Abstract
Background
Decorin is a multifunctional molecule of the extracellular matrix and impedes different kinds of tumor cell growth, but the role and molecular mechanism by which decorin inhibits HepG2 cells is not fully understood. Our objective was to construct recombinant human decorin (pcDNA3.1-DCN) and to explore the mechanism by which it inhibits HepG2 cells.
Methods
This experiment was divided into three groups, ie, a control group, an empty vector group, and a pcDNA3.1-DCN group. pcDNA3.1-DCN was constructed using recombinant DNA technology, and the vector for pcDNA3.1-DCN and pcDNA3.1 was then transfected into HepG2 cells using Lipofectamine 2000.
Results
Compared with cells in the control group and in the empty vector group, growth of cells in the pcDNA3.1-DCN group was significantly suppressed, the ratios of cells in the G0/G1 phases and proportion of early apoptotic cells were significantly increased, and the level of p21WAF1/CIP1 (p21) protein was markedly upregulated (P < 0.05). However, there was no significant difference among the three groups in p53 protein expression (P > 0.05).
Conclusion
The pcDNA3.1-DCN vector was successfully constructed and transfected into HepG2 cells, and decorin overexpression suppressed the growth of HepG2 cells by upregulation of p21 via a p53-independent pathway.
Introduction
Hepatocellular carcinoma is the seventh most common malignancy and the third leading cause of cancer-related deaths worldwide.Citation1 Hepatocellular carcinoma causes 662,000 deaths globally per year, about half of which occur in China. Special areas, such as Asia and sub-Saharan Africa, have high incidence rates of hepatocellular carcinoma. Despite the recent advances in diagnosis and treatment of hepatocellular carcinoma, it remains a highly lethal disease. The main cause of death in patients with hepatocellular carcinoma is tumor progression with metastasis. Therefore, there is an urgent need for improved treatment modalities in liver cancer.
Decorin is a small leucine-rich proteoglycan secreted mainly by cells of mesenchymal origin, where it plays an important role in intercellular contact modulation, migration, and cellular proliferation by modulating the interactions of cell surface receptors with extracellular matrix proteins.Citation2 Decorin has been shown to inhibit the growth of cancer cells, and it has now become a focus in several areas of cancer research. In vivo, although decorin-null animals do not develop spontaneous tumors, double-mutant mice, lacking both decorin and the tumor suppressor gene p53, develop lymphomas at accelerated rates as compared with p53-null animals, indicating that the absence of decorin enables tumor development.Citation3 Another report shows that about 30% of decorin-null mice develop spontaneous intestinal tumors.Citation4 Furthermore, increased decorin expression in vivo, achieved by adenovirus-mediated gene transfer techniques, causes growth inhibition in various tumors.Citation5 Moreover, an experiment by Reed et al demonstrated that decorin prevents metastatic spread of breast cancer.Citation6 In vitro, a number of human cancer lines do not express decorin,Citation7,Citation8 and forced expression of decorin in cancer cell lines in vitro causes a severe cytostatic effect.
At present, the molecular mechanisms for the antioncogenic activity of decorin include the following several aspects. Some experiments have showed that decorin can interact with epidermal growth factorCitation9 and erb2,Citation10 causing transient receptor phosphorylation, which is followed by induction of endogenous cyclin-dependent kinase inhibitor p21 expression. Citation11 Another experiment demonstrates that decorin antagonizes met receptor activity and downregulates beta-catenin and myc levels.Citation12 Recently, research by Iozzo et al has shown that decorin antagonizes insulin-like growth factor-1 (IGF1) receptor function by interfering with IGF1 receptor activity and attenuating downstream signaling.Citation13
Although many studies have shown that decorin has a role in tumor inhibition, very few studies have indicated its expression is suppressed in hepatocellular carcinomaCitation14,Citation15 and yet the molecular mechanism of inhibition of hepatocellular carcinoma is not fully understood. Thus, we hypothesize that decorin may have an inhibitory role in hepatocellular carcinoma, and targeted decorin upregulation may emerge as a feasible therapeutic modality in the disease. Nothing is known about the role of recombinant human decorin in HepG2 cells, so the aim of our study was to examine the inhibitory effects of recombinant human decorin on the growth of liver carcinoma (HepG2) cells in vitro, and explore possible mechanisms of decorin-induced inhibition of HepG2 cell growth.
