Abstract
Background
Previous studies have shown the central role of 78 kDa glucose-regulated protein (GRP78) in colorectal cancer (CRC) survival and chemoresistance. In the present study, we aimed to design a GRP78 inhibitor and test its potential to inhibit CRC cells growth.
Materials and methods
Computer-aided drug design was used to establish novel compounds as potential inhibitors of GRP78. Discovery Studio 3.5 software was used to evaluate a series of designed compounds and assess their mode of binding to the active site of the protein. The cytotoxicity of the designed compounds was evaluated using the MTT assay and the propidium iodide method. The effect of the inhibitor on the expression of GRP78 was evaluated by immunoblotting.
Results
Among the designed compounds, only potassium-3-beta-hydroxy-20-oxopregn-5-en-17-alpha-yl sulfate (PHOS) has a potential to inhibit the growth of CRC cells. Inhibition of cellular growth was largely attributed to downregulation of GRP78 and induction of apoptotic cell death.
Conclusion
These results introduce PHOS as a promising GRP78 inhibitor that could be used in future studies as a combination with chemotherapy in the treatment of CRC patients. Our ongoing studies aim to characterize PHOS safety profile as well as its mechanism of action.
Introduction
Cellular conditions such as hypoxia, alterations in glycosylation status, and disturbances of calcium flux can lead to the accumulation of unfolded and/or misfolded proteins in the endoplasmic reticulum (ER) lumen and challenge the function of the ER–Golgi network, resulting in ER stress.Citation1–Citation3 The ER responds to stress conditions by unfolded protein response (UPR), which involves activation of a range of signaling pathways that couple ER protein folding load with ER protein folding capacity.Citation1–Citation3
The UPR is of fundamental importance for the survival of all eukaryotic cells under conditions of ER stress. The 78 kDa glucose-regulated protein (GRP78), also called heat shock protein A5 (HSPA5) or binding immunoglobulin protein (Bip), mediates UPR by sequestration of unfolded proteins, which activate three ER transmembrane protein sensors: activating transcription factor 6 (ATF6), inositol-requiring enzyme 1 (IRE1), and double stranded RNA-activated protein kinase-like ER kinase (PERK).Citation1–Citation3 These activations involve phosphorylation and homodimerization of IRE1 and PERK, and relocation and proteolytic cleavage of ATF6.Citation1–Citation3
In addition to the UPR sensing, GRP78 protein acts as ER chaperone that facilitates proper protein folding and targets misfolded proteins for degradation by proteasome. It is also involved in ER calcium binding.Citation1–Citation3 Elevated expression of GRP78 has been reported in several cancers, such as breast cancer, colorectal cancer (CRC), and prostate cancer.Citation4,Citation5 Moreover, GRP78 expression has been associated with tumor development and growth, vascularization, metastasis, and positively correlated with cancer resistance to chemotherapy including CRC cells.Citation4,Citation6,Citation7 It has been shown that cancer cells may have adapted to ER stress by activation of the UPR without resulting in apoptosis.Citation8,Citation9 The central feature of this adaptive response is the maintenance of the expression of proteins that facilitate survival, such as GRP78.Citation9
Structurally, GRP78 is a monomer that is composed of 654 amino acids and contains an active site that is fit for ATP binding (ATPase pocket). This pocket can be exploited to design inhibitors which possess an affinity to this active site more than ATP, to be successful.Citation10
Our previous studies have shown that CRC cells differentially express GRP78 protein, thus variably respond to chemotherapy. For example, the constitutive expression of GRP78 in SW480 cells was higher than that in the chemotherapy sensitive HCT116 cells.Citation11,Citation12 The rationale of the present study was to study the active site of GRP78 inhibitors utilizing computer-aided drug design techniques. Based on computer modeling, the active site will be subjected to mapping procedure using structure-based pharmacophore modeling in Discovery Studio 3.5 based on commercially available compound database. Molecules showing the best scoring rate will be tested against both sensitive (HCT116) and resistant (SW480) CRC cells and their toxicity will be determined as a possible future use in chemotherapy combination against CRC.
