Abstract
Cluster of differentiation 44 (CD44) is a transmembrane glycoprotein that serves as the receptor for the extracellular matrix component hyaluronic acid. CD44 has been reported to play key roles in cell proliferation, motility, and survival, but its role in breast cancer remains controversial. In this study, we conducted a meta-analysis. A total of 23 published Gene Expression Omnibus databases were included to evaluate the association between CD44 mRNA expression and clinicopathological characteristics or prognosis of the patients with breast cancer. Our analysis revealed that CD44 expression was associated with clinicopathological features, including the histological grade, estrogen receptor status, progesterone receptor status, and human epidermal growth factor receptor-2 status. Higher levels of CD44 expression were observed in the basal subtype of breast cancer both at the mRNA and protein levels (odds ratio [OR] =2.08, 95% confidence interval [CI]: 1.72–2.52; OR =2.11, 95% CI: 1.67–2.68). Patients with CD44 overexpression exhibited significantly worse overall survival (hazard ratio =1.27; 95% CI: 1.04–1.55). Whole gene profile analysis revealed that CD44 expression was enriched in basal-type breast cancer and correlated with epithelial–mesenchymal transition and cancer stem cell gene profiles. In summary, our analyses indicated that CD44 potentially might be a prognostic marker for breast cancer and thus can serve as a therapeutic target for basal-type breast cancer.
Introduction
Breast cancer is one of the most common female cancers, accounting for approximately 28% of all female cancers and the second leading cause of cancer-related deaths in women.Citation1 Progress has been made to the earlier diagnosis and better treatment of breast cancer during the past few decades, leading to the 5-year survival rates of breast cancer patients at approximately 85%. However, distant metastasis and recurrence still occur and result in poor prognosis. Therefore, there is an urgent need for identifying novel biomarkers that can be used to screen high-risk patients and help predict the progression and prognosis of breast cancer.Citation2–Citation4
Cluster of differentiation 44 (CD44) is a complex transmembrane glycoprotein that is encoded by the CD44 gene on chromosome 11p13.Citation5 CD44 consists of seven extracellular domains, a transmembrane domain, and a cytoplasmic domain.Citation6 CD44 has several isoforms, including CD44s and CD44v.Citation7,Citation8 Functionally, CD44 was initially identified as the receptor for the extracellular matrix component, hyaluronic acid (HA), and was involved in multiple physiological and pathological processes, such as angiogenesis, cell adhesion, inflammation, and cancer development.Citation9 In addition, CD44 has been reported to play important roles in cell proliferation, motility, and survival.Citation9,Citation10 A recent study indicated that CD44 expression was elevated in tumor-initiating cells in many kinds of cancer.Citation11 Thus, CD44 is thought to be a biomarker for cancer stem cells (CSCs).Citation12 Subsequent functional studies have shown that CD44 is involved in tumorigenesis and metastasis in many cancer types such as colon,Citation13–Citation15 bladder,Citation16 gastric,Citation17 and breast cancers.Citation18–Citation20 Studies on CD44 expression have suggested a correlation between it and clinical outcome in patients with breast cancer. It has been shown that the overexpression of CD44 has a bad impact on survival of breast cancer patients,Citation21 but different results were also reported.Citation22 Currently, the role of CD44 in breast cancer has not been clearly defined. To investigate the role of CD44 in breast cancer, a meta-analysis was performed. Our analysis indicated that CD44 expression was elevated in basal-type breast cancer. Currently, there are no effectively targeted therapies for patients with this subtype of breast cancer and prognosis is poor compared with other subtypes.Citation23 Since CD44 expression is associated with mesenchymal and CSC signature and predicts poor prognosis,Citation24,Citation25 our study indicates that CD44 may represent a potential therapeutic target for basal-type breast cancer.
Materials and methods
Database and literature search
We performed a comprehensive search of relevant Gene Expression Omnibus (GEO) databases for CD44 mRNA expression and literatures for CD44 protein level. First, we searched the ArrayExpress for uploaded databases within the topic of interest, using the search terms “breast cancer” by filtering Homo sapiens, RNA array, array assay, and all arrays. We also searched Oncomine for databases of breast cancer with mRNA information of CD44. Second, PubMed was reviewed to identify potentially relevant literatures using the search terms associated with CD44 (“CD44 antigen”, “hyaluronan-binding protein”, “receptors”, “hyaluronan”) and breast cancer (“breast neoplasm”, “breast tumor”, “breast carcinoma”, “mammary cancer”). The references were also searched to discover additional relevant publications.
Inclusion and exclusion criteria
This meta-analysis collected data aimed at evaluating the role of CD44 expression in breast cancer at both mRNA and protein levels. Databases that met the following criteria were included: 1) the datasets were about breast cancer; 2) CD44 expression was measured in these databases; 3) the sample capacity was >50; and 4) clinical information of patients was showed in these databases. The exclusion criteria were as follows: 1) the datasets were about animals such as mice and rabbits and 2) the datasets were about DNA, rather than RNA. When several databases shared the same patient population, only the latest and most complete datasets were included. Literature that met the following criteria were included: 1) patients recruited in the study were pathologically diagnosed with breast cancer; 2) CD44 expression was measured in breast cancer tissues; and 3) the hazard ratio (HR)/odds ratio (OR) and corresponding 95% confidence interval (CI) were reported or could be statistically extracted from the study. The exclusion criteria were as follows: 1) reviews, case reports, comments, letters, and conference abstract and 2) ineligible samples or those where the required data were not available. When several articles were from the same patient population, the latest or most complete article was included.