Materials and methods
Cell lines
HepG2 cells were obtained from the American Type Culture Collection (Manassas, VA), maintained in Dulbecco’s modified Eagle’s medium, supplemented with 10% fetal bovine serum, and grown in 5% CO2 at 37°C.
Construction of recombinant human decorin and stably transfected HepG2 cells
Decorin cDNA was kindly provided by Professor Sun Yin, King’s College London, UK. The full length of the human decorin gene (1080 bp) was amplified by polymerase chain reaction (PCR) using a designed primer pair (Supplementary Table 1). The PCR conditions used were 94°C for 5 minutes followed by 94°C for one minute, 58°C for 45 seconds and 72°C for 1.5 minutes for 30 cycles, and a final extension of 72°C for 10 minutes. PCR products were analyzed by electrophoresis on 1% agarose gel with ethidium bromide. After a pcDNA3.1 vector and decorin gene were digested using the EcoR I and Xba I restriction enzymes, the human-specific decorin gene was ligated with the pcDNA3.1 vector to obtain pcDNA3.1-DCN. The pcDNA3.1-DCN was identified by digestion of EcoR I and Xba I and sequencing.
Transfection of pcDNA3.1-DCN and vector plasmid pcDNA3.1 into HepG2 cells was performed by Lipofectamine 2000 (Invitrogen Life Technologies, Carlsbad, CA). Selection of HepG2 cells containing the decorin gene and pcDNA3.1 vector was carried out by adding G418. Briefly, about 1 × 106 cells were transfected with 0.8 μg of purified pcDNA3.1-DCN and pcDNA3.1 vector, respectively. After 48 hours of incubation in nonselective medium to allow expression of the decorin gene, the cells were trypsinized and passaged at a 1:10 dilution. Within 24 hours, G418 (800 μg/mL) was added to the medium and the medium was replaced every 3 days. Colonies were detected after 14 days in the selective medium and about 7 days later, independent colonies were trypsinized and transferred to 96-well plates, and the cells were then cultured routinely with G418 (400 μg/mL). Expression of decorin protein was measured as described previously.Citation7 In this study, Western blotting was performed using the mouse monoclonal antihuman decorin antibody (R&D Systems, Minneapolis, MN). Our experiment was divided into three groups, ie, a control group (HepG2 cells), an empty vector group (HepG2 cells stably transfected with vector plasmid pcDNA3.1), and a pcDNA3.1-DCN group (HepG2 cells stably transfected with pcDNA3.1-DCN).
MTT assay
The three groups of cells were seeded into 96-well plates at a density of 1 × 104 cells/well; 20 μL of MTT solution (Sigma, St Louis, MO, 5 mg/mL) was added into each well daily from days 1–6, and plates were incubated for 4 hours at 37°C. After removal of the supernatant, 100 μL of dimethyl sulfoxide (Sigma) was added to dissolve the crystals. Absorbance values were measured at a wavelength of 490 nm with a microplate reader (Biotek Synergy HT). A growth curve was made according to the values of 490 nm wavelength light absorption in the three groups. Values represent the mean ± standard error of the mean of three independent experiments run in triplicate. Based on the MTT results, three groups of cells on the third day were chosen for performing the following experiment.
Cell growth cycle analysis by flow cytometry
HepG2 cells from each group were seeded in 6-well plates at a density of 5 × 104 cells/well, with each well containing 2 mL of Dulbecco’s modified Eagle’s medium with 10% fetal bovine serum. After the cells were cultured for 72 hours, three groups of cells were trypsinized to prepare a single cell suspension. The cells were fixed with 70% ice-cold ethanol at −20°C overnight, and 0.01% RNase and 0.5% propidium iodide (Sigma) were then added to the cells. The cell growth cycle and proliferative index were measured by flow cytometry (FC500, Beckman Coulter Inc, Fullerton, CA) and the cell growth cycle was analyzed by Multicycle software. The proliferative index is defined as the percentage of cells in the S + G2/M phases divided by the total number of cells in the G0/G1 + S + G2/M phases × 100. Values represent the mean ± the standard error of the mean of three independent experiments run in triplicate.