Materials and methods
Computer-aided drug design Pharmacophore design
Protein GRP78 was searched for any crystallized structures in the Protein Data Bank, and this resulted in identifying a series of crystallized proteins that vary in the nature of the ligand crystallized inside the active site and the resolution of the crystallization process ().Citation13 After manual investigation of the crystallized proteins using Discovery Studio 3.5 (BIOVIA, San Diego, CA, USA), the selection was finalized by choosing a protein that has an ATP molecule in the active site and with a good resolution, namely, 3LDL.Citation10 The crystal structure of 3LDL has a resolution power of 2.3 Å, which is considered an acceptable resolution for the structure-based drug design strategy (). The 406 amino acids which are the nucleotide-binding domain of this protein have been downloaded from the Protein Data Bank and cleaned according to Prepare Protein protocol. The whole protein structure is not available in the Protein Data Bank.Citation14 The protein structure was then minimized to local minima to ensure the protein has been relaxed and all the strain that was found during the process of crystallization has been alleviated. In this step, a strategic step by drawing the attached ligand (ATP) was performed. Next, possible interactions of ATP were studied to design a molecule that has similar properties to ATP. This step was performed using Receptor–Ligand Interaction Generation protocol.Citation14 Thereafter and using Ligand Pharmacophore Mapping, the selection of a potent inhibitor was confirmed.Citation15 Finally, screening of commercially available chemical compounds, namely ALDRICHCPR (Sigma-Aldrich Co., St Louis, MO, USA), was performed as described in previous studies.Citation16,Citation17
Figure 1 A 3D structure of 78 kDa glucose-regulated protein where the active site is occupied with an ATP molecule.
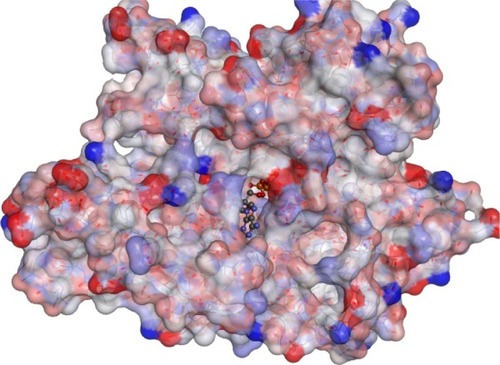
Table 1 A sample of four crystal structures found in the Protein Data Bank which show their PBB codes, resolution they were crystallized upon, and type of ligand attached to each of them
Antitumor activity
The acute cytotoxic effect of the compounds on CRC cell lines (American Type Culture Collection, Manassas, VA, USA) was determined using the MTT assays. This study was approved by the Scientific Research Committee at the Deanship of Research, Jordan University of Science and Technology, and the Scientific Research Funds at the Ministry of Higher Education and Scientific Research, Amman, Jordan. Further ethical approval was not required for this study as it did not involve human- or vertebrate animal-related biological materials or data. Briefly, cells were seeded at 5,000/well onto flat-bottomed 96-well culture plates and allowed to grow for 24 hours before the desired treatment. Cells were then incubated with a wide range of compound concentrations (0–100 µM) for 72 hours. Cells were then labeled with MTT from the Vybrant MTT Cell proliferation assay kit (Molecular Probes, Eugene, OR, USA) according to the manufacturer’s instruction and the resulting formazan was solubilized in dimethylsulfoxide (DMSO). Absorbance was read using a microplate reader at 540 nm.
Apoptosis
Apoptotic cells were determined by the propidium iodide method.Citation18 In brief, CRC cells were adhered overnight in a 24-well plate at a concentration of 1×105/well in 10% fetal calf serum (FCS). Cells in suspension were added on the day of the assay. The medium was removed and 1,000 µL of fresh medium and 10% FCS containing one of the proposed GRP78 inhibitors at 50 µM was added. Then, cells were incubated for 72 hours at 37°C, followed by medium removal. Adhered and suspended cells were washed once with phosphate-buffered saline (PBS). The medium and PBS were placed in a 12×75 mm Falcon polystyrene tube and centrifuged at 200× g for 5 minutes. A hypotonic buffer 1 mL (propidium iodide, 50 µg/mL, in 0.1% sodium citrate plus 0.1% Triton X-100; Sigma-Aldrich Co) was added directly to the cells adhered in the 24-well plate. The buffer was gently pipetted off, and added to the appropriate cell pellet. The tubes were placed at 4°C in the dark overnight before flow cytometric analyses. The propidium iodide fluorescence of individual nuclei was measured in the red fluorescence using a Facscan flow cytometer (Becton Dickinson, Mountain View, CA, USA) and the data were registered in a logarithmic scale. At least 104 cells of each sample were analyzed. Apoptotic nuclei appeared as a broad hypodiploid DNA peak, which was easily distinguished from the narrow hyperdiploid peak of nuclei in the CRC cells.