Data extraction
Data were abstracted in a standardized collection form, with information recorded as follows: last name of first author, publication year, country, duration, tumor–node–metastasis (TNM) stage, quality score, detection, and cutoff values for CD44. We reviewed ArrayExpress and Oncomine and identified 23 independent human breast cancer microarray datasets with CD44 mRNA expression and clinical data. Overall survival (OS), recurrence-free survival (RFS), and metastasis-free survival (MFS) were evaluated by Cox proportional HRs and 95% CIs using these numerical data. If HRs were not given in an article, we used the methods described by Tierney et al to calculate the statistical variables from published survival curves.Citation26 The quality of observational studies was evaluated according to the Newcastle–Ottawa Quality Assessment Scale. This scale reflects patient selection, study comparability, and outcomes and is based on the identification of eight sources of potential study bias. Two reviewers performed the literature search, study selection, and data abstraction independently, and disagreements between the reviewers were solved by discussion.
Statistical analysis
Statistical analysis was performed based on the requirements of the meta-analysis of observational studies. The STATA software package (Version 12.0; StataCorp LP, College Station, TX, USA) was utilized to perform the meta-analysis. The random-effect model was employed when heterogeneity was present, and the fixed-effect model was used when homogeneity was demonstrated. The heterogeneity of publication was evaluated by means of the chi-square-based Q statistic and inconsistency index (I2) statistic. Begg’s and Egger’s tests were employed to assess the publication bias. HRs were employed to assess the survival outcome of patients with breast cancer who had high CD44 expression, and HR >1 indicated that high expression of CD44 predicted worse survival of patients. The OR and 95% CI were used to evaluate the association between CD44 expression and clinicopathological parameters.
Results
Search results
The flow diagram for the identification of relevant studies is shown in . A total of 1,472 datasets and 1,147 literatures were initially identified by our search approach. For GEO databases, after the sample capacity and clinical information were checked, 23 datasetsCitation21,Citation27–Citation48 met the criteria for this analysis. For 1,147 literatures, after title/abstract scanning and full-text reading, 12 eligible articlesCitation22,Citation49–Citation59 were included. shows the features of these 23 studies. Four Gene Expression Omnibus series (GSE) datasets were analyzed for finding the difference in CD44 mRNA expression between breast tumors and normal breast tissues. For finding the association between CD44 mRNA expression and TNM stage, tumor grade, estrogen receptor (ER) status, progesterone receptor (PR) status, human epidermal growth factor receptor-2 (HER2) status, and basal-like breast cancer, four, 13, eleven, four, six, and seven GSE datasets, respectively, were analyzed. To estimate the prognostic role of CD44 mRNA expression in OS, RFS, and MFS, eleven, ten, and nine, respectively, GSE datasets were adopted. Three GSE datasets were analyzed for the association between CD44 mRNA expression and the RFS in basal-like breast cancer. shows the characteristics of 12 studies. A total of nine, eight, seven, and five articles were assessed for the correlation between CD44 protein abundance and ER status, PR status, HER2 status, and basal-like breast cancer, respectively. Clinical stages I and II were grouped as early-stage disease, whereas stages III and IV were grouped as late-stage disease. Clinical T stages 1 and 2 were identified as early T stage, and 3 and 4 were identified as late T stage. Clinical N stages 1 and 2 were classified into early N stage, and 3 and 4 were classified into late N stage. Histological grades I and II were pooled as low-grade disease, and III and IV were pooled as high-grade disease.
Figure 1 Flow diagram of article selection.
Abbreviation: CD44, cluster of differentiation 44.
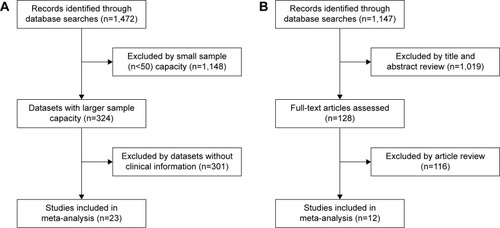
Table 1 Characteristics of the included studies by CD44 mRNA expression in the meta-analysis
Table 2 Characteristics of the included studies by CD44 protein abundance in the meta-analysis
CD44 expression correlates with clinicopathological features of breast cancer
Eighteen studies assessed the association between CD44 mRNA expression and tumor clinicopathological features. Our meta-analysis indicated that CD44 expression in breast cancer tissues was increased when compared with that in normal breast tissues (pooled OR =1.15, 95% CI: 1.02–1.31, Cochran’s Q test P=0.070, and I2=57.5%; ). However, there was no statistically significant correlation between CD44 expression and tumor TNM stage (pooled OR =1.10, 95% CI: 0.94–1.29, Cochran’s Q test P=0.039, and I2=64.1%; ), T stage (pooled OR =1.00, 95% CI: 0.84–1.19, Cochran’s Q test P=0.137, and I2=33.9%; ), and N status (pooled OR =0.98, 95% CI: 0.91–1.06, Cochran’s Q test P=0.006, and I2=57.8%; ). Patients with breast cancer with higher histological grade were likely to have a higher content of CD44 at both mRNA (pooled OR =1.15, 95% CI: 1.06–1.25, Cochran’s Q test P=0.582, and I2=0.0%; ) and protein levels (pooled OR =1.11, 95% CI: 1.02–1.20, Cochran’s Q test P=0.055, and I2=45.8%; ).
Figure 2 Correlation between CD44 mRNA expression or CD44 protein and breast cancer development and progression as evaluated by the OR.
Abbreviations: CD44, cluster of differentiation 44; CI, confidence interval; OR, odds ratio; TNM, tumor–node–metastasis.