Early apoptosis detection by flow cytometry
HepG2 cells from each group were seeded in 6-well plates at a density of 5 × 104 cells/well, with each well containing 2 mL of Dulbecco’s modified Eagle’s medium with 10% fetal bovine serum. After the cells were cultured for 72 hours, cells from the three groups were trypsinized to prepare a single cell suspension. The cells were collected, washed with cold phosphate-buffered solution (4°C) three times, and floated with 0.3 mL of binding buffer. The cells were stained using 5 μL of Annexin V-FITC (Kaiji Bio Co, Nanjing, China) and then by 5 μL of propidium iodide stain for 5 minutes. Finally, the cells were analyzed by flow cytometry. The signals of early apoptotic cells were localized in the lower right quadrant of the resulting dot-plot graph. Values represent the mean ± the standard error of the mean of three independent experiments run in triplicate.
p21 and p53 protein expression by Western blotting
HepG2 cells from each group were seeded in 6-well plates at a density of 5 × 104 cells/well, with each well containing 2 mL of Dulbecco’s modified Eagle’s medium with 10% fetal bovine serum. After the cells were cultured for 72 hours, cells from the three groups were lysed with cell lysate solution (including aprotinin and phenylmethyl sulfonyl fluoride), and the lysates were collected. Protein concentrations were determined by Bradford protein assay (Beyotime Institute of Biotechnology, Haimen, China). Equal amounts of protein were separated on 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis and transferred onto polyvinylidene fluoride membranes. Membranes were blocked in 5% skim milk dissolved in 10 mmol/L Tris-HCl, 100 mmol/L NaCl, and 0.1% Tween-20, and incubated with 1:500 dilution of anti-p21 and anti-p53 antibody (Santa Cruz Biotechnology, Santa Cruz, CA) overnight at 4°C. The following day, the membranes were washed three times and incubated with a 1:2000 dilution of horseradish peroxide-conjugated goat antimouse secondary antibody (Santa Cruz Biotechnology). Proteins were detected using enhanced chemiluminescence detection (Santa Cruz Biotechnology). The protein bands were quantified by the average ratios of integral optic density following normalization to the housekeeping gene.
Statistical analysis
All the data were analyzed by SPSS version 12.0 (SPSS Inc, Chicago, IL). Experiments were performed in triplicate. The unpaired or paired Student’s t-test was used where appropriate to compare two groups. Analysis of variance was used to compare multiple groups. Results were expressed as the mean ± standard error of the mean. P < 0.05 was regarded as statistically significant.
Results
Recombinant human decorin vector and its expression in HepG2 cells
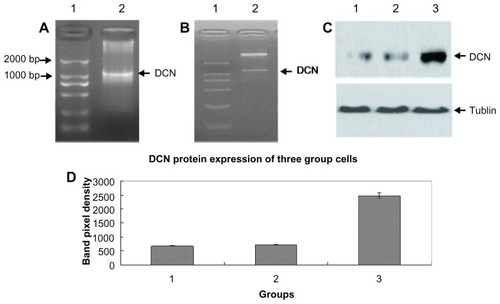
After conducting the PCR reaction using a designed primer pair, the PCR product was run on agarose gel to perform electrophoresis. As shown in , a special DNA band of about 1000 bp size was successfully cloned. The special DNA was then inserted into the vector of pcDNA3.1. The results of digestion by double restriction enzyme (shown in ) and sequencing (Supplementary Figure 1) demonstrated that the pcDNA3.1-DCN vector was successfully constructed. When HepG2 cells were transfected with pcDNA3.1-DCN, stably transfected clones were selected by G418. Compared with the control group and the empty vector group, the pcDNA3.1-DCN group expressed a significantly higher amount of decorin by Western blotting (shown as lane 3 in ), demonstrating that the recombinant decorin expression vector was successfully transfected into HepG2 cells. However, there was no significant difference in decorin expression between the control group and the empty vector group (P > 0.05, shown ). In fact, decorin was expressed by HepG2 cells at low levels (shown as lane 1 in ), and our results also showed that the empty vector did not influence decorin expression (shown as lane 2 in ). Therefore, decorin overexpression in the pcDNA3.1-DCN group must have been plasmid-initiated, not endogenous.