Protein expression analysis
Cell extracts were prepared as described previouslyCitation19 and the protein content was determined by the Bradford assay. Briefly, a total of 30 µg of total protein was added to each well of 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gels. Electrophoresis was carried out. Then, proteins on the gels were transferred onto nitrocellulose membranes. Membranes were blocked using 5% skim milk before overnight incubation with the primary antibodies at a concentration of 1:100 v/v. Incubation of membranes with horseradish peroxidase–conjugated goat antirabbit IgG or goat antimouse IgG (1:2,000 v/v; Bio-Rad Laboratories Inc., Hercules, CA, USA) was performed at room temperature with shaking for 1 hour. Labeled bands were detected with Immuno-Star HRP Chemiluminescent kit and the images were captured and quantified using the VersaDoc image system (Bio-Rad). The relative expression of detected proteins was determined as shown previously.Citation20 Briefly, the relative expression of GRP78 was determined by dividing the densitometric value of GRP78 by that of the glyceraldehyde-3-phosphate dehydrogenase.
Statistical analysis
Data are expressed as mean ± standard error. The statistical significance of intergroup differences in normally distributed continuous variables was determined using Student’s t-test. P-values ≤0.05 were considered statistically significant. P-values <0.05 and <0.001 are indicated by * and **, respectively.
Results
Computer-aided drug design pharmacophore design
The protein preparation step produced a protein cleaned from the common problems in the input protein structure for further processing by other protocols in Discovery Studio software. The software standardized atom names, inserted missing atoms in residues, and removed alternate conformations. It also removed water and ligand molecules as required by the setting of Advanced-Keep Waters and Advanced-Keep Ligands. It inserted missing loop regions based on SEQRES data. It optimized short and medium size loop regions with the LOOPER algorithm, minimized the remaining loop regions, and calculated the pKa and protonated the structure. The prepared protein is ready for processing to the next steps which depend mainly on its quality.Citation14
The ATP molecule was used as a scaffold to find the most important interactions with the active site; these interactions were used to design a drug that could bind to the active site instead of ATP (). The next step was to generate a pharmacophore that reflects the best interactions of ATP with active site and use it to find new inhibitors. A set of features from the binding ligand were identified such as hydrogen bond acceptor, hydrogen bond donor, hydrophobic, negative ionizable, positive ionizable, and ring aromatic. The features that match the interactions between the binding ligand and the protein were used to build the pharmacophore models. Excluded volumes were added based on the locations of atoms on the protein, which can optionally add a shape constraint to the pharmacophores using Add Shape Constraint. The generated pharmacophore models were ranked based on a measure of sensitivity and specificity and the top models were selected. The pharmacophore models were enumerated and then the selectivity was estimated based on a Genetic Function Approximation model.Citation21
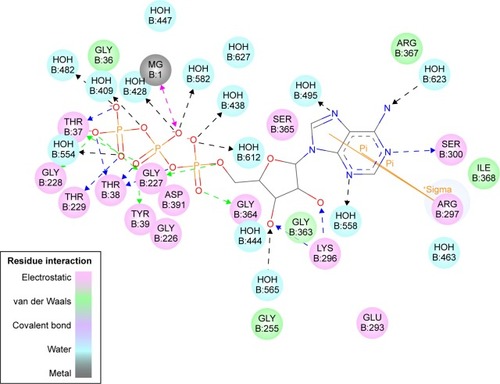
Figure 2 The 2D structure of the ATP molecule and type of interactions performed in the active site.
The automated Ligand Pharmacophore Mapping protocol produced ten pharmacophores that could be important for ligand binding (). A final pharmacophore that was built based on matching the most important interactions has been sketched in . The pharmacophore contains many features and we left this large number intentionally to find many molecules, which have the possibility to bind as many features as possible to compete with ATP for the active site.
Figure 3 A generated pharmacophore showing the most important features ATP can perform; this figure shows only three pharmacophores of the ten generated.