CD44 expression correlates with molecular subtypes of breast cancer
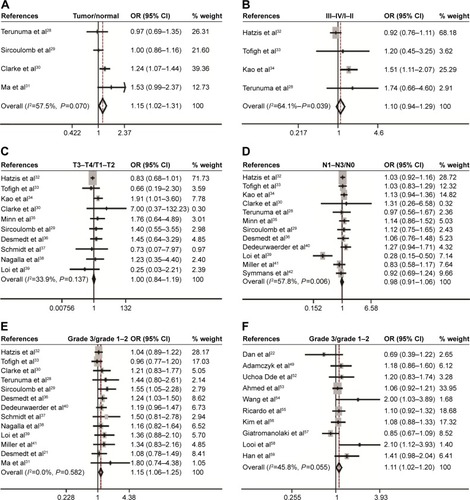
The association of CD44 expression with ER, PR, HER2 status, and basal-like breast cancer was also analyzed. At the mRNA level, CD44 was inversely correlated with ER status (pooled OR =1.93, 95% CI: 1.69–2.20, Cochran’s Q test P=0.002, and I2=63.9%; ), PR status (pooled OR =1.31, 95% CI: 1.14–1.51, Cochran’s Q test P=0.173, and I2=39.8%; ), and HER2 status (pooled OR =1.05, 95% CI: 1.00–1.10, Cochran’s Q test P=0.000, and I2=82.4%; ). Interestingly, CD44 mRNA expression was higher in basal-like tumors than in the luminal subtype of breast cancer (pooled OR =2.08, 95% CI: 1.72–2.52, Cochran’s Q test P=0.001, and I2=72.1%; ). At the protein level, CD44 expression was conversely linked to ER status (pooled OR =1.31, 95% CI: 1.15–1.48, Cochran’s Q test P=0.329, and ICitation2=12.7%; ). However, there is no statistical significance in terms of an association between CD44 expression and PR status (pooled OR =0.99, 95% CI: 0.90–1.08, Cochran’s Q test P=0.816, and I2=0.0%; ) or HER2 status (pooled OR =1.03, 95% CI: 0.98–1.08, Cochran’s Q test P=0.008, and ICitation2=65.5%; ) at protein level. Moreover, CD44 protein abundance in basal-like tumors was much higher than in the luminal subtype of breast cancer (pooled OR =2.11, 95% CI: 1.67–2.68, Cochran’s Q test P=0.017, and I2=66.9%; ).
Figure 3 Association between CD44 expression and molecular subtype.
Abbreviations: CD44, cluster of differentiation 44; CI, confidence interval; ER, estrogen receptor; HER2, human epidermal growth factor receptor-2; OR, odds ratio; PR, progesterone receptor.
CD44 mRNAexpression correlates with breast cancer survival
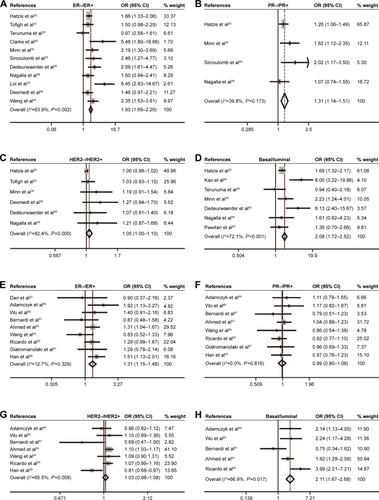
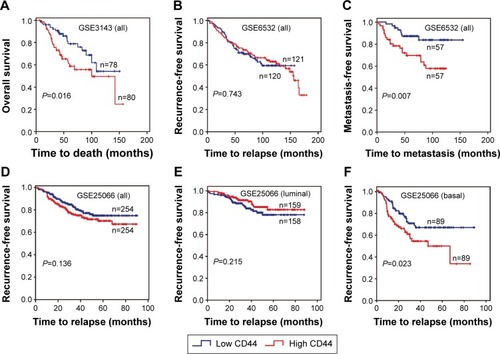
The association between CD44 expression level and breast cancer patient survival was analyzed. Our analysis indicated that there was a significant correlation between CD44 overexpression and the poor OS rate (pooled OR =1.27, 95% CI: 1.04–1.55, Cochran’s Q test P=0.505, and I2=0.0%; ). However, CD44 expression was not statistically significant in terms of an association between the RFS rate (pooled OR =1.04, 95% CI: 0.89–1.23, Cochran’s Q test P=0.417, and I2=2.4%; ) and the MFS rate (pooled OR =1.30, 95% CI: 0.89–1.90, Cochran’s Q test P=0.010, and I2=60.2%; ). Subcategory analyses according to the molecular classification of breast cancer were also performed. We found that higher CD44 mRNA expression correlated with worse RFS in patients with basal-like breast cancer (pooled OR =1.84, 95% CI: 1.17–2.87, Cochran’s Q test P=0.574, and I2=0.0%; ). However, there was no statistically significant correlation between CD44 mRNA expression and the survival performance of patients with luminal subtype of breast cancer. The latter included the OS rate (pooled OR =1.14, 95% CI: 0.73–1.79, Cochran’s Q test P=0.296, and I2=17.8%), the RFS rate (pooled OR =0.99, 95% CI: 0.75–1.31, Cochran’s Q test P=0.258, and I2=23.5%), and the MFS rate (pooled OR =1.25, 95% CI: 0.65–2.38, Cochran’s Q test P=0.010, and I2=69.7%). Kaplan–Meier survival analysis of GSE3143 demonstrated that there was a significant effect of CD44 on OS (P=0.016; ). Kaplan–Meier survival analysis of GSE6532 showed that there were no significant effects of CD44 on the RFS in all population of breast cancer (P=0.743; ), but it was inversely associated with the MFS rate (P=0.007; ). Kaplan–Meier survival analysis of GSE25066 demonstrated that there was a significant effect of CD44 on the RFS in basal-like breast cancer (P=0.023; ) but no significant association between CD44 mRNA expression and the RFS in all molecular subtypes (P=0.136; ) or in luminal breast cancer (P=0.215; ). In all, the results from the CD44 mRNA profile indicated that higher CD44 expression predicted a poorer prognosis in patients with breast cancer subtype.