Figure 1 Construction and identification of pcDNA3.1-DCN and its expression in HepG2 cells. (A) Agarose gel electrophoresis of polymerase chain reaction product. Lane 1 represents DNA marker DL 2000 and lane 2, the decorin gene. (B) Detection of pcDNA3.1-DCN on gel electrophoresis after digestion by double restriction enzyme EcoRI and Xba I, lane 1 represents DNA marker DL 2000, and lane 2, double enzyme digestion results. (C) Decorin expression of three groups. Lane 1, lane 2, and lane 3, respectively, showed the decorin expression of the control, empty vector, and pcDNA3.1-DCN groups. The results were shown to be representative in three repeated experiments. (D) The decorin protein expression results were quantified based on three experiments and are represented as the mean ± standard deviation.
Abbreviation: DCN, decorin.
Recombinant human decorin overexpression inhibits HepG2 cell growth
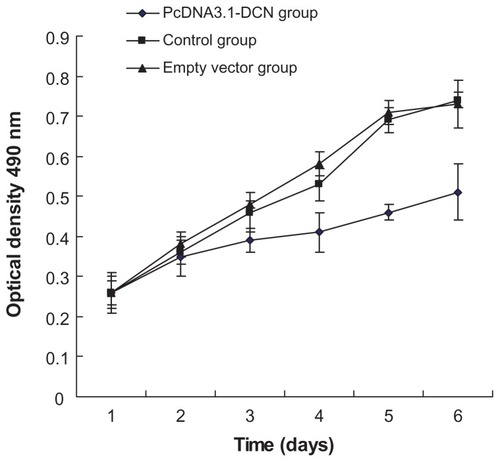
To investigate whether recombinant human decorin played any role in development of HepG2 cells, we firstly examined HepG2 cell growth in vitro after transfecting the cells with pcDNA3.1-DCN vector. Compared with the control group and empty vector group, our results show that recombinant human decorin overexpression could suppress the growth of HepG2 cells from the third day (shown in ). Therefore, according to the MTT results, three groups of cells were chosen after culturing for 72 hours to perform the following experiment.
Figure 2 Cell proliferation in the three groups.
Abbreviation: DCN, decorin.
Influence of recombinant human decorin overexpression on HepG2 cell cycle
Next, we determined whether the inhibitory effects of recombinant human decorin were due to a block in the onset of DNA synthesis. The cell cycle of HepG2 cells after culturing for 72 hours was tested by flow cytometry. As shown in and , the ratios of cells in the G0/G1 phases were significantly increased in the pcDNA3.1-DCN group compared with those in the control group and empty vector group (P < 0.05). There was no significant difference in cell cycle distribution between control and empty vector group (P > 0.05). The proliferation index was remarkably lower in the pcDNA3.1-DCN group (34.764% ± 1.231%) than in the control group (52.897% ± 2.031%) and empty vector group (51.354% ± 1.981%, P < 0.05).
Figure 3 Cell cycles of cells from the three groups. Histograms are representative of three independent experiments. (A) Control group, (B) empty vector group, and (C) pcDNA3.1-DCN group.
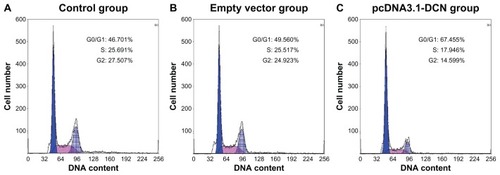
Table 1 Cell cycle distribution in different groups of HepG2 cells (n = 3)
Recombinant human decorin overexpression and early HepG2 apoptosis
Then, the effect of decorin overexpression on early apoptosis of HepG2 cells after culturing for 72 hours was performed by flow cytometry. As shown in and , the pcDNA3.1-DCN group exhibited a significantly higher proportion of early apoptotic cells (P < 0.05) compared to the control and empty vector group. There was no significant difference in apoptotic distribution between control and empty vector group (P > 0.05). Our results suggested that recombination human decorin overexpression induced early apoptosis of HepG2 cells.
Figure 4 The early apoptosis results for cells from the three groups. Flow cytometry was performed for HepG2 cells in each group by double staining with Annexin V and propidium iodide after culturing the cells for 72 hours. The lower left quadrant showed viable cells and the lower right quadrant showed early apoptotic cells. The results for apoptosis shown are representative of three repeated experiments. (A) Control group, (B) empty vector group, and (C) pcDNA3.1-DCN group.