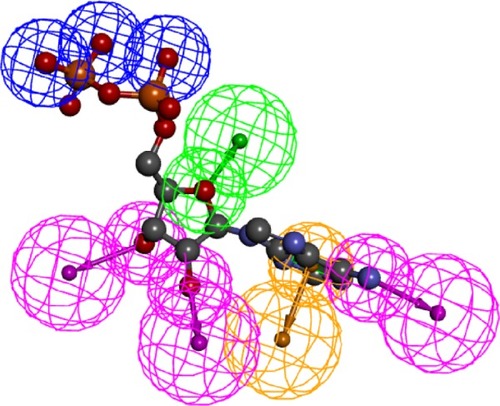
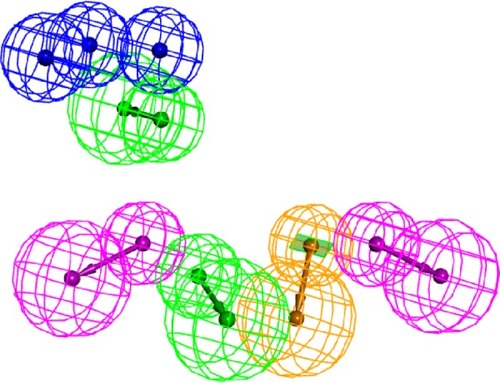
Figure 4 The final pharmacophore used for extracting the potential 78 kDa glucose-regulated protein inhibitors from the Protein Data Bank (blue, negative ionizable feature; green, hydrogen bond acceptor; purple, hydrogen bond donor; and light brown, vectored aromatic feature).
The generated pharmacophore was exploited to extract possible inhibitors from a commercially available compound library named ALDRICHCPR. The structures in this library were screened over the accepted pharmacophore and then the compound that has the highest possibility to match the pharmacophore was selected. This process has generated nine candidate compounds () which scored the best in which the structures were shown. Four of the nine compounds have been purchased according to the availability of stock from Sigma-Aldrich Co. They were R270776, R285986, S781819, and R214035. The most active compounds were mapped on the generated pharmacophore ().
Figure 5 The structure of the candidate for 78 kDa glucose-regulated protein inhibitors named R214035 mapped over the generated pharmacophore.
Table 2 The codes and structure of the nine candidate compounds that have been selected as GRP78 competitive inhibitors
To explore the cytotoxicity of these designed GRP78 inhibitors, the most active compounds were tested at different concentrations (0–100 µM) against HCT116 and SW480 CRC cell lines using the MTT assay (). These cells were chosen based on their different expression of the GRP78 levels and variable sensitivity degrees to chemotherapy (5, 9). Results shown revealed that the only active compound was R214035 (potassium-3-beta-hydroxy-20-oxopregn-5-en-17-alpha-yl sulfate [PHOS]). The optimal concentration of PHOS to induce cell growth inhibition was 50 µM. It has a molecular weight of 450.644 Da and a cholesterol-based scaffold, which bears a big possibility to be a safe compound for human use.
Table 3 Percentage of cell viability for HCT116 and SW480 treated with different GRP78 inhibitors
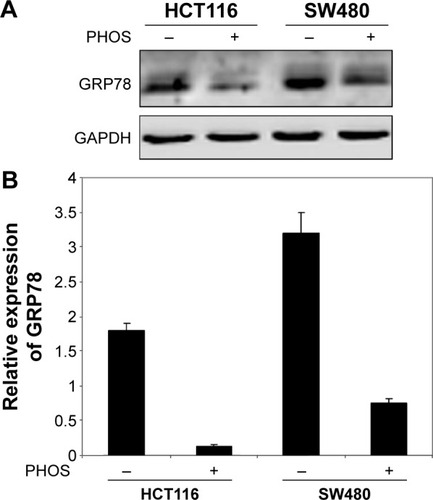
Next we studied whether the reduction in cell viability was due to induction of apoptosis. HCT116 and SW480 CRC cells were incubated with PHOS at 50 µM for 72 hours. Apoptotic cells were measured by the propidium iodide method. Results in show that PHOS significantly (P=0.041) induces apoptosis in HCT116 cells but not in SW480. The potential of PHOS to induce apoptosis was largely attributed to its effect in downregulation of the antiapoptotic protein GRP78, particularly in HCT116 cells ().
Figure 6 Potassium-3-beta-hydroxy-20-oxopregn-5-en-17-alpha-yl sulfate (PHOS)-induced apoptosis in colorectal cancer cell lines.