Figure 4 Forest plot for the correlation of CD44 mRNA expression with breast cancer survival.
Abbreviations: CD44, cluster of differentiation 44; CI, confidence interval; HR, hazard ratio; MFS, metastasis-free survival; OS, overall survival; RFS, recurrence-free survival.
Figure 5 Kaplan–Meier survival curves for the correlation of CD44 mRNA expression with breast cancer.
Abbreviations: CD44, cluster of differentiation 44; MFS, metastasis-free survival; OS, overall survival; RFS, recurrence-free survival.
CD44 correlates with epithelial–mesenchymal transition and CSC markers
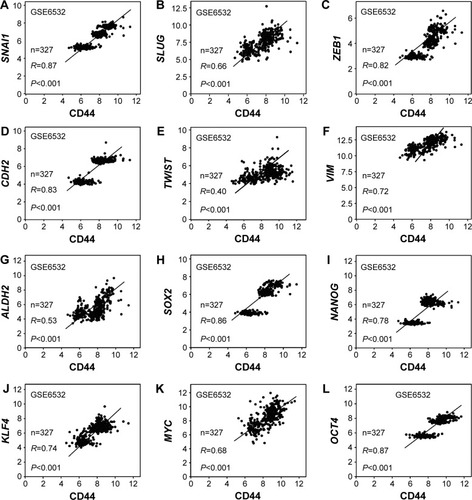
The association between CD44 and epithelial–mesenchymal transition (EMT) or CSC-related genes was also assessed. The results indicated that there was a positive relation between CD44 and SNAI1 (R=0.87, P<0.001; ), SLUG (R=0.66, P<0.001; ), ZEB1 (R=0.82, P<0.001; ), CDH2 (R=0.83, P<0.001; ), TWIST (R=0.40, P<0.001; ), and VIM (R=0.72, P<0.001; ). The association between CD44 and CSC markers was also evaluated. It was shown that CD44 was positively associated with ALDH1 (R=0.53, P<0.001; ), SOX2 (R=0.86, P<0.001; ), NANOG (R=0.78, P<0.001; ), KLF4 (R=0.74, P<0.001; ), MYC (R=0.68, P<0.001; ), and OCT4 (R=0.87, P<0.001; ).
Figure 6 CD44 expression was associated with stem cell and EMT markers.
Abbreviations: CD44, cluster of differentiation 44; EMT, epithelial–mesenchymal transition.
Publication bias
Publication bias statistics were obtained using Begg’s and Egger’s tests, and did not indicate any significant publication bias; CD44 mRNA expression: breast cancer: Begg’s test P=0.734, Egger’s test P=0.905; TNM stage: Begg’s test P=1, Egger’s test P=0.796; tumor size: Begg’s test P=0.466, Egger’s test P=0.362; lymph node metastasis: Begg’s test P=0.945, Egger’s test P=0.097; histological grade: Begg’s test P=0.246, Egger’s test P=0.948; expression of ER: Begg’s test P=0.640, Egger’s test P=0.313; expression of PR: Begg’s test P=1, Egger’s test P=0.809; expression of Her2: Begg’s test P=0.260, Egger’s test P=0.494; basal-like breast cancer: Begg’s test P=1, Egger’s test P=0.77; OS: Begg’s test P=0.436, Egger’s test P=0.436; RFS: Begg’s test P=0.592, Egger’s test P=0.612; MFS: Begg’s test P=0.251, Egger’s test P=0.146; OS of luminal breast cancer: Begg’s test P=1, Egger’s test P=0.642; RFS of luminal breast cancer: Begg’s test P=0.260, Egger’s test P=0.436; MFS of luminal breast cancer: Begg’s test P=0.806, Egger’s test P=0.528; RFS of basal-like breast cancer: Begg’s test P=1, Egger’s test P=0.698. Protein level: histological grade: Begg’s test P=0.917, Egger’s test P=0.911; expression of ER: Begg’s test P=0.917, Egger’s test P=0.009; expression of PR: Begg’s test P=0.266, Egger’s test P=0.743; expression of HER2: Begg’s test P=1, Egger’s test P=0.434; and basal-like breast cancer: Begg’s test P=0.462, Egger’s test P=0.065.