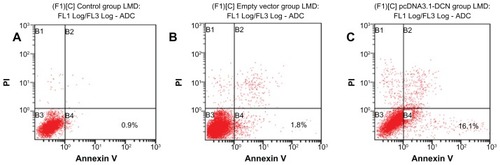
Table 2 Effect of decorin overexpression on early apoptosis in different groups of HepG2 cells (n = 3)
pcDNA3.1-DCN-induced HepG2 cell growth arrest and p21 protein expression
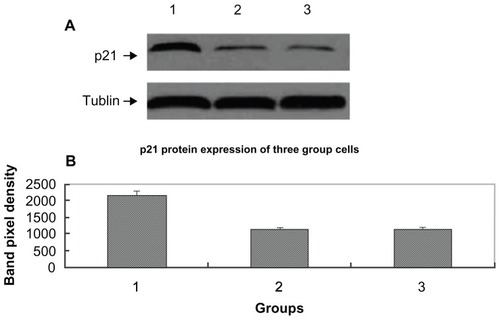
We assessed if the HepG2 cell growth-suppressive properties of pcDNA3.1-DCN might be mediated by p21, a potent inhibitor of G1 cyclin-dependent kinase. The level of p21 protein in the three group HepG2 cells after culturing for 72 hours was measured by Western blotting. Our experiment demonstrated that the level of p21 protein in the pcDNA3.1-DCN group was relatively higher compared with the control group and empty vector group (shown in ). There was no significant difference in p21 protein expression between control and empty vector group (P > 0.05). These results show that decorin overexpression inhibited the growth of HepG2 via upregulating the expression of p21 protein.
Figure 5 p21 protein expression for cells from three groups. Western blotting of equal amounts of cell lysates probed with the monoclonal antibody directed against the human p21 protein. (A) There was a marked upregulation of p21 in the pcDNA3.1-DCN group as compared with the control group and the empty vector group. Lane 1, lane 2, and lane 3, respectively, show p21 expression for the pcDNA3.1-DCN, control, and empty vector groups. The results are representative of three repeated experiments. (B) Densitometric analysis was done using Lab image software.
Abbreviation: DCN, decorin.
Recombinant human decorin overexpression might increase p21 protein via p53-independent pathway
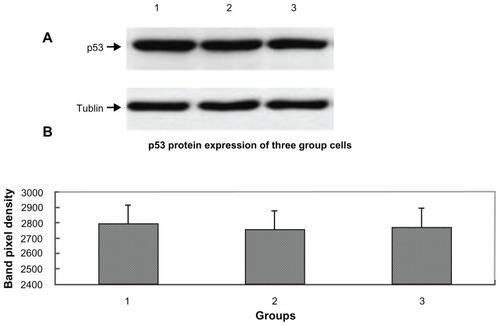
Finally, we determined whether the increasing of p21 protein induced by decorin overexpression was controlled by the tumor suppressor protein p53. The expression of p53 was performed by Western blotting as shown in , and there was no significant difference in p53 protein expression between the three groups of cells (P > 0.05). These results showed that decorin overexpression in HepG2 cells might increase p21 protein expression through an 53-independent pathway.
Figure 6 p53 protein expression for the cells from the three groups. (A) Lane 1, lane 2, and lane 3, respectively, show the p53 expression of pcDNA3.1-DCN, control, and empty vector groups. The results were representative of three repeated experiments. (B) Densitometric analysis was done using Lab image software.
Abbreviation: DCN, decorin.
Discussion
Decorin, a member of the small leucine-rich proteoglycan gene family that harbors one chondroitin/dermatan sulfate side chain at its N terminus, is 359 amino acids in length.Citation16 In our study, a full length decorin gene (1080 bp) was cloned using designed primers. In order to enhance the translation efficiency of the decorin gene, a Kozak consensus sequence is added into the upstream primer. In our study, we first constructed a pcDNA3.1-DCN vector which is a basis for further study of decorin.