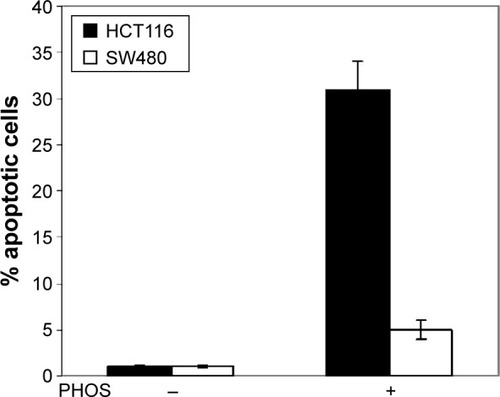
Figure 7 Potassium-3-beta-hydroxy-20-oxopregn-5-en-17-alpha-yl sulfate (PHOS) significantly inhibited the expression of enhanced 78 kDa glucose-regulated protein (GRP78).
Discussion
Resistance of human CRC cells to the available chemotherapeutic agents is considered a major obstacle for successful treatment. The GRP78 is one of the most abundant chaperones in the ER lumen. It is a 78 kDa protein, also known as binding immunoglobulin protein, with antiapoptotic properties.Citation22 Under physiological conditions, GRP78 binds to transmembrane proteins that form the UPR arm. Upon ER stress and initiation of UPR, GRP78 dissociates from transmembrane proteins and its expression increases to regulate the ER homeostasis and increase the folding capacity of the ER.Citation23
GRP78 is believed to play a major role in the resistance of CRC to the available treatment modalities. Elevated expression of GRP78 has been reported in several cancers, such as breast cancer and prostate cancer.Citation4,Citation5 Moreover, GRP78 expression has been associated with tumor development and growth, vascularization, metastasis, and correlated with cancer resistance to chemotherapy.Citation5–Citation7 It seems that some cancer cells may have adapted to ER stress by activation of the UPR without ending in apoptosisCitation8,Citation9 via maintaining the expression of proteins that facilitate survival, such as GRP78.Citation9 Previous studies have shown that GRP78 is associated with the enhanced proliferation of colorectal carcinoma cell lines and their resistance to apoptosis.Citation24 Overexpression of GRP78 protein was shown in colon carcinoma tissue of Chinese patients and suggested as a biomarker for their malignant transformation.Citation25
Consistently, our studies showed that GRP78 expression increased with the progression from early to advanced CRC stages.Citation11 In addition, a significant association was found between GRP78 expression and response to chemotherapy.Citation12 Moreover, GRP78 was found to be constitutively activated in CRC cell lines and CRC tissues isolated from patients.Citation26 Furthermore, levels of GRP78 were inversely associated with sensitivity of CRC cells to chemotherapy-induced apoptosis. Inhibition of GRP78 by small interfering RNA (siRNA) resulted in increased sensitivity of CRC cells to chemotherapeutic agents. Based on the above findings, development of an effective inhibitor of GRP78 might be considered an innovative approach in the treatment of CRC patients.
Results in the present study indicate that PHOS, out of four used compounds, has the potential to induce apoptosis in the HCT116 CRC cell line. Induction of apoptosis is largely attributed to the inhibition of GRP78 levels as shown by immunoblotting. To a very small and insignificant degree, PHOS inhibits proliferation of SW480 cells. This might be explained by the high constitutive expression of GRP78 in SW480 cells in which the decrease in GRP78 levels induced by PHOS was insufficient to modulate signaling pathways downstream of GRP78 involved in misfolded proteins response. Further investigations are required to explore this mechanism.