Discussion
Molecular characterization contributes to the discovery of biomarkers and potential targets for anticancer therapy, which is the basis of precise medicine.Citation60 Accumulating evidence suggests that CD44 is a marker of tumor-initiating cells, plays a role in tumorigenesis, and linked to the progression of breast cancer.Citation15,Citation61–Citation63 CD44 was also reported to have an impact on the prognosis of breast cancer including recurrenceCitation64 and chemoresistance.Citation65 Uchino et al found that the upregulation of CD44 represented an aggressive subtype in noninvasive breast cancer cell.Citation19 The blockade of CD44 intracellular domain (CD44ICD) cleavage and nuclear translocation have been shown in cancer cells. The activation of CD44 by HA promoted the chemoresistance in breast cancer cells.Citation66 CD44/cellular prion protein interaction has an effect on the responses to neoadjuvant chemotherapy in patients with breast cancer and exhibits aggressive behaviors of breast cancer cells.Citation67 CD44–STAT3 interaction plays an important role in breast cancer invasion.Citation64 Moreover, Cox regression analysis showed that ezrin and CD44 co-expression were independent prognostic factors of breast cancer.Citation68
In our meta-analysis, the role of CD44 in breast cancer, at both mRNA and protein levels, was investigated. We found that the mRNA level of CD44 was higher in breast tumor tissues than in normal breast tissues, indicating that CD44 might participate in the tumorigenesis of specific subtypes of breast cancer. Moreover, our meta-analysis suggests a positive association between histological grade and the CD44 levels. This would indicate that patients with high expression of CD44 mRNA might have poor prognosis, because high-grade tumor tends to be more aggressive and tends toward early recurrence. It has been shown that CD44 was activated in breast cancer cells but inactivated in normal cells in vitro and in vivo.Citation69 However, the association between CD44 mRNA expression and TNM stage, T stage, and N stage was not statistically significant.
Based on the status of ER, PR, and HER2, breast cancer could be divided into five molecular subtypes, including normal-like, luminal A, luminal B, HER2-overexpressing, and basal-like breast cancer.Citation70,Citation71 Each subtype exhibits distinctive expression patterns of specific molecules, clinical outcomes, and responses to adjuvant chemotherapy.Citation33,Citation72,Citation73 Some studies indicated that CD44 expression was negatively associated with the status of ER,Citation49,Citation53,Citation74,Citation75 PR,Citation74 and HER2.Citation53,Citation74 At the mRNA level, our meta-analysis showed that CD44 expression was significantly inversely associated with the status of ER, PR, and HER2. Consistently, significant correlation between CD44 expression and ER status was found at its protein level. Among the five molecular subtypes, basal-like breast cancer tends to be more aggressive and there is a lack of effective therapy, resulting in poorer outcomes.Citation76 Jang et al showed that CD44(+)/CD24(−) subpopulation was much higher in basal-like breast cancer than that in non-basal-like cancer,Citation24 and that CD44(+)/CD24(−) cells had a high capacity of proliferation, migration, invasion, and tumorigenesis.Citation25 By providing a highly hydrated environment favoring cellular invasion, HA–CD44 interaction contributed to the progression of basal-like breast cancer.Citation77 Consistently, our results showed that CD44 expression was higher in basal-like breast cancers than in luminal breast cancer or all other subtypes.
CD44 is critical for regulating EMT.Citation19 CD44 activation can lead to the expression of epithelial growth factor receptor and the activation of phosphoinositide-3 kinase/Akt. CD44 also upregulates N-cadherin, α-actin, vimentin, fibronectin, and other EMT markers. The latter is involved in cell invasion and migration.Citation78 By knocking down CD44 expression in human hepatoma cell line HLE, the levels of snail and vimentin were decreased, which was correlated with a less-mesenchymal-like phenotype.Citation79 Consistently, our analysis indicated that CD44 expression was significantly associated with mesenchymal gene SNAI1, SLUG, ZEB1, CDH2, and TWIST.
CD44 is a well-known breast CSC marker that plays a role in promoting tumorigenesis of breast cancer through interaction with its intracellular domain and stemness factors such as NANOG, OCT4, and SOX2.Citation20 Analysis of gene expression profiles revealed that CD44 is closely associated with key stem cell genes ALDH1, SOX2, NANOG, KLF4, OCT4, and MYC. Since CSC is thought to be a major cause for cancer progression and therapeutic resistance,Citation80,Citation81 the role of CD44 in breast cancer might be attributable to those stem cell factors.
Studies identified several genes that might have prognostic values for breast cancer, including urokinase plasminogen activator and its inhibitorCitation82 and the genes in the DACH–EYA–SIX pathway.Citation83–Citation85 Interestingly, insulin-like growth factor 1 receptor expression showed different prognostic values for patients with different subtypes of breast cancer.Citation86 Ubiquitin protein D and KLF4 have been reported to predict the response to chemotherapy.Citation87,Citation88 Accumulating evidence indicates that CD44 could be a prognostic biomarker for breast cancer.Citation12 Our meta-analysis suggested that CD44 high expression could be a prognostic marker for OS. Although there was no association between CD44 expression and RFS in the whole population of breast cancer, a significant association between CD44 mRNA expression and RFS in patients with basal-like breast cancer was identified. This agrees with a previous study showing that patients with CSC markers CD44(+)/CD24(−) had a lower survival rate, while patients without this subpopulation had a higher survival rate in basal-like breast cancer.Citation89 Some studies showed that CD44 expression was positively correlated with the metastasis of breast carcinoma,Citation75 but others reported opposite results.Citation90 Martin and Jiang found that CD44 was markedly reduced in patients with ductal breast cancer with metastasis.Citation91 Our meta-analysis showed that CD44 expression has no significant effect on the MFS (), but some GSE data did demonstrate that CD44 was correlated with the MFS (). Breast cancer metastasis is a complicated process which is involved in the alteration of a number of proteins, including epithelial growth factor receptor and transforming growth factor-β.Citation92 Considering the complex regulation of the metastasis process of breast cancer, the effects of CD44 on the MFS might be covered by other factors.