Decorin is of special interest due to its ability to inhibit tumor growth. Many studies have shown that decorin expression is downregulated in tumors of the colon, prostate, pancreas, breast, and liver in vivo.Citation14,Citation15,Citation17–Citation19 However, its presence in the tumor microenvironment is proposed to reflect an attempt by the stroma to wall off the tumor.Citation20,Citation21 In vitro, analysis of cancer cell lines from the skin, uterus, soft tissues, and bone marrows showed no expression of decorin.Citation7,Citation8 Hence, low levels of decorin may be associated with tumorigenesis, progression, early recurrence, and poor survival. In contrast, upregulation of decorin in tumor cells and/or extracellular tissue could reduce cancer cell proliferation. Moreover, over-expressed decorin in vitro or provided as a recombinant decorin protein also inhibits the growth of various tumor cell lines. Thus, our hypothesis is that decorin gene therapy may help in retarding the growth of human HepG2 liver carcinoma cells. In our present study, HepG2 cells were stably transfected by the decorin gene using a plasmid transfection method, and our results show that recombinant human decorin overexpression can suppress growth of HepG2 cells.
Although the decorin gene is downregulated in hepatocellular carcinoma and can inhibit cancer growth and invasion, the mechanisms by which decorin impedes liver carcinoma (HepG2) cells are not understood. Our results have demonstrated for the first time that decorin overexpression induces upregulation of p21 protein in HepG2 cells and increases the ratio of early apoptotic cells and arrest of HepG2 cells in the G1 phase of the cell cycle, with similar effects in other malignancies.Citation11,Citation22–Citation24 Tumor suppressor protein p53 is involved in control of the G1 and G2 phase of the cell cycle, and p21, a cyclin-dependent kinase inhibitor, is a downstream target and effector of p53 to induce G1 arrest. The generally accepted dogma for p53-controlled cell fate holds that cell cycle arrest is predominantly mediated by expression and activation of p21.Citation25 There is some work showing that decorin can interact with the epidermal growth factor receptor,Citation9 causing transient receptor phosphorylation followed by induction of p21 expression.Citation11 Other researchers have demonstrated that decorin induces p21 upregulation involving the Akt/protein kinase B pathway.Citation26 Although our results show that decorin overexpression may increase the expression of p21 protein in HepG2 cells, levels of p53 protein expression in the three cell groups did not change. The fact that decorin-induced HepG2 cell growth suppression may not require p53 protein suggests that decorin overexpression may increase p21 protein levels via a p53-independent pathway. Some of the known mechanisms for cell growth inhibition by decorin were demonstrated probably by interacting with multiple receptors, including the epidermal growth factor receptor, MET receptor, IGF receptor 1, vascular endothelial growth factor, and transforming growth factor beta.Citation10,Citation12,Citation13,Citation27,Citation28 Therefore, the exact mechanisms by which decorin inhibits liver cancer cells need further study.
Conclusion
In conclusion, we have constructed pcDNA3.1-DCN and established HepG2 cells with overexpression of the decorin gene, and our results show that decorin overexpression can inhibit growth of HepG2 liver carcinoma cells in vitro by upregulating p21 protein expression via a p53-independent pathway, arresting cells in the G1 phase of the cell cycle and inducing increased ratios of early apoptotic cells. Our results suggest the potential for decorin to represent a novel therapeutic modality for hepatocellular carcinoma. We believe that additional studies are needed to determine whether decorin might have a role in the treatment of patients with liver cancer.
Acknowledgments
This study was supported by grants from the Development and Reform Commission (20061550) and Department of Public Health (2009Z080) in Jilin Province, China.
Supplementary data
Table 1 Primer sequences used in the study
Disclosure
The authors report no conflicts of interest in this work.