It has been shown that transcriptional inhibition of GRP78 by siRNA increases in vitro and in vivo chemotherapy-induced cell death.Citation12,Citation27 In addition, inhibition of GRP78 by using nucleoside analogs has been shown to inhibit cancer cell proliferation.Citation10 This was due to binding of these analogs to the ATPase domain of GRP78 and resulted in enhanced synthesis of procaspases, thus enhancing apoptotic cell death. Other studies have shown that treatment of cancer cells with natural products such as epigallocatechin gallate and honokiol might inhibit cancer cell growth by binding and inhibiting GRP78.Citation28,Citation29
The structure of PHOS is a steroidal one. Steroids are a group of lipids that comprise a wide variety of structurally-related compounds acting as physiological regulators as well as hormones in vivo. Various steroidal hormones including estrogens, androgens, progestins, glucocorticoids, and mineralocorticoids are chemically synthesized and have vital roles inside human bodies.Citation30 There are several anticancer drugs that are steroidal in nature and used mainly in the treatment of several cancer types. Among these are exemestane, cyproterone acetate, and formestane which act as aromatase inhibitors used in the treatment of breast and prostate cancers.Citation31,Citation32
The mechanism by which GRP78 mediates tumor proliferation and progression is not clear. However, GRP78 has been shown to activate different signaling pathways by acting as a cell surface coreceptor. A study by Zhang et al showed that resistance of breast and prostate cancer cells to chemotherapy is associated with relocalization of GRP78 to the cell surface, binding of GRP78 to PI3K, and activation of PI(3,4,5)P3 production.Citation33
Another mechanism by which GRP78 might antagonize apoptotic cell death is by modulating the activity of Bcl-2 family proteins. GRP78 has been found to inhibit the proapoptotic protein Bax and caspase-7.Citation34 In addition, GRP78 was shown to bind physically to caspase-7 and prevent its activation.Citation35 Moreover, GRP78 might promote cancer growth by enhancing the expression levels of antiapoptotic proteins, particularly Mcl-1 (unpublished data). Indeed, further investigations are needed in order to clarify the relationship between the steroidal structure of PHOS and its potential to inhibit GRP78 activity.
In summary, among the designed compounds as potential inhibitors of GRP78, only PHOS inhibited proliferation and induces apoptosis in CRC cells. Moreover, the inhibition of cellular growth and apoptotic cell death was largely attributed to downregulation of GRP78. Further experimental work is recommended to shed light on the mechanism by which PHOS can modulate the expression of GRP78 to be utilized in chemotherapy combination regimens in the treatment of CRC patients.
Acknowledgments
We would like to acknowledge the Scientific Research Support Fund at the Ministry of Higher Education and Scientific Research, Amman, Jordan, for the financial support (Grant number 118–2011).
Disclosure
The authors report no conflicts of interest in this work.
References
- HardingHPCalfonMUranoFNovoaIRonDTranscriptional and translational control in the mammalian unfolded protein responseAnnu Rev Cell Dev Biol20021857559912142265
- ZhangKKaufmanRJSignaling the unfolded protein response from the endoplasmic reticulumJ Biol Chem200427925259352593815070890
- SchroderMKaufmanRJThe mammalian unfolded protein responseAnnu Rev Biochem20057473978915952902
- ErhardtPSchremserEJCooperGMB-Raf inhibits programmed cell death downstream of cytochrome c release from mitochondria by activating the MEK/Erk pathwayMol Cell Biol19991985308531510409722
- AdeyinkaANuiYCherletTSnellLWatsonPHMurphyLCActivated mitogen-activated protein kinase expression during human breast tumorigenesis and breast cancer progressionClin Cancer Res2002861747175312060612
- PyrkoPSchonthalAHHofmanFMChenTCLeeASThe unfolded protein response regulator GRP78/BiP as a novel target for increasing chemosensitivity in malignant gliomasCancer Res200767209809981617942911
- ReddyRKMaoCBaumeisterPAustinRCKaufmanRJLeeASEndoplasmic reticulum chaperone protein GRP78 protects cells from apoptosis induced by topoisomerase inhibitors: role of ATP binding site in suppression of caspase-7 activationJ Biol Chem200327823209152092412665508
- RutkowskiDTKaufmanRJThat which does not kill me makes me stronger: adapting to chronic ER stressTrends Biochem Sci2007321046947617920280