Heterogeneity tests are essential to a meta-analysis. In this study, the evidence of minor heterogeneities was observed with respect to TNM stage, ER status, molecular subtypes, and the MFS. However, there was substantial heterogeneity with respect to HER2 status. This result might be due to the following aspects: 1) The sample size is limited, indicating that multicenter prospective studies are needed. 2) The variations in assessing CD44 mRNA expression might also contribute to heterogeneity. The cutoff value was estimated in 23 studies using the median CD44 level measured by gene microarray. 3) Publication bias is worth considering in meta-analyses. This study was a meta-analysis based on GEO datasets and published studies. Thus, our analysis has the following limitations: 1) we cannot exclude the publication bias; 2) the relevant papers were limited; and 3) methods and cutoff values used to assess CD44 expression were different.
Conclusion
In conclusion, our meta-analysis suggests that CD44 might be a prognostic factor for patients with breast cancer, particularly for the basal-like breast cancer. Since CD44 expression was elevated in basal-type breast cancer and its expression levels were correlated with EMT and CSC signatures, these considerations might partially explain why patients with basal-type breast cancer have a high risk of metastasis and relapse. Moreover, our meta-analysis might help identify subpopulation of patients with breast cancer for CD44-based therapy in the future.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (Nos 81572608, 81172422, and 81072169).
Disclosure
The authors report no conflicts of interest in this work.
References
- SiegelRLMillerKDJemalACancer statistics, 2015CA Cancer J Clin201565152925559415
- RadenkovicSMilosevicZKonjevicGLactate dehydrogenase, catalase, and superoxide dismutase in tumor tissue of breast cancer patients in respect to mammographic findingsCell Biochem Biophys201366228729523197387
- RadenkovicSKonjevicGJurisicVValues of MMP-2 and MMP-9 in tumor tissue of basal-like breast cancer patientsCell Biochem Biophys201468114315223812723
- StankovicSKonjevicGGopcevicKActivity of MMP-2 and MMP-9 in sera of breast cancer patientsPathol Res Pract2010206424124720092959
- ScreatonGRCaceresJFMayedaAIdentification and characterization of three members of the human SR family of pre-mRNA splicing factorsEMBO J19951417433643497556075
- van der WindtGJSchoutenMZeerlederSCD44 is protective during hyperoxia-induced lung injuryAm J Respir Cell Mol Biol201144337738320463290
- XuHTianYYuanXThe role of CD44 in epithelial-mesenchymal transition and cancer developmentOnco Targets Ther201583783379226719706
- ErbUMegaptcheAPGuXCD44 standard and CD44v10 isoform expression on leukemia cells distinctly influences niche embedding of hematopoietic stem cellsJ Hematol Oncol201472924684724
- NaorDSionovRVIsh-ShalomDCD44: structure, function, and association with the malignant processAdv Cancer Res1997712413199111868
- PontaHShermanLHerrlichPACD44: from adhesion molecules to signalling regulatorsNat Rev Mol Cell Biol200341334512511867
- ZollerMCD44: can a cancer-initiating cell profit from an abundantly expressed molecule?Nat Rev Cancer201111425426721390059
- BasakranNSCD44 as a potential diagnostic tumor markerSaudi Med J201536327327925737167
- DalerbaPDyllaSJParkIKPhenotypic characterization of human colorectal cancer stem cellsProc Natl Acad Sci U S A200710424101581016317548814
- DuLWangHHeLCD44 is of functional importance for colorectal cancer stem cellsClin Cancer Res200814216751676018980968
- Santoyo-RamosPLikhatchevaMGarcia-ZepedaEAHypoxia-inducible factors modulate the stemness and malignancy of colon cancer cells by playing opposite roles in canonical Wnt signalingPLoS One2014911e11258025396735
- WuKNingZZengJSilibinin inhibits beta-catenin/ZEB1 signaling and suppresses bladder cancer metastasis via dual-blocking epithelial-mesenchymal transition and stemnessCell Signal201325122625263324012496
- YuDShinHSLeeYSmiR-106b modulates cancer stem cell characteristics through TGF-beta/Smad signaling in CD44-positive gastric cancer cellsLab Invest201494121370138125286029
- XieGYaoQLiuYIL-6-induced epithelial-mesenchymal transition promotes the generation of breast cancer stem-like cells analogous to mammosphere culturesInt J Oncol20124041171117922134360
- UchinoMKojimaHWadaKNuclear beta-catenin and CD44 upregulation characterize invasive cell populations in non-aggressive MCF-7 breast cancer cellsBMC Cancer20101041420696077
- ChoYLeeHWKangHGCleaved CD44 intracellular domain supports activation of stemness factors and promotes tumorigenesis of breast cancerOncotarget20156118709872125909162
- DesmedtCPietteFLoiSStrong time dependence of the 76-gene prognostic signature for node-negative breast cancer patients in the TRANSBIG multicenter independent validation seriesClin Cancer Res200713113207321417545524
- DanTHewittSMOhriNCD44 is prognostic for overall survival in the NCI randomized trial on breast conservation with 25 year follow-upBreast Cancer Res Treat20141431111824276281
- TomaoFPapaAZaccarelliETriple-negative breast cancer: new perspectives for targeted therapiesOnco Targets Ther2015817719325653541
- JangMHKimHJKimEJExpression of epithelial-mesenchymal transition-related markers in triple-negative breast cancer: ZEB1 as a potential biomarker for poor clinical outcomeHum Pathol20154691267127426170011
