. doi:10.1016/j.jhep.2018.01.012
- Wang S, Zhang Y, Cai Q, et al. Circular RNA FOXP1 promotes tumor progression and Warburg effect in gallbladder cancer by regulating PKLR expression. 2019;18(1):145.
- Li J, Tan S, Kooger R, Zhang C, Zhang YJ. MicroRNAs as novel biological targets for detection and regulation. Chem Soc Rev. 2014;43(2):506–517. doi:10.1039/c3cs60312a
- Rupaimoole R, Slack F. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat Rev Drug Discovery. 2017;16(3):203–222. doi:10.1038/nrd.2016.246
- Bracken C, Scott H, Goodall G. A network-biology perspective of microRNA function and dysfunction in cancer. Nat Rev Genetics. 2016;17(12):719–732. doi:10.1038/nrg.2016.134
- Sandbothe M, Buurman R, Reich N, et al. The microRNA-449 family inhibits TGF-β-mediated liver cancer cell migration by targeting SOX4. J Hepatol. 2017;66(5):1012–1021. doi:10.1016/j.jhep.2017.01.004
- Hydbring P, Wang Y, Fassl A, et al. Cell-cycle-targeting MicroRNAs as therapeutic tools against refractory cancers. Cancer Cell. 2017;31(4):576–590.e8. doi:10.1016/j.ccell.2017.03.004
- Kristensen LS, Andersen MS, Stagsted LVW, Ebbesen KK, Hansen TB, Kjems J. The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet. 2019;20(11):675–691. doi:10.1038/s41576-019-0158-7
- Yao R, Wang Y, Chen L. Cellular functions of long noncoding RNAs. Nat Cell Biol. 2019;21(5):542–551. doi:10.1038/s41556-019-0311-8
- Jonas S, Izaurralde G. Towards a molecular understanding of microRNA-mediated gene silencing. Nat Rev Genetics. 2015;16(7):421–433. doi:10.1038/nrg3965
- Wong N, Wang X. miRDB: an online resource for microRNA target prediction and functional annotations. Nucl Acids Res. 2015;43:D146–52. doi:10.1093/nar/gku1104
- Agarwal V, Bell G, Nam J, Bartel DJ. Predicting effective microRNA target sites in mammalian mRNAs. 2015;4.
- Miranda KC, Huynh T, Tay Y, et al. A pattern-based method for the identification of MicroRNA binding sites and their corresponding heteroduplexes. Cell. 2006;126(6):1203–1217. doi:10.1016/j.cell.2006.07.031
- Salzman J, Gawad C, Wang P, Lacayo N, Brown P. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. 2012;7(2):e30733.
- Kristensen L, Hansen T, Venø M, Kjems JJO. Circular RNAs in cancer: opportunities and challenges in the field. Oncogene. 2018;37(5):555–565. doi:10.1038/onc.2017.361
- Wei Y, Chen X, Liang C, et al. A noncoding regulatory RNAs network driven by Circ-CDYL acts specifically in the early stages Hepatocellular carcinoma. Hepatology. 2020;71(1):130–147. doi:10.1002/hep.30795
- Wang D, Cai L, Tian X, Li W. MiR-543 promotes tumorigenesis and angiogenesis in non-small cell lung cancer via modulating metastasis associated protein 1. Mol Med. 2020;26(1):44. doi:10.1186/s10020-020-00175-1
- Shi Y, Yang Z, Zhang T, Shen L, Li Y, Ding S. SIRT1-targeted miR-543 autophagy inhibition and epithelial-mesenchymal transition promotion in Helicobacter pylori CagA-associated gastric cancer. Cell Death Dis. 2019;10(9):625. doi:10.1038/s41419-019-1859-8
- Du Y, Liu X, Zhu H, Wang L, Ning J. MiR-543 promotes proliferation and epithelial-mesenchymal transition in prostate cancer via targeting RKIP. biochemistry. 2017;41(3):1135–1146.
- Yu L, Zhou L, Cheng Y, et al. MicroRNA-543 acts as an oncogene by targeting PAQR3 in hepatocellular carcinoma. Am J Cancer Res. 2014;4(6):897–906.
- Luo Q, Wang C, Jin G, et al. LIFR functions as a metastasis suppressor in hepatocellular carcinoma by negatively regulating phosphoinositide 3-kinase/AKT pathway. Carcinogenesis. 2015;36(10):1201–1212. doi:10.1093/carcin/bgv108