- RutkowskiDTArnoldSMMillerCNAdaptation to ER stress is mediated by differential stabilities of pro-survival and pro-apoptotic mRNAs and proteinsPLoS Biol2006411e37417090218
- MaciasATWilliamsonDSAllenNAdenosine-derived inhibitors of 78 kDa glucose regulated protein (Grp78) ATPase: insights into isoform selectivityJ Med Chem201154124034404121526763
- MhaidatNMAlzoubiKHAlmomaniNKhabourOFExpression of glucose regulated protein 78 (GRP78) determines colorectal cancer response to chemotherapyCancer Biomark201515219720325519021
- MhaidatNMAlzoubiKHKhabourOFBanihaniMNAl-BalasQASwaidanSGRP78 regulates sensitivity of human colorectal cancer cells to DNA targeting agentsCytotechnology Epub20141116
- BermanHMBattistuzTBhatTNThe protein data bankActa Crystallogr D Biol Crystallogr200258Pt 6189990712037327
- SpassovVZFlookPKYanLLOOPER: a molecular mechanics-based algorithm for protein loop predictionProtein Eng Des Sel20082129110018194981
- Al-BalasQASowailehMFHassanMANovel N-substituted aminobenzamide scaffold derivatives targeting the dipeptidyl peptidase-IV enzymeDrug Des Devel Ther20148129163
- LipinskiCALombardoFDominyBWFeeneyPJExperimental and computational approaches to estimate solubility and permeability in drug discovery and development settingsAdv Drug Deliv Rev2001461–332611259830
- Al-BalasQAAmawiHAHassanMAQandilAMAlmaaytahAMMhaidatNMVirtual lead identification of farnesyltransferase inhibitors based on ligand and structure-based pharmacophore techniquesPharmaceuticals (Basel)20136670071524276257
- MhaidatNMZhangXDJiangCCHerseyPDocetaxel-induced apoptosis of human melanoma is mediated by activation of c-Jun NH2-terminal kinase and inhibited by the mitogen-activated protein kinase extracellular signal-regulated kinase 1/2 pathwayClin Cancer Res20071341308131417317842
- MhaidatNMAbdul-RazzakKKAlkofahiASAlsarhanAMAldaherANThorneRFAltholactone induces apoptotic cell death in human colorectal cancer cellsPhytother Res201226692693122105918
- MhaidatNMThorneRFde BockCEZhangXDHerseyPMelanoma cell sensitivity to Docetaxel-induced apoptosis is determined by class III beta-tubulin levelsFEBS Lett2008582226727218086570
- MaSLvMDengFZhangXZhaiHLvWPredicting the ecotoxicity of ionic liquids towards Vibrio fischeri using genetic function approximation and least squares support vector machineJ Hazard Mater201528359159825464300
- SatoMYaoVJArapWPasqualiniRGRP78 signaling hub a receptor for targeted tumor therapyAdv Genet2010699711420807604
- ApostolouAShenYLiangYLuoJFangSArmet, a UPR-upregulated protein, inhibits cell proliferation and ER stress-induced cell deathExp Cell Res2008314132454246718561914
- XingXLiYLiuHWangLSunLGlucose regulated protein 78 (GRP78) is overexpressed in colorectal carcinoma and regulates colorectal carcinoma cell growth and apoptosisActa Histochem2011113877778221156321
- XingXLaiMWangYXuEHuangQOverexpression of glucose-regulated protein 78 in colon cancerClin Chim Acta20063641–230831516182273
- MhaidatNMAlzoubiKHAbushbakAX-box binding protein 1 (XBP-1) enhances colorectal cancer cell invasionJ Chemother201527316717325692573
- VirreyJJDongDStilesCStress chaperone GRP78/BiP confers chemoresistance to tumor-associated endothelial cellsMol Cancer Res2008681268127518708359
- ErmakovaSPKangBSChoiBY(-)-Epigallocatechin gallate overcomes resistance to etoposide-induced cell death by targeting the molecular chaperone glucose-regulated protein 78Cancer Res200666189260926916982771
- LinJWChenJTHongCYHonokiol traverses the blood-brain barrier and induces apoptosis of neuroblastoma cells via an intrinsic bax-mitochondrion-cytochrome c-caspase protease pathwayNeuro Oncol201214330231422259050
- GuptaAKumarBSNegiASCurrent status on development of steroids as anticancer agentsJ Steroid Biochem Mol Biol201313724227023727548
- DeeksEDScottLJExemestane: a review of its use in postmenopausal women with breast cancerDrugs200969788991819441873
- BajettaEZilemboNBuzzoniRFormestane: effective therapy in postmenopausal women with advanced breast cancerAnn Oncol19945Suppl 7S15S177873456
- ZhangYTsengCCTsaiYLFuXSchiffRLeeASCancer cells resistant to therapy promote cell surface relocalization of GRP78 which complexes with PI3K and enhances PI(3,4,5)P3 productionPLoS One2013811e8007124244613
- LeeENicholsPSpicerDGroshenSYuMCLeeASGRP78 as a novel predictor of responsiveness to chemotherapy in breast cancerCancer Res200666167849785316912156
- ZhangLJChenSWuPInhibition of MEK blocks GRP78 up-regulation and enhances apoptosis induced by ER stress in gastric cancer cellsCancer Lett20092741404618829155