- MaFLiHWangHEnriched CD44(+)/CD24(−) population drives the aggressive phenotypes presented in triple-negative breast cancer (TNBC)Cancer Lett2014353215315925130168
- TierneyJFStewartLAGhersiDPractical methods for incorporating summary time-to-event data into meta-analysisTrials200781617555582
- HeikkinenTGrecoDPelttariLMVariants on the promoter region of PTEN affect breast cancer progression and patient survivalBreast Cancer Res2011136R13022171747
- TerunumaAPutluriNMishraPMYC-driven accumulation of 2-hydroxyglutarate is associated with breast cancer prognosisJ Clin Invest2014124139841224316975
- SircoulombFBekhoucheIFinettiPGenome profiling of ERBB2-amplified breast cancersBMC Cancer20101053920932292
- ClarkeCMaddenSFDoolanPCorrelating transcriptional networks to breast cancer survival: a large-scale coexpression analysisCarcinogenesis201334102300230823740839
- MaXJDahiyaSRichardsonEGene expression profiling of the tumor microenvironment during breast cancer progressionBreast Cancer Res2009111R719187537
- HatzisCPusztaiLValeroVA genomic predictor of response and survival following taxane-anthracycline chemotherapy for invasive breast cancerJAMA2011305181873188121558518
- TofighASudermanMPaquetERThe prognostic ease and difficulty of invasive breast carcinomaCell Rep20149112914225284793
- KaoKJChangKMHsuHCCorrelation of microarray-based breast cancer molecular subtypes and clinical outcomes: implications for treatment optimizationBMC Cancer20111114321501481
- MinnAJGuptaGPSiegelPMGenes that mediate breast cancer metastasis to lungNature2005436705051852416049480
- DesmedtCDi LeoAde AzambujaEMultifactorial approach to predicting resistance to anthracyclinesJ Clin Oncol201129121578158621422418
- SchmidtMBohmDvon TorneCThe humoral immune system has a key prognostic impact in node-negative breast cancerCancer Res200868135405541318593943
- NagallaSChouJWWillinghamMCInteractions between immunity, proliferation and molecular subtype in breast cancer prognosisGenome Biol2013144R3423618380
- LoiSHaibe-KainsBDesmedtCDefinition of clinically distinct molecular subtypes in estrogen receptor-positive breast carcinomas through genomic gradeJ Clin Oncol200725101239124617401012
- DedeurwaerderSDesmedtCCalonneEDNA methylation profiling reveals a predominant immune component in breast cancersEMBO Mol Med201131272674121910250
- MillerLDSmedsJGeorgeJAn expression signature for p53 status in human breast cancer predicts mutation status, transcriptional effects, and patient survivalProc Natl Acad Sci U S A200510238135501355516141321
- SymmansWFHatzisCSotiriouCGenomic index of sensitivity to endocrine therapy for breast cancerJ Clin Oncol201028274111411920697068
- PawitanYBjohleJAmlerLGene expression profiling spares early breast cancer patients from adjuvant therapy: derived and validated in two population-based cohortsBreast Cancer Res200576R953R96416280042
- WangYKlijnJGZhangYGene-expression profiles to predict distant metastasis of lymph-node-negative primary breast cancerLancet2005365946067167915721472
- HuZFanCLivasyCA compact VEGF signature associated with distant metastases and poor outcomesBMC Med20097919291283
- HennessyBTGonzalez-AnguloAMStemke-HaleKCharacterization of a naturally occurring breast cancer subset enriched in epithelial-to-mesenchymal transition and stem cell characteristicsCancer Res200969104116412419435916
- BildAHYaoGChangJTOncogenic pathway signatures in human cancers as a guide to targeted therapiesNature2006439707435335716273092
- MinnAJGuptaGPPaduaDLung metastasis genes couple breast tumor size and metastatic spreadProc Natl Acad Sci U S A2007104166740674517420468
- AdamczykANiemiecJAAmbickaACD44/CD24 as potential prognostic markers in node-positive invasive ductal breast cancer patients treated with adjuvant chemotherapyJ Mol Histol2014451354523835592
- WuYSarkissyanMElshimaliYTriple negative breast tumors in African–American and Hispanic/Latina women are high in CD44+, low in CD24+, and have loss of PTENPLoS One2013810e7825924167614
- BernardiMALogulloAFPasiniFSPrognostic significance of CD24 and claudin-7 immunoexpression in ductal invasive breast cancerOncol Rep2012271283821956537
- Uchoa DdeMGraudenzMSCallegari-JacquesSMExpression of cancer stem cell markers in basal and penta-negative breast carcinomas – a study of a series of triple-negative tumorsPathol Res Pract2014210743243924726267
- AhmedMAAleskandaranyMARakhaEAA CD44(−)/CD24(+) phenotype is a poor prognostic marker in early invasive breast cancerBreast Cancer Res Treat2012133397999522119938
- WangZShiQWangZClinicopathologic correlation of cancer stem cell markers CD44, CD24, VEGF and HIF-1alpha in ductal carcinoma in situ and invasive ductal carcinoma of breast: an immunohistochemistry-based pilot studyPathol Res Pract2011207850551321802218
- RicardoSVieiraAFGerhardRBreast cancer stem cell markers CD44, CD24 and ALDH1: expression distribution within intrinsic molecular subtypeJ Clin Pathol2011641193794621680574
- KimHJKimMJAhnSHDifferent prognostic significance of CD24 and CD44 expression in breast cancer according to hormone receptor statusBreast2011201788520810282
- GiatromanolakiASivridisEFiskaAThe CD44+/CD24− phenotype relates to ‘triple-negative’ state and unfavorable prognosis in breast cancer patientsMed Oncol201128374575220405247
- LooiLMCheahPLZhaoWCD44 expression and axillary lymph node metastasis in infiltrating ductal carcinoma of the breastMalays J Pathol2006282838618376796