References
- YangJDRobertsLRHepatocellular carcinoma: a global viewNat Rev Gastroenterol Hepatol20107844845820628345
- TralhaoJGSchaeferLMicegovaMIn vivo selective and distant killing of cancer cells using adenovirus-mediated decorin gene transferFASEB J200317346446612631584
- IozzoRVChakraniFPerrottiDCooperative action of germ-line mutations in decorin and p53 accelerates lymphoma tumorigenesisProc Natl Acad Sci U S A19999663092309710077642
- BiXTongCDockendorffAGenetic deficiency of decorin causes intestinal tumor formation through disruption of intestinal cell maturationCarcinogenesis20082971435144018550571
- ReedCCGauldieJIozzoRVSuppression of tumorigenicity by adenovirus-mediated gene transfer of decorinOncogene200221233688369512032837
- ReedCCWaterhouseAKirbySDecorin prevents metastatic spreading of breast cancerOncogene20052461104111015690056
- SantraMSkorskiTCalabrettaBLattimeECIozzoRVDe novo decorin gene expression suppresses the malignant phenotype in human colon cancer cellsProc Natl Acad Sci U S A19959215701670207624361
- SantraMMannDMMercerEWSkorskiTCalabrettaBIozzoRVEctopic expression of decorin protein core causes a generalized growth suppression in neoplastic cells of various histogenetic origin and requires endogenous p21, an inhibitor of cyclin-dependent kinasesJ Clin Invest199710011491579202067
- MoscatelloDKSantraMMannDMMcQuillanDJWongAJIozzoRVDecorin suppresses tumor cell growth by activating the epidermal growth factor receptorJ Clin Invest199810124064129435313
- SantraMEichstetterIIozzoRVAn anti-oncogenic role for decorin. Down-regulation of ErbB2 leads to growth suppression and cytodifferentiation of mammary carcinoma cellsJ Biol Chem200027545351533516110942781
- De LucaASantraMBaldiAGiordanoAIozzoRVDecorin-induced growth suppression is associated with up-regulation of p21, an inhibitor of cyclin-dependent kinasesJ Biol Chem19962713118961189658702560
- BuraschiSPalNTyler-RubinsteinNOwensRTNeillTIozzoRVDecorin antagonizes Met receptor activity and down-regulates {beta}-catenin and Myc levelsJ Biol Chem201028553420754208520974860
- IozzoRVBuraschiSGenuaMDecorin antagonizes IGF receptor I (IGF-IR) function by interfering with IGF-IR activity and attenuating downstream signalingJ Biol Chem201128640347123472121840990
- MiyasakaYEnomotoNNagayamaKAnalysis of differentially expressed genes in human hepatocellular carcinoma using suppression subtractive hybridizationBr J Cancer200185222823411461082
- ChungEJSungYKFarooqMGene expression profile analysis in human hepatocellular carcinoma by cDNA microarrayMol Cell2002143382387
- KrusiusTRuoslahtiEPrimary structure of an extracellular matrix proteoglycan core protein deduced from cloned cDNAProc Natl Acad Sci U S A19868320768376873484330
- McDoniels-SilversALNimriCFStonerGDLubetRAYouMDifferential gene expression in human lung adenocarcinomas and squamous cell carcinomasClin Cancer Res2002841127113811948124
- ShridharVLeeJPanditaAGenetic analysis of early-versus late-stage ovarian tumorsCancer Res200161155895590411479231
- TroupSNjueCKliewerEVReduced expression of the small leucine-rich proteoglycans, lumican, and decorin is associated with poor outcome in node-negative invasive breast cancerClin Cancer Res20039120721412538471
- IozzoRVThe family of the small leucine-rich proteoglycans: key regulators of matrix assembly and cellular growthCrit Rev Biochem Mol Biol19973221411749145286
- GoldoniSIozzoRVTumor microenvironment: modulation by decorin and related molecules harboring leucine-rich tandem motifsInt J Cancer2008123112473247918798267
- ZhuJXGoldoniSBixGDecorin evokes protracted internalization and degradation of the epidermal growth factor receptor via caveolar endocytosisJ Biol Chem200528037324683247915994311
- CsordasGSantraMReedCCSustained down-regulation of the epidermal growth factor receptor by decorin. A mechanism for controlling tumor growth in vivoJ Biol Chem200027542328793288710913155
- SeidlerDGGoldoniSAgnewCDecorin protein core inhibits in vivo cancer growth and metabolism by hindering epidermal growth factor receptor function and triggering apoptosis via caspase-3 activationJ Biol Chem200628136264082641816835231
- HillRMadureiraPAWaismanDMLeePWDNA-PKCS binding to p53 on the p21 WAF1/CIP1 promoter blocks transcription resulting in cell deathOncotarget20112121094110822190353
- SchonherrELevkauBSchaeferLKresseHWalshKDecorin-mediated signal transduction in endothelial cells. Involvement of Akt/protein kinase B in up-regulation of p21(WAF1/CIP1) but not p27(KIP1)J Biol Chem200127644406874069211546775
- StanderMNaumannUWickWWellerMTransforming growth factor-beta and p-21: multiple molecular targets of decorin-mediated suppression of neoplastic growthCell Tissue Res1999296222122710382266
- GrantDSYeniseyCRoseRWTootellMSantraMIozzoRVDecorin suppresses tumor cell-mediated angiogenesisOncogene200221314765477712101415