- HanZChenZZhengRClinicopathological significance of CD133 and CD44 expression in infiltrating ductal carcinoma and their relationship to angiogenesisWorld J Surg Oncol2015135625889325
- SmithADRodaDYapTAStrategies for modern biomarker and drug development in oncologyJ Hematol Oncol201477025277503
- MontgomeryNHillAMcFarlaneSCD44 enhances invasion of basal-like breast cancer cells by upregulating serine protease and collagen-degrading enzymatic expression and activityBreast Cancer Res2012143R8422621373
- ParkJSchledererMSchreiberMAF1q is a novel TCF7 co-factor which activates CD44 and promotes breast cancer metastasisOncotarget2015624206972071026079538
- NamKOhSLeeKMCD44 regulates cell proliferation, migration, and invasion via modulation of c-Src transcription in human breast cancer cellsCell Signal20152791882189425979842
- SoJYSmolarekAKSalernoDMTargeting CD44-STAT3 signaling by Gemini vitamin D analog leads to inhibition of invasion in basal-like breast cancerPLoS One201381e5402023326564
- BoulbesDRChauhanGBJinQCD44 expression contributes to trastuzumab resistance in HER2-positive breast cancer cellsBreast Cancer Res Treat2015151350151325971596
- WeiXXuMWeiYThe addition of rituximab to CHOP therapy alters the prognostic significance of CD44 expressionJ Hematol Oncol201473424739401
- ChengYTaoLXuJCD44/cellular prion protein interact in multidrug resistant breast cancer cells and correlate with responses to neoadjuvant chemotherapy in breast cancer patientsMol Carcinog201453968669723681900
- MaLJiangTClinical implications of Ezrin and CD44 coexpression in breast cancerOncol Rep20133041899190523900701
- YangCHeYZhangHSelective killing of breast cancer cells expressing activated CD44 using CD44 ligand-coated nanoparticles in vitro and in vivoOncotarget2015617152831529625909172
- PerouCMSorlieTEisenMBMolecular portraits of human breast tumoursNature2000406679774775210963602
- SorlieTPerouCMTibshiraniRGene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implicationsProc Natl Acad Sci U S A20019819108691087411553815
- CameriniADonatiSViacavaPEvaluation of HER2 and p53 expression in predicting response to docetaxel-based first-line chemotherapy in advanced breast cancerJ Exp Clin Canc Res20113038
- Cancer Genome Atlas NetworkComprehensive molecular portraits of human breast tumoursNature20124907418617023000897
- McFarlaneSCoulterJATibbitsPCD44 increases the efficiency of distant metastasis of breast cancerOncotarget2015613114651147625888636
- BolodeokuJYoshidaKSuginoTCD44 expression in human breast cancer cell lines is related to oestrogen receptor (ER) status and confluency in vitroBiochem Soc Trans1997252356S9191401
- SorlieTTibshiraniRParkerJRepeated observation of breast tumor subtypes in independent gene expression data setsProc Natl Acad Sci U S A2003100148418842312829800
- HeldinPBasuKKozlovaIHAS2 and CD44 in breast tumorigenesisAdv Cancer Res201412321122925081531
- ChoSHParkYSKimHJCD44 enhances the epithelial-mesenchymal transition in association with colon cancer invasionInt J Oncol201241121121822552741
- FernandoJMalfettoneACepedaEBA mesenchymal-like phenotype and expression of CD44 predict lack of apoptotic response to sorafenib in liver tumor cellsInt J Cancer20151364E161E17225053293
- SchulenburgABlattKCerny-ReitererSCancer stem cells in basic science and in translational oncology: can we translate into clinical application?J Hematol Oncol2015811625886184
- YinXZhangBHZhengSSCoexpression of gene Oct4 and Nanog initiates stem cell characteristics in hepatocellular carcinoma and promotes epithelial-mesenchymal transition through activation of Stat3/Snail signalingJ Hematol Oncol201582325879771
- WitzelIMilde-LangoschKSchmidtMRole of urokinase plasminogen activator and plasminogen activator inhibitor mRNA expression as prognostic factors in molecular subtypes of breast cancerOnco Targets Ther201472205221325506225
- WuKChenKWangCCell fate factor DACH1 represses YB-1- mediated oncogenic transcription and translationCancer Res201474382983924335958
- WuKLiZCaiSEYA1 phosphatase function is essential to drive breast cancer cell proliferation through cyclin D1Cancer Res201373144488449923636126
- LiuYHanNZhouSThe DACH/EYA/SIX gene network and its role in tumor initiation and progressionInt J Cancer Epub201549
- YanSJiaoXLiKThe impact of IGF-1R expression on the outcomes of patients with breast cancer: a meta-analysisOnco Targets Ther2015827928725674003
- HanTLiuZLiHHigh expression of UBD correlates with epirubicin resistance and indicates poor prognosis in triple-negative breast cancerOnco Targets Ther201581643164926185453
- DongMJWangLBJiangZNThe transcription factor KLF4 as an independent predictive marker for pathologic complete remission in breast cancer neoadjuvant chemotherapy: a case-control studyOnco Targets Ther201471963196925368523
- ChekhunSVZadvornyTVTymovskaYOCyrillicD44+/CD24− markers of cancer stem cells in patients with breast cancer of different molecular subtypesExp Oncol2015371586325804234
- LohTJMoonHChoSCD44 alternative splicing and hnRNP A1 expression are associated with the metastasis of breast cancerOncol Rep20153431231123826151392
- MartinTAJiangWGEvaluation of the expression of stem cell markers in human breast cancer reveals a correlation with clinical progression and metastatic disease in ductal carcinomaOncol Rep201431126227224173498
- De CraeneBBerxGRegulatory networks defining EMT during cancer initiation and progressionNat Rev Cancer20131329711023344542